Current Status of Regulatory Non-Coding RNAs Research in the Tritryp
Abstract
:1. Introduction
| RNA Biotype (Acronym Used) | Derived Fragments/Small RNA (Acronym Used) | Canonical Process Involved |
|---|---|---|
| Transfer RNA (tRNA) | tRNA-derived RNA fragments (tsRNAs) [42] | Translation |
| Ribosomal RNA (rRNA) | rRNA-derived fragments (sdrRNA) [40] | Translation |
| small nuclear RNAs (snRNAs) | snRNA-derived fragments (snsRNA) [40] | RNA processing |
| Small nucleolar RNAs (snoRNAs) | snoRNA-derived fragments (sdRNAs) [40] | RNA processing |
| Long non-coding RNAs (lncRNAs) | lncRNAs-derived fragments (lncRNAs) [43] | Gene expression |
| Vault RNA (vtRNA) | Vault RNA-derived (svRNAs) [44] | Vault particle |
| Splice leader RNA (SL-RNA) | RNA processing | |
| Editing guide RNAs (gRNAs) | RNA Editing | |
| Small interfering RNAs (siRNAs) | Gene expression | |
| Telomerase template bearing RNA (TR) | Chromosome maintenance |
2. ncRNAs Annotations in the TriTryps Genomes
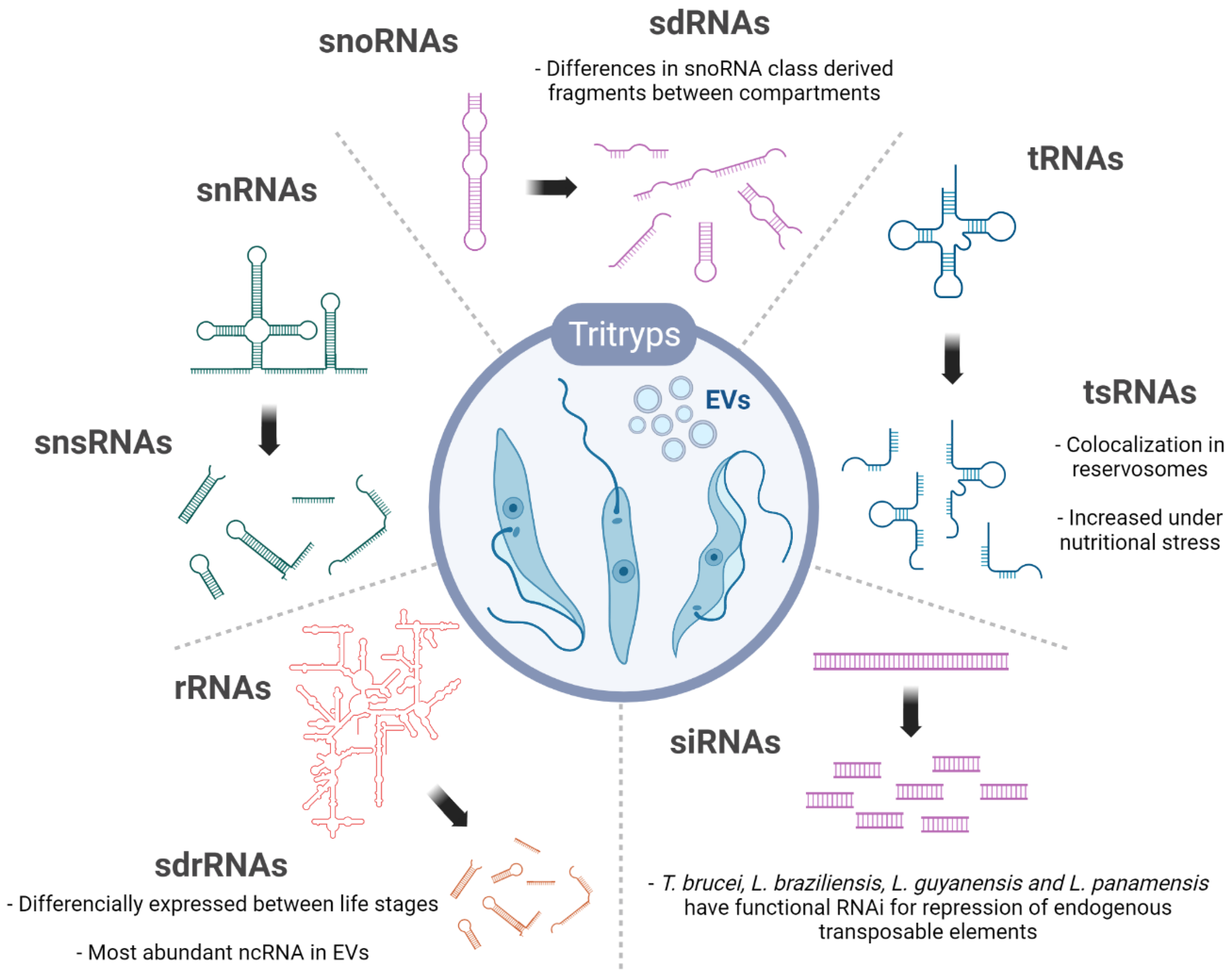
3. ncRNA Biotypes
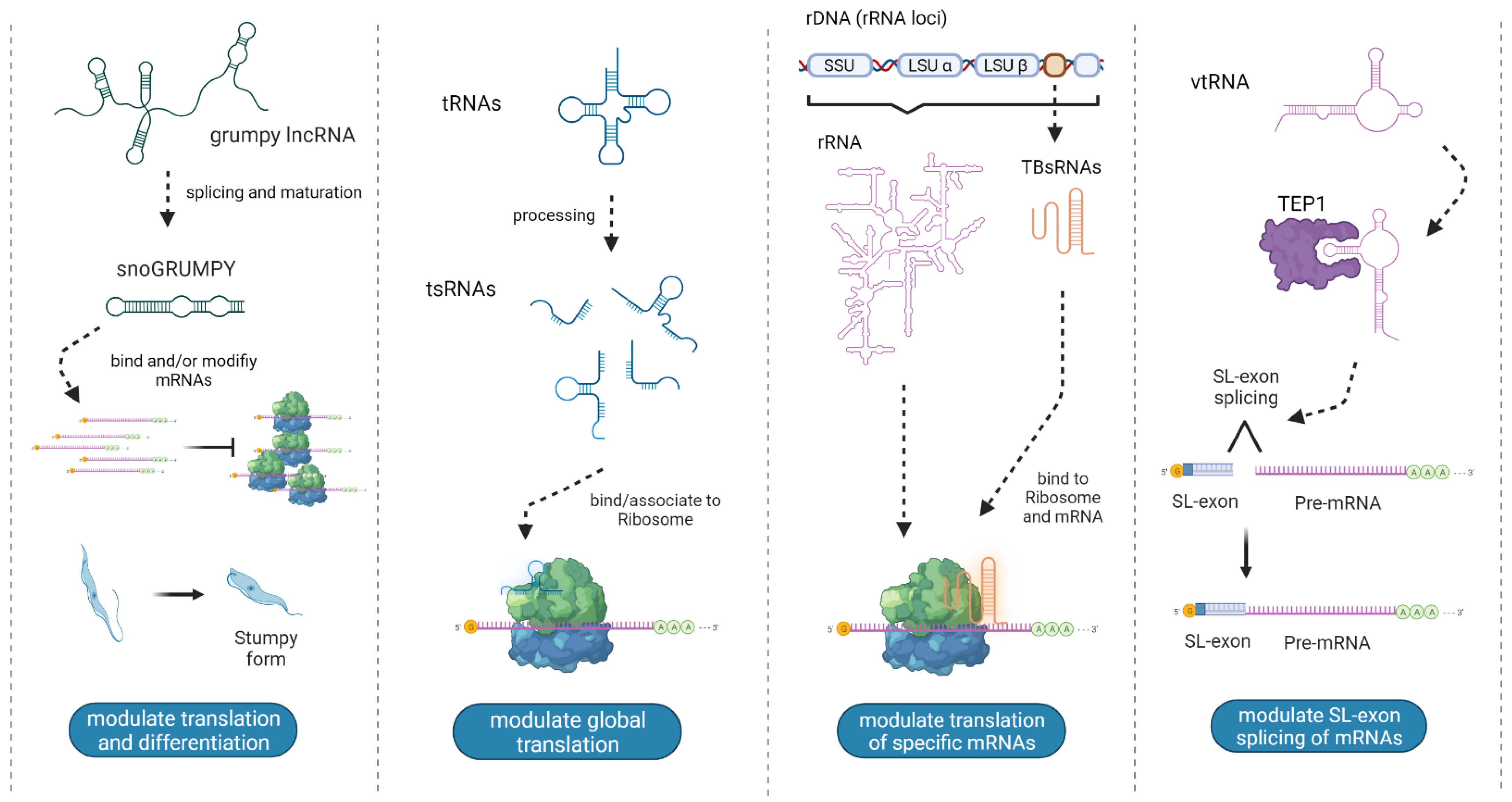
3.1. tRNA and tsRNAs
3.2. snRNA and snsRNAs
3.3. snoRNAs and sdRNAs
3.4. rRNA and sdrRNA
3.5. siRNAs
3.6. vtRNA
3.7. Editing gRNAs
3.8. Splice Leader RNA
3.9. Telomerase RNA
3.10. lncRNAs
4. Conclusions and Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Burki, F. Progress towards the Tree of Eukaryotes. Curr. Biol. 2019, 29, R808–R817. [Google Scholar] [CrossRef] [PubMed]
- Cavalcanti, D.P.; de Souza, W. The Kinetoplast of Trypanosomatids: From Early Studies of Electron Microscopy to Recent Advances in Atomic Force Microscopy. Scanning 2018, 2018, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Büscher, P.; Cecchi, G.; Jamonneau, V.; Priotto, G. Human African Trypanosomiasis. Lancet 2017, 390, 2397–2409. [Google Scholar] [CrossRef]
- Pérez-Molina, J.A.; Molina, I. Chagas Disease. Lancet 2018, 391, 82–94. [Google Scholar] [CrossRef]
- Burza, S.; Croft, S.L.; Boelaert, M. Leishmaniasis. Lancet 2018, 392, 951–970. [Google Scholar] [CrossRef]
- Barrett, M.P.; Burchmore, R.J.S.; Stich, A.; Lazzari, J.O.; Frasch, A.C.; Cazzulo, J.J.; Krishna, S. The Trypanosomiases. Lancet 2003, 362, 1469–1480. [Google Scholar] [CrossRef]
- Giordani, F.; Morrison, L.J.; Rowan, T.G.; De Koning, H.P.; Barrett, M.P. The Animal Trypanosomiases and Their Chemotherapy: A Review. Parasitology 2016, 143, 1862–1889. [Google Scholar] [CrossRef]
- Martínez-Calvillo, S.; Vizuet-de-Rueda, J.C.; Florencio-Martínez, L.E.; Manning-Cela, R.G.; Figueroa-Angulo, E.E. Gene Expression in Trypanosomatid Parasites. J. Biomed. Biotechnol. 2010, 2010, 525241. [Google Scholar] [CrossRef]
- LeBowitz, J.H.; Smith, H.Q.; Rusche, L.; Beverley, S.M. Coupling of Poly(A) Site Selection and Trans-Splicing in Leishmania. Genes Dev. 1993, 7, 996–1007. [Google Scholar] [CrossRef]
- Berberof, M.; Vanhamme, L.; Pays, E. Trypanosoma Brucei: A Preferential Splicing at the Inverted Polyadenylation Site of the VSG MRNA Provides Further Evidence for Coupling between Trans-Splicing and Polyadenylation. Exp. Parasitol. 1995, 80, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Mair, G.; Shi, H.; Li, H.; Djikeng, A.; Aviles, H.O.; Bishop, J.R.; Falcone, F.H.; Gavrilescu, C.; Montgomery, J.L.; Santori, M.I.; et al. A New Twist in Trypanosome RNA Metabolism: Cis-Splicing of Pre-MRNA. RNA 2000, 6, 163. [Google Scholar] [CrossRef] [PubMed]
- De Gaudenzi, J.G.; Noé, G.; Campo, V.A.; Frasch, A.C.; Cassola, A. Gene Expression Regulation in Trypanosomatids. Essays Biochem. 2011, 51, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Vickers, K.C.; Roteta, L.A.; Hucheson-Dilks, H.; Han, L.; Guo, Y. Mining Diverse Small RNA Species in the Deep Transcriptome. Trends Biochem. Sci. 2015, 40, 4–7. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Boivin, V.; Faucher-Giguère, L.; Scott, M.; Abou-Elela, S. The Cellular Landscape of Mid-Size Noncoding RNA. Wiley Interdiscip. Rev. RNA 2019, 10, e1530. [Google Scholar] [CrossRef]
- Chen, C.J.; Heard, E. Small RNAs Derived from Structural Non-Coding RNAs. Methods 2013, 63, 76–84. [Google Scholar] [CrossRef]
- Wajahat, M.; Bracken, C.P.; Orang, A. Emerging Functions for SnoRNAs and SnoRNA-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 10193. [Google Scholar] [CrossRef]
- Lambert, M.; Benmoussa, A.; Provost, P. Small Non-Coding RNAs Derived From Eukaryotic Ribosomal RNA. Non-Coding RNA 2019, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Martens-Uzunova, E.S.; Olvedy, M.; Jenster, G. Beyond MicroRNA—Novel RNAs Derived from Small Non-Coding RNA and Their Implication in Cancer. Cancer Lett. 2013, 340, 201–211. [Google Scholar] [CrossRef]
- Pan, Q.; Han, T.; Li, G. Novel Insights into the Roles of TRNA-Derived Small RNAs. RNA Biol. 2021, 18, 2157–2167. [Google Scholar] [CrossRef] [PubMed]
- Guan, L.; Grigoriev, A. Computational Meta-Analysis of Ribosomal RNA Fragments: Potential Targets and Interaction Mechanisms. Nucleic Acids Res. 2021, 49, 4085–4103. [Google Scholar] [CrossRef]
- Li, S.; Xu, Z.; Sheng, J. TRNA-Derived Small RNA: A Novel Regulatory Small Non-Coding RNA. Genes 2018, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G. TRNA-Derived Small RNAs: New Players in Genome Protection against Retrotransposons. RNA Biol. 2018, 15, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Fuchs, G.; Wang, S.; Wei, W.; Zhang, Y.; Park, H.; Roy-Chaudhuri, B.; Li, P.; Xu, J.; Chu, K.; et al. A Transfer-RNA-Derived Small RNA Regulates Ribosome Biogenesis. Nature 2017, 552, 57–62. [Google Scholar] [CrossRef]
- Fricker, R.; Brogli, R.; Luidalepp, H.; Wyss, L.; Fasnacht, M.; Joss, O.; Zywicki, M.; Helm, M.; Schneider, A.; Cristodero, M.; et al. A TRNA Half Modulates Translation as Stress Response in Trypanosoma Brucei. Nat. Commun. 2019, 10, 118. [Google Scholar] [CrossRef]
- Pircher, A.; Gebetsberger, J.; Polacek, N. Ribosome-Associated NcRNAs: An Emerging Class of Translation Regulators. RNA Biol. 2014, 11, 1335–1339. [Google Scholar] [CrossRef]
- Rajan, K.S.; Doniger, T.; Cohen-Chalamish, S.; Rengaraj, P.; Galili, B.; Aryal, S.; Unger, R.; Tschudi, C.; Michaeli, S. Developmentally Regulated Novel Non-Coding Anti-Sense Regulators of MRNA Translation in Trypanosoma Brucei. iScience 2020, 23, 101780. [Google Scholar] [CrossRef]
- Pagès, A.; Dotu, I.; Pallarès-Albanell, J.; Martí, E.; Guigó, R.; Eyras, E. The Discovery Potential of RNA Processing Profiles. Nucleic Acids Res. 2018, 46, e15. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhou, T.; Chen, Q. Exploring the Expanding Universe of Small RNAs. Nat. Cell Biol. 2022, 24, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ender, C.; Meister, G.; Moore, P.S.; Chang, Y.; John, B. Extensive Terminal and Asymmetric Processing of Small RNAs from RRNAs, SnoRNAs, SnRNAs, and TRNAs. Nucleic Acids Res. 2012, 40, 6787–6799. [Google Scholar] [CrossRef]
- Blum, B.; Bakalara, N.; Simpson, L. A Model for RNA Editing in Kinetoplastid Mitochondria: “Guide” RNA Molecules Transcribed from Maxicircle DNA Provide the Edited Information. Cell 1990, 60, 189–198. [Google Scholar] [CrossRef]
- Boothroyd, J.C.; Cross, G.A.M. Transcripts Coding for Variant Surface Glycoproteins of Trypanosoma Brucei Have a Short, Identical Exon at Their 5’ End. Gene 1982, 20, 281–289. [Google Scholar] [CrossRef]
- Michaeli, S.; Doniger, T.; Gupta, S.K.; Wurtzel, O.; Romano, M.; Visnovezky, D.; Sorek, R.; Unger, R.; Ullu, E. RNA-Seq Analysis of Small RNPs in Trypanosoma Brucei Reveals a Rich Repertoire of Non-Coding RNAs. Nucleic Acids Res. 2012, 40, 1282–1298. [Google Scholar] [CrossRef]
- Kolev, N.G.; Rajan, K.S.; Tycowski, K.T.; Toh, J.Y.; Shi, H.; Lei, Y.; Michaeli, S.; Tschudi, C. The Vault RNA of Trypanosoma Brucei Plays a Role in the Production of Trans-Spliced MRNA. J. Biol. Chem. 2019, 294, 15559–15574. [Google Scholar] [CrossRef]
- Shikha, S.; Brogli, R.; Schneider, A.; Polacek, N. TRNA Biology in Trypanosomes. Chimia 2019, 73, 395–405. [Google Scholar] [CrossRef]
- Peng, R.; Santos, H.J.; Nozaki, T. Transfer RNA-Derived Small RNAs in the Pathogenesis of Parasitic Protozoa. Genes 2022, 13, 286. [Google Scholar] [CrossRef]
- Franzén, O.; Arner, E.; Ferella, M.; Nilsson, D.; Respuela, P.; Carninci, P.; Hayashizaki, Y.; Åslund, L.; Andersson, B.; Daub, C.O. The Short Non-Coding Transcriptome of the Protozoan Parasite Trypanosoma Cruzi. PLoS Negl. Trop. Dis. 2011, 5, e1283. [Google Scholar] [CrossRef]
- Fernandez-Calero, T.; Garcia-Silva, R.; Pena, A.; Robello, C.; Persson, H.; Rovira, C.; Naya, H.; Cayota, A. Profiling of Small RNA Cargo of Extracellular Vesicles Shed by Trypanosoma Cruzi Reveals a Specific Extracellular Signature. Mol. Biochem. Parasitol. 2015, 199, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Frugier, M.; Tosar, J.P.; Correa-Dominguez, A.; Ronalte-Alves, L.; Parodi-Talice, A.; Rovira, C.; Robello, C.; Goldenberg, S.; Cayota, A. A Population of TRNA-Derived Small RNAs Is Actively Produced in Trypanosoma Cruzi and Recruited to Specific Cytoplasmic Granules. Mol. Biochem. Parasitol. 2010, 171, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Jalali, S.; Jayaraj, G.G.; Scaria, V. Integrative Transcriptome Analysis Suggest Processing of a Subset of Long Non-Coding RNAs to Small RNAs. Biol. Direct 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Persson, H.; Kvist, A.; Vallon-Christersson, J.; Medstrand, P.; Borg, A.; Rovira, C. The Non-Coding RNA of the Multidrug Resistance-Linked Vault Particle Encodes Multiple Regulatory Small RNAs. Nat. Cell Biol. 2009, 11, 1268–1271. [Google Scholar] [CrossRef]
- Berriman, M.; Ghedin, E.; Hertz-Fowler, C.; Blandin, G.; Renauld, H.; Bartholomeu, D.C.; Lennard, N.J.; Caler, E.; Hamlin, N.E.; Haas, B.; et al. The Genome of the African Trypanosome Trypanosoma Brucei. Science 2005, 309, 416–422. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, N.M.; Myler, P.J.; Bartholomeu, D.C.; Nilsson, D.; Aggarwal, G.; Tran, A.-N.N.; Ghedin, E.; Worthey, E.A.; Delcher, A.L.; Blandin, G.; et al. The Genome Sequence of Trypanosoma Cruzi, Etiologic Agent of Chagas Disease. Science 2005, 309, 409–415. [Google Scholar] [CrossRef]
- Ivens, A.C.; Peacock, C.S.; Worthey, E.A.; Murphy, L.; Aggarwal, G.; Berriman, M.; Sisk, E.; Rajandream, M.-A.; Adlem, E.; Aert, R.; et al. The Genome of the Kinetoplastid Parasite, Leishmania Major. Science 2005, 309, 436–442. [Google Scholar] [CrossRef]
- El-Sayed, N.M.; Myler, P.J.; Blandin, G.; Berriman, M.; Crabtree, J.; Aggarwal, G.; Caler, E.; Renauld, H.; Worthey, E.A.; Hertz-Fowler, C.; et al. Comparative Genomics of Trypanosomatid Parasitic Protozoa. Science 2005, 309, 404–409. [Google Scholar] [CrossRef]
- Aslett, M.; Aurrecoechea, C.; Berriman, M.; Brestelli, J.; Brunk, B.P.; Carrington, M.; Depledge, D.P.; Fischer, S.; Gajria, B.; Gao, X.; et al. TriTrypDB: A Functional Genomic Resource for the Trypanosomatidae. Nucleic Acids Res. 2010, 38, D457–D462. [Google Scholar] [CrossRef]
- Berná, L.; Rodriguez, M.; Chiribao, M.L.; Parodi-Talice, A.; Pita, S.; Rijo, G.; Alvarez-Valin, F.; Robello, C. Expanding an Expanded Genome: Long-Read Sequencing of Trypanosoma Cruzi. Microb. Genom. 2018, 4, e0001779. [Google Scholar] [CrossRef]
- Garcia-Silva, M.R.; Cura Das Neves, R.F.; Cabrera-Cabrera, F.; Sanguinetti, J.; Medeiros, L.C.; Robello, C.; Naya, H.; Fernandez-Calero, T.; Souto-Padron, T.; De Souza, W.; et al. Extracellular Vesicles Shed by Trypanosoma Cruzi Are Linked to Small RNA Pathways, Life Cycle Regulation, and Susceptibility to Infection of Mammalian Cells. Parasitol. Res. 2014, 113, 285–304. [Google Scholar] [CrossRef] [PubMed]
- Reifur, L.; Garcia-Silva, M.R.; Poubel, S.B.; Alves, L.R.; Arauco, P.; Buiar, D.K.; Goldenberg, S.; Cayota, A.; Dallagiovanna, B. Distinct Subcellular Localization of TRNA-Derived Fragments in the Infective Metacyclic Forms of Trypanosoma Cruzi. Mem. Inst. Oswaldo Cruz 2012, 107, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Bayer-Santos, E.; Lima, F.M.; Ruiz, J.C.; Almeida, I.C.; Da Silveira, J.F. Characterization of the Small RNA Content of Trypanosoma Cruzi Extracellular Vesicles. Mol. Biochem. Parasitol. 2014, 193, 71–74. [Google Scholar] [CrossRef]
- Garcia-Silva, M.R.; Cabrera-Cabrera, F.; Cura das Neves, R.F.; Souto-Padrón, T.; de Souza, W.; Cayota, A. Gene Expression Changes Induced by Trypanosoma Cruzi Shed Microvesicles in Mammalian Host Cells: Relevance of TRNA-Derived Halves. Biomed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.L.; Wen, Y.Z.; Yang, J.H.; Liao, J.Y.; Shao, P.; Xu, H.; Zhou, H.; Wen, J.Z.; Lun, Z.R.; Ayala, F.J.; et al. Comparative Transcriptome Analysis of Small Noncoding RNAs in Different Stages of Trypanosoma Brucei. RNA 2013, 19, 863–875. [Google Scholar] [CrossRef]
- Lambertz, U.; Oviedo Ovando, M.E.; Vasconcelos, E.J.R.; Unrau, P.J.; Myler, P.J.; Reiner, N.E. Small RNAs Derived from TRNAs and RRNAs Are Highly Enriched in Exosomes from Both Old and New World Leishmania Providing Evidence for Conserved Exosomal RNA Packaging. BMC Genomics 2015, 16. [Google Scholar] [CrossRef]
- Guegan, F.; Rajan, K.S.; Bento, F.; Pinto-Neves, D.; Sequeira, M.; Gumińska, N.; Mroczek, S.; Dziembowski, A.; Cohen-Chalamish, S.; Doniger, T.; et al. A Long Noncoding RNA Promotes Parasite Differentiation in African Trypanosomes. Sci. Adv. 2022, 8, eabn2706. [Google Scholar] [CrossRef]
- Hoskins, A.A.; Moore, M.J. The Spliceosome: A Flexible, Reversible Macromolecular Machine. Trends Biochem. Sci. 2012, 37, 179–188. [Google Scholar] [CrossRef]
- Turunen, J.J.; Niemelä, E.H.; Verma, B.; Frilander, M.J. The Significant Other: Splicing by the Minor Spliceosome. Wiley Interdiscip. Rev. RNA 2013, 4, 61–76. [Google Scholar] [CrossRef]
- Morais, P.; Adachi, H.; Yu, Y.-T. Spliceosomal SnRNA Epitranscriptomics. Front. Genet. 2021, 12, 652129. [Google Scholar] [CrossRef]
- Ambrósio, D.L.; Badjatia, N.; Günzl, A. The Spliceosomal PRP19 Complex of Trypanosomes. Mol. Microbiol. 2015, 95, 885–901. [Google Scholar] [CrossRef] [PubMed]
- Preußer, C.; Jaé, N.; Bindereif, A. MRNA Splicing in Trypanosomes. Int. J. Med. Microbiol. 2012, 302, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, J.D.; Wimberger, P.; Wilsch, K.; Fluck, M.; Suter, L.; Brunner, G. Increased Level of Circulating U2 Small Nuclear RNA Fragments Indicates Metastasis in Melanoma Patients. Clin. Chem. Lab. Med. 2015, 53, 605–611. [Google Scholar] [CrossRef]
- Mesquita-Ribeiro, R.; Fort, R.S.; Rathbone, A.; Farias, J.; Lucci, C.; James, V.; Sotelo-Silveira, J.; Duhagon, M.A.; Dajas-Bailador, F. Distinct Small Non-Coding RNA Landscape in the Axons and Released Extracellular Vesicles of Developing Primary Cortical Neurons and the Axoplasm of Adult Nerves. RNA Biol. 2021, 18, 832–855. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.L.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Deep-Sequencing of Human Argonaute-Associated Small RNAs Provides Insight into MiRNA Sorting and Reveals Argonaute Association with RNA Fragments of Diverse Origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Pillman, K.A.; Anderson, M.L.; Lawrence, D.M.; Toubia, J.; Goodall, G.J.; Bracken, C.P. Assessing the Gene Regulatory Properties of Argonaute-Bound Small RNAs of Diverse Genomic Origin. Nucleic Acids Res. 2015, 43, 470–481. [Google Scholar] [CrossRef]
- Baraniskin, A.; Nöpel-Dünnebacke, S.; Ahrens, M.; Jensen, S.G.; Zöllner, H.; Maghnouj, A.; Wos, A.; Mayerle, J.; Munding, J.; Kost, D.; et al. Circulating U2 Small Nuclear RNA Fragments as a Novel Diagnostic Biomarker for Pancreatic and Colorectal Adenocarcinoma. Int. J. Cancer 2013, 132. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M. From SnoRNA to MiRNA: Dual Function Regulatory Non-Coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef]
- Rajan, K.S.; Chikne, V.; Decker, K.; Waldman Ben-Asher, H.; Michaeli, S. Unique Aspects of RRNA Biogenesis in Trypanosomatids. Trends Parasitol. 2019, 35, 778–794. [Google Scholar] [CrossRef]
- Rajan, K.S.; Doniger, T.; Cohen-Chalamish, S.; Chen, D.; Semo, O.; Aryal, S.; Saar, E.G.; Chikne, V.; Gerber, D.; Unger, R.; et al. Pseudouridines on Trypanosoma Brucei Spliceosomal Small Nuclear RNAs and Their Implication for RNA and Protein Interactions. Nucleic Acids Res. 2019, 47, 7633–7647. [Google Scholar] [CrossRef]
- Chikne, V.; Rajan, K.S.; Shalev-Benami, M.; Decker, K.; Cohen-Chalamish, S.; Madmoni, H.; Biswas, V.K.; Gupta, S.K.; Doniger, T.; Unger, R.; et al. Small Nucleolar RNAs Controlling RRNA Processing in Trypanosoma Brucei. Nucleic Acids Res. 2019, 47, 2609–2629. [Google Scholar] [CrossRef] [PubMed]
- Rajan, K.S.; Zhu, Y.; Adler, K.; Doniger, T.; Cohen-Chalamish, S.; Srivastava, A.; Shalev-Benami, M.; Matzov, D.; Unger, R.; Tschudi, C.; et al. The Large Repertoire of 2’-O-Methylation Guided by C/D SnoRNAs on Trypanosoma Brucei RRNA. RNA Biol. 2020, 17, 1018–1039. [Google Scholar] [CrossRef]
- Barth, S.; Shalem, B.; Hury, A.; Tkacz, I.D.; Liang, X.H.; Uliel, S.; Myslyuk, I.; Doniger, T.; Salmon-Divon, M.; Unger, R.; et al. Elucidating the Role of C/D SnoRNA in RRNA Processing and Modification in Trypanosoma Brucei. Eukaryot. Cell 2008, 7, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Ender, C.; Krek, A.; Friedländer, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A Human SnoRNA with MicroRNA-like Functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Taft, R.J.; Glazov, E.A.; Lassmann, T.; Hayashizaki, Y.; Carninci, P.; Mattick, J.S. Small RNAs Derived from SnoRNAs. RNA 2009, 15, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Nakamura, M.; Takahashi, Y.; Sandelin, A.; Katayama, S.; Fukuda, S.; Daub, C.O.; Kai, C.; Kawai, J.; Yasuda, J.; et al. Hidden Layers of Human Small RNAs. BMC Genomics 2008, 9, 157. [Google Scholar] [CrossRef]
- Brameier, M.; Herwig, A.; Reinhardt, R.; Walter, L.; Gruber, J. Human Box C/D SnoRNAs with MiRNA like Functions: Expanding the Range of Regulatory RNAs. Nucleic Acids Res. 2011, 39, 675–686. [Google Scholar] [CrossRef]
- Saraiya, A.A.; Wang, C.C. SnoRNA, a Novel Precursor of MicroRNA in Giardia Lamblia. PLoS Pathog. 2008, 4, e1000224. [Google Scholar] [CrossRef]
- Mleczko, A.M.; Machtel, P.; Walkowiak, M.; Wasilewska, A.; Pietras, P.J.; Bąkowska-Żywicka, K. Levels of SdRNAs in Cytoplasm and Their Association with Ribosomes Are Dependent upon Stress Conditions but Independent from SnoRNA Expression. Sci. Rep. 2019, 9, 18397. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-Coding RNA: What Is Functional and What Is Junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef]
- Zywicki, M.; Bakowska-Zywicka, K.; Polacek, N. Revealing Stable Processing Products from Ribosome-Associated Small RNAs by Deep-Sequencing Data Analysis. Nucleic Acids Res. 2012, 40, 4013–4024. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Zhou, B.; Zhang, F.; Tu, Y.; Hu, Y.; Zhang, B.; Zhai, Q. Profiling and Identification of Small RDNA-Derived RNAs and Their Potential Biological Functions. PLoS ONE 2013, 8, e56842. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-C.; Chang, S.-S.; Choudhary, S.; Aalto, A.P.; Maiti, M.; Bamford, D.H.; Liu, Y. QiRNA Is a New Type of Small Interfering RNA Induced by DNA Damage. Nature 2009, 459, 274–277. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Sun, Y.; Yang, X.; Wu, Z.; Guo, K.; Niu, X.; Wang, Q.; Ruan, J.; Bu, W.; Gao, S. Two Featured Series of RRNA-Derived RNA Fragments (RRFs) Constitute a Novel Class of Small RNAs. PLoS ONE 2017, 12, e0176458. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Sanguinetti, J.; Cabrera-Cabrera, F.; Franzén, O.; Cayota, A. A Particular Set of Small Non-Coding RNAs Is Bound to the Distinctive Argonaute Protein of Trypanosoma Cruzi: Insights from RNA-Interference Deficient Organisms. Gene 2014, 538, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Trocoli Torrecilhas, A.C.; Tonelli, R.R.; Pavanelli, W.R.; da Silva, J.S.; Schumacher, R.I.; de Souza, W.; E Silva, N.C.; de Almeida Abrahamsohn, I.; Colli, W.; Manso Alves, M.J. Trypanosoma Cruzi: Parasite Shed Vesicles Increase Heart Parasitism and Generate an Intense Inflammatory Response. Microbes Infect. 2009, 11, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Shabalina, S.A.; Koonin, E.V. Origins and Evolution of Eukaryotic RNA Interference. Trends Ecol. Evol. 2008, 23, 578. [Google Scholar] [CrossRef]
- Cerutti, H.; Casas-Mollano, J.A. On the Origin and Functions of RNA-Mediated Silencing: From Protists to Man. Curr. Genet. 2006, 50, 81–99. [Google Scholar] [CrossRef]
- Ngô, H.; Tschudi, C.; Gull, K.; Ullu, E. Double-Stranded RNA Induces MRNA Degradation in Trypanosoma Brucei. Proc. Natl. Acad. Sci. USA 1998, 95, 14687–14692. [Google Scholar] [CrossRef]
- Bråte, J.; Neumann, R.S.; Fromm, B.; Haraldsen, A.A.B.; Tarver, J.E.; Suga, H.; Donoghue, P.C.J.; Peterson, K.J.; Ruiz-Trillo, I.; Grini, P.E.; et al. Unicellular Origin of the Animal MicroRNA Machinery. Curr. Biol. 2018, 28, 3288–3295.e5. [Google Scholar] [CrossRef]
- Barnes, R.L.; Shi, H.; Kolev, N.G.; Tschudi, C.; Ullu, E. Comparative Genomics Reveals Two Novel RNAi Factors in Trypanosoma Brucei and Provides Insight into the Core Machinery. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef]
- Durand-Dubief, M.; Bastin, P. TbAGO1, an Argonaute Protein Required for RNA Interference, Is Involved in Mitosis and Chromosome Segregation in Trypanosoma Brucei. BMC Biol. 2003, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Chamond, N.; Tschudi, C.; Ullu, E. Selection and Characterization of RNA Interference-Deficient Trypanosomes Impaired in Target MRNA Degradation. Eukaryot. Cell 2004, 3, 1445–1453. [Google Scholar] [CrossRef]
- Patrick, K.L.; Shi, H.; Kolev, N.G.; Ersfeld, K.; Tschudi, C.; Ullu, E. Distinct and Overlapping Roles for Two Dicer-like Proteins in the RNA Interference Pathways of the Ancient Eukaryote Trypanosoma Brucei. Proc. Natl. Acad. Sci. USA 2009, 106, 17933–17938. [Google Scholar] [CrossRef]
- Shi, H.; Djikeng, A.; Tschudi, C.; Ullu, E. Argonaute Protein in the Early Divergent Eukaryote Trypanosoma Brucei: Control of Small Interfering RNA Accumulation and Retroposon Transcript Abundance. Mol. Cell. Biol. 2004, 24, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Djikeng, A.; Shi, H.; Tschudi, C.; Ullu, E. RNA Interference in Trypanosoma Brucei: Cloning of Small Interfering RNAs Provides Evidence for Retroposon-Derived 24-26-Nucleotide RNAs. RNA 2001, 7, 1522–1530. [Google Scholar]
- Tschudi, C.; Shi, H.; Franklin, J.B.; Ullu, E. Small Interfering RNA-Producing Loci in the Ancient Parasitic Eukaryote Trypanosoma Brucei. BMC Genom. 2012, 13, 427. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.-Z.; Zheng, L.-L.; Liao, J.-Y.; Wang, M.-H.; Wei, Y.; Guo, X.-M.; Qu, L.-H.; Ayala, F.J.; Lun, Z.-R. Pseudogene-Derived Small Interference RNAs Regulate Gene Expression in African Trypanosoma Brucei. Proc. Natl. Acad. Sci. USA 2011, 108, 8345–8350. [Google Scholar] [CrossRef]
- Reynolds, D.; Hofmeister, B.T.; Cliffe, L.; Alabady, M.; Siegel, T.N.; Schmitz, R.J.; Sabatini, R. Histone H3 Variant Regulates RNA Polymerase II Transcription Termination and Dual Strand Transcription of SiRNA Loci in Trypanosoma Brucei. PLOS Genet. 2016, 12, e1005758. [Google Scholar] [CrossRef]
- Lye, L.-F.; Owens, K.; Shi, H.; Murta, S.M.F.; Vieira, A.C.; Turco, S.J.; Tschudi, C.; Ullu, E.; Beverley, S.M. Retention and Loss of RNA Interference Pathways in Trypanosomatid Protozoans. PLoS Pathog. 2010, 6, e1001161. [Google Scholar] [CrossRef]
- Matveyev, A.V.; Alves, J.M.P.; Serrano, M.G.; Lee, V.; Lara, A.M.; Barton, W.A.; Costa-Martins, A.G.; Beverley, S.M.; Camargo, E.P.; Teixeira, M.M.G.; et al. The Evolutionary Loss of RNAi Key Determinants in Kinetoplastids as a Multiple Sporadic Phenomenon. J. Mol. Evol. 2017, 84, 104. [Google Scholar] [CrossRef] [PubMed]
- Atayde, V.D.; Shi, H.; Franklin, J.B.; Carriero, N.; Notton, T.; Lye, L.F.; Owens, K.; Beverley, S.M.; Tschudi, C.; Ullu, E. The Structure and Repertoire of Small Interfering RNAs in Leishmania (Viannia) Braziliensis Reveal Diversification in the Trypanosomatid RNAi Pathway. Mol. Microbiol. 2013, 87, 580–593. [Google Scholar] [CrossRef]
- Grimson, A.; Srivastava, M.; Fahey, B.; Woodcroft, B.J.; Chiang, H.R.; King, N.; Degnan, B.M.; Rokhsar, D.S.; Bartel, D.P. Early Origins and Evolution of MicroRNAs and Piwi-Interacting RNAs in Animals. Nature 2008, 455, 1193–1197. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.L.; Rome, L.H. Isolation and Characterization of a Novel Ribonucleoprotein Particle: Large Structures Contain a Single Species of Small RNA. J. Cell Biol. 1986, 103, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Vilalta, A.; Kickhoefer, V.A.; Rome, L.H.; Johnson, D.L. The Rat Vault RNA Gene Contains a Unique RNA Polymerase III Promoter Composed of Both External and Internal Elements That Function Synergistically. J. Biol. Chem. 1994, 269, 29752–29759. [Google Scholar] [CrossRef]
- Kedersha, N.L.; Heuser, J.E.; Chugani, D.C.; Rome, L.H. Vaults. III. Vault Ribonucleoprotein Particles Open into Flower-like Structures with Octagonal Symmetry. J. Cell Biol. 1991, 112, 225–235. [Google Scholar] [CrossRef]
- Kickhoefer, V.A.; Rajavel, K.S.; Scheffer, G.L.; Dalton, W.S.; Scheper, R.J.; Rome, L.H. Vaults Are Up-Regulated in Multidrug-Resistant Cancer Cell Lines. J. Biol. Chem. 1998, 273, 8971–8974. [Google Scholar] [CrossRef] [PubMed]
- Nandy, C.; Mrázek, J.; Stoiber, H.; Grässer, F.A.; Hüttenhofer, A.; Polacek, N. Epstein-Barr Virus-Induced Expression of a Novel Human Vault RNA. J. Mol. Biol. 2009, 388, 776–784. [Google Scholar] [CrossRef]
- van Zon, A.; Mossink, M.H.; Schoester, M.; Scheffer, G.L.; Scheper, R.J.; Sonneveld, P.; Wiemer, E.A. Multiple Human Vault RNAs. Expression and Association with the Vault Complex. J. Biol. Chem. 2001, 276, 37715–37721. [Google Scholar] [CrossRef]
- Sajini, A.A.; Choudhury, N.R.; Wagner, R.E.; Bornelöv, S.; Selmi, T.; Spanos, C.; Dietmann, S.; Rappsilber, J.; Michlewski, G.; Frye, M. Loss of 5-Methylcytosine Alters the Biogenesis of Vault-Derived Small RNAs to Coordinate Epidermal Differentiation. Nat. Commun. 2019, 10, 2550. [Google Scholar] [CrossRef]
- Miñones-Moyano, E.; Friedländer, M.R.; Pallares, J.; Kagerbauer, B.; Porta, S.; Escaramís, G.; Ferrer, I.; Estivill, X.; Martí, E. Upregulation of a Small Vault RNA (SvtRNA2-1a) Is an Early Event in Parkinson Disease and Induces Neuronal Dysfunction. RNA Biol. 2013, 10, 1093–1106. [Google Scholar] [CrossRef] [PubMed]
- Fort, R.S.; Garat, B.; Sotelo-Silveira, J.R.; Duhagon, M.A. VtRNA2-1/Nc886 Produces a Small RNA That Contributes to Its Tumor Suppression Action through the MicroRNA Pathway in Prostate Cancer. Non-Coding RNA 2020, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Benne, R.; Van Den Burg, J.; Brakenhoff, J.P.J.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major Transcript of the Frameshifted CoxII Gene from Trypanosome Mitochondria Contains Four Nucleotides That Are Not Encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef]
- Shaw, J.M.; Feagin, J.E.; Stuart, K.; Simpson, L. Editing of Kinetoplastid Mitochondrial MRNAs by Uridine Addition and Deletion Generates Conserved Amino Acid Sequences and AUG Initiation Codons. Cell 1988, 53, 401–411. [Google Scholar] [CrossRef]
- van der Spek, H.; van den Burg, J.; Croiset, A.; van den Broek, M.; Sloof, P.; Benne, R. Transcripts from the Frameshifted MURF3 Gene from Crithidia Fasciculata Are Edited by U Insertion at Multiple Sites. EMBO J. 1988, 7, 2509–2514. [Google Scholar] [CrossRef]
- Harris, M.E.; Moore, D.R.; Hajduk, S.L. Addition of Uridines to Edited RNAs in Trypanosome Mitochondria Occurs Independently of Transcription. J. Biol. Chem. 1990, 265, 11368–11376. [Google Scholar] [CrossRef]
- Gray, M.W. Evolutionary Origin of RNA Editing. Biochemistry 2012, 51, 5235–5242. [Google Scholar] [CrossRef]
- Shlomai, J. The Structure and Replication of Kinetoplast DNA. Curr. Mol. Med. 2004, 4, 623–647. [Google Scholar] [CrossRef]
- Blum, B.; Simpson, L. Guide RNAs in Kinetoplastid Mitochondria Have a Nonencoded 3’ Oligo(U) Tail Involved in Recognition of the Preedited Region. Cell 1990, 62, 391–397. [Google Scholar] [CrossRef]
- Madina, B.R.; Kumar, V.; Metz, R.; Mooers, B.H.M.; Bundschuh, R.; Cruz-Reyes, J. Native Mitochondrial RNA-Binding Complexes in Kinetoplastid RNA Editing Differ in Guide RNA Composition. RNA 2014, 20, 1142–1152. [Google Scholar] [CrossRef]
- Aphasizhev, R.; Sbicego, S.; Peris, M.; Jang, S.H.; Aphasizheva, I.; Simpson, A.M.; Rivlin, A.; Simpson, L. Trypanosome Mitochondrial 3′ Terminal Uridylyl Transferase (TUTase): The Key Enzyme in U-Insertion/Deletion RNA Editing. Cell 2002, 108, 637–648. [Google Scholar] [CrossRef]
- Seiwert, S.D.; Heidmann, S.; Stuart, K. Direct Visualization of Uridylate Deletion In Vitro Suggests a Mechanism for Kinetoplastid RNA Editing. Cell 1996, 84, 831–841. [Google Scholar] [CrossRef]
- Aphasizhev, R.; Aphasizheva, I. Mitochondrial RNA Editing in Trypanosomes: Small RNAs in Control. Biochimie 2014, 100, 125. [Google Scholar] [CrossRef] [PubMed]
- Koslowsky, D.; Sun, Y.; Hindenach, J.; Theisen, T.; Lucas, J. The Insect-Phase GRNA Transcriptome in Trypanosoma Brucei. Nucleic Acids Res. 2014, 42, 1873–1886. [Google Scholar] [CrossRef] [PubMed]
- Kirby, L.E.; Sun, Y.; Judah, D.; Nowak, S.; Koslowsky, D. Analysis of the Trypanosoma Brucei EATRO 164 Bloodstream Guide RNA Transcriptome. PLoS Negl. Trop. Dis. 2016, 10, e0004793. [Google Scholar] [CrossRef] [PubMed]
- Simpson, L.; Douglass, S.M.; Lake, J.A.; Pellegrini, M.; Li, F. Comparison of the Mitochondrial Genomes and Steady State Transcriptomes of Two Strains of the Trypanosomatid Parasite, Leishmania Tarentolae. PLoS Negl. Trop. Dis. 2015, 9, e0003841. [Google Scholar] [CrossRef]
- van der Ploeg, L.H.T.; Liu, A.Y.C.; Michels, P.A.M.; de Lange, T.; Borst, P.; Majumder, H.K.; Weber, H.; Veeneman, G.H.; Boom, J. Van RNA Splicing Is Required to Make the Messenger RNA for a Variant Surface Antigen in Trypanosomes. Nucleic Acids Res. 1982, 10, 3591–3604. [Google Scholar] [CrossRef]
- Agabian, N. Trans Splicing of Nuclear Pre-MRNAs. Cell 1990, 61, 1157–1160. [Google Scholar] [CrossRef]
- Tan, T.H.P.; Pach, R.; Crausaz, A.; Ivens, A.; Schneider, A. TRNAs in Trypanosoma Brucei: Genomic Organization, Expression, and Mitochondrial Import. Mol. Cell. Biol. 2002, 22, 3707–3717. [Google Scholar] [CrossRef]
- Perry, K.L.; Watkins, K.P.; Agabian, N. Trypanosome MRNAs Have Unusual “Cap 4” Structures Acquired by Addition of a Spliced Leader. Proc. Natl. Acad. Sci. USA 1987, 84, 8190–8194. [Google Scholar] [CrossRef]
- Gilinger, G.; Bellofatto, V. Trypanosome Spliced Leader RNA Genes Contain the First Identified RNA Polymerase II Gene Promoter in These Organisms. Nucleic Acids Res. 2001, 29, 1556–1564. [Google Scholar] [CrossRef] [PubMed]
- Milhausen, M.; Nelson, R.G.; Sather, S.; Selkirk, M.; Agabian, N. Identification of a Small Rna Containing the Trypanosome Spliced Leader: A Donor of Shared 5′ Sequences of Trypanosomatid MRNAs? Cell 1984, 38, 721–729. [Google Scholar] [CrossRef]
- Podlevsky, J.D.; Chen, J.J.L. Evolutionary Perspectives of Telomerase RNA Structure and Function. RNA Biol. 2016, 13, 720–732. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, R.; Sanford, S.; Basu, S.; Park, M.; Pandya, U.M.; Li, B.; Chakrabarti, K. A Trans-Spliced Telomerase RNA Dictates Telomere Synthesis in Trypanosoma Brucei. Cell Res. 2013, 23, 537–551. [Google Scholar] [CrossRef]
- Davis, J.A.; Chakrabarti, K. Telomerase Ribonucleoprotein and Genome Integrity—An Emerging Connection in Protozoan Parasites. Wiley Interdiscip. Rev. RNA 2021, 1–20. [Google Scholar] [CrossRef]
- Gupta, S.K.; Kolet, L.; Doniger, T.; Biswas, V.K.; Unger, R.; Tzfati, Y.; Michaeli, S. The Trypanosoma Brucei Telomerase RNA (TER) Homologue Binds Core Proteins of the C/D SnoRNA Family. FEBS Lett. 2013, 587, 1399–1404. [Google Scholar] [CrossRef]
- Logeswaran, D.; Li, Y.; Podlevsky, J.D.; Chen, J.J.-L. Monophyletic Origin and Divergent Evolution of Animal Telomerase RNA. Mol. Biol. Evol. 2021, 38, 215–228. [Google Scholar] [CrossRef]
- Dey, A.; Chakrabarti, K. Current Perspectives of Telomerase Structure and Function in Eukaryotes with Emerging Views on Telomerase in Human Parasites. Int. J. Mol. Sci. 2018, 19, 333. [Google Scholar] [CrossRef]
- Alcaraz-Pérez, F.; García-Castillo, J.; García-Moreno, D.; López-Muñoz, A.; Anchelin, M.; Angosto, D.; Zon, L.I.; Mulero, V.; Cayuela, M.L. A Non-Canonical Function of Telomerase RNA in the Regulation of Developmental Myelopoiesis in Zebrafish. Nat. Commun. 2014, 5, 3228. [Google Scholar] [CrossRef]
- Raghunandan, M.; Decottignies, A. The Multifaceted HTR Telomerase RNA from a Structural Perspective: Distinct Domains of HTR Differentially Interact with Protein Partners to Orchestrate Its Telomerase-Independent Functions. BioEssays 2021, 43, 2100099. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, J.; Li, Y.; Song, T.; Wu, Y.; Fang, S.; Bu, D.; Li, H.; Sun, L.; Pei, D.; et al. NONCODEV6: An Updated Database Dedicated to Long Non-Coding RNA Annotation in Both Animals and Plants. Nucleic Acids Res. 2021, 49, D165–D171. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Kolev, N.G.; Franklin, J.B.; Carmi, S.; Shi, H.; Michaeli, S.; Tschudi, C. The Transcriptome of the Human Pathogen Trypanosoma Brucei at Single-Nucleotide Resolution. PLOS Pathog. 2010, 6, e1001090. [Google Scholar] [CrossRef]
- Kim, H.C.; Jolly, E.R. LncRNAs Are Differentially Expressed between Wildtype and Cell Line Strains of African Trypanosomes. Non-Coding RNA 2022, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Madej, M.J.; Alfonzo, J.D.; Hüttenhofer, A. Small NcRNA Transcriptome Analysis from Kinetoplast Mitochondria of Leishmania Tarentolae. Nucleic Acids Res. 2007, 35, 1544–1554. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Silva, M.R.; Tosar, J.P.; Frugier, M.; Pantano, S.; Bonilla, B.; Esteban, L.; Serra, E.; Rovira, C.; Robello, C.; Cayota, A. Cloning, Characterization and Subcellular Localization of a Trypanosoma Cruzi Argonaute Protein Defining a New Subfamily Distinctive of Trypanosomatids. Gene 2010, 466, 26–35. [Google Scholar] [CrossRef]

| Organism | RNA Biotypes Studies | Sample Origin | RNA Size Selection | Library Type | Sequencing Technology | Reference |
|---|---|---|---|---|---|---|
| T. brucei | siRNAs | High-speed pellet and supernatant fractions | 20–30 nt | Cloned | Sanger sequencing | [96] |
| L. tarentolae | gRNAs | Kinetoplast RNA | 15–600 nt | 5′/3′RACE | Sanger sequencing | [145] |
| T. cruzi | tRNAs | Epimastigotes normal growth conditions or under nutritional stress | 18–40 nt | Cloned | Sanger sequencing | [146] |
| T. brucei | siRNAs | Bloodstream | 18–30-nt | Ligation and cDNA | Illumina sequencing | [98] |
| T. cruzi | tRNAs, rRNAs, snRNAs and snoRNAs | Epimastigotes | 16–60 nt | Ligation and cDNA | 454 sequencing | [40] |
| T. brucei | tRNAs, rRNAs, snRNAs and snoRNAs | Procyclic small RNPs complexes | 20–30 nt | Ligation and cDNA | Illumina sequencing | [36] |
| T. cruzi | tRNAs | Metacyclic trypomastigotes | 18–40 nt | Cloned | Sanger sequencing | [52] |
| T. brucei | siRNAs | Epimastigotes TbAGO1 immunoprecipitates mut Dicer 1/2 | 18–30 nt | Ligation and cDNA | Illumina sequencing | [97] |
| L. braziliensis | siRNAs | Promastigotes RNA immunoprecipitated with BB2-LbrAGO1 | 18–30 nt | Ligation and cDNA | Illumina sequencing | [102] |
| T. brucei | siRNAs, tRNAs, rRNAs, snRNAs and snoRNAs | Slender form and procyclic form | 18–30 nt | Ligation and cDNA | Illumina sequencing | [55] |
| T. cruzi | tRNAs, rRNAs, snRNAs and snoRNAs | Epimastigotes TcPIWI-tryp bound sRNAs | 16–50 nt | Ligation and cDNA | Illumina sequencing | [85] |
| T. cruzi | siRNAs, tRNAs, rRNAs, snRNAs and snoRNAs | EVs/parental cells from epimastigotes and metacyclic trypomastigotes | 16–40 nt | Ligation and cDNA | Illumina sequencing | [53] |
| T. brucei | gRNA | Procyclic form enriched mitochondrial vesicles | 40–80 nt | Ligation and cDNA (PNK treatment) | Illumina sequencing | [124] |
| L. donovani and L. braziliensis | siRNAs, tRNAs, rRNAs, snRNAs and snoRNAs | Exosomes from early axenic amastigotes | 20–250 nt | Ligation and cDNA (CIP and TAP treatment) | Illumina sequencing | [56] |
| L. tarentolae | gRNA | Purified kinetoplast mitochondrial | <200 nt | Ligation and cDNA (PNK treatment) | Illumina sequencing | [126] |
| T. cruzi | tRNAs, rRNAs, snRNAs and snoRNAs | Epimastigotes EVs and intracellular | <60 nt | Ligation and cDNA | Illumina sequencing | [41] |
| T. brucei | gRNA | Bloodstream form enriched mitochondrial vesicles | 40–80 nt | Ligation and cDNA (PNK treatment) | Illumina sequencing | [125] |
| T. brucei | siRNAs | Epimastigotes WT and H3.VKO | 18–30 nt | Ligation and cDNA | Illumina sequencing | [99] |
| T. brucei | tRNAs, rRNAs, snRNAs and snoRNAs | RNAs co-purify with cytosolic ribosomes of parasites grown under different conditions | 20–300 nt | Ligation and cDNA | Illumina sequencing | [28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fort, R.S.; Chavez, S.; Trinidad Barnech, J.M.; Oliveira-Rizzo, C.; Smircich, P.; Sotelo-Silveira, J.R.; Duhagon, M.A. Current Status of Regulatory Non-Coding RNAs Research in the Tritryp. Non-Coding RNA 2022, 8, 54. https://doi.org/10.3390/ncrna8040054
Fort RS, Chavez S, Trinidad Barnech JM, Oliveira-Rizzo C, Smircich P, Sotelo-Silveira JR, Duhagon MA. Current Status of Regulatory Non-Coding RNAs Research in the Tritryp. Non-Coding RNA. 2022; 8(4):54. https://doi.org/10.3390/ncrna8040054
Chicago/Turabian StyleFort, Rafael Sebastián, Santiago Chavez, Juan M. Trinidad Barnech, Carolina Oliveira-Rizzo, Pablo Smircich, José Roberto Sotelo-Silveira, and María Ana Duhagon. 2022. "Current Status of Regulatory Non-Coding RNAs Research in the Tritryp" Non-Coding RNA 8, no. 4: 54. https://doi.org/10.3390/ncrna8040054
APA StyleFort, R. S., Chavez, S., Trinidad Barnech, J. M., Oliveira-Rizzo, C., Smircich, P., Sotelo-Silveira, J. R., & Duhagon, M. A. (2022). Current Status of Regulatory Non-Coding RNAs Research in the Tritryp. Non-Coding RNA, 8(4), 54. https://doi.org/10.3390/ncrna8040054






