Abstract
The use of lactic acid bacteria (LAB) in the fermentation process to produce fermented foods has a long history. Furthermore, LAB are beneficial microorganisms known for their health-promoting characteristics. During fermentation, LAB have the capacity to produce significant amounts of bioactive substances, such as peptides, bacteriocins, lactic acid, exopolysaccharides (EPSs), enzymes, and others. Lactococcus lactis as one of the best-known and well-characterized species of LAB serves as a model organism for studying LAB. For a very long time, L. lactis has been used in milk fermentation, both in well-monitored industrial settings and on a small scale in traditional operations. Furthermore, L. lactis is a vital microorganism in the dairy food fermentation industry due to its role in acidification, flavor development, and the creation of various dairy products, including cheese, fermented butter, and others. The novelty of this review is the comprehensive and organized presentation of the main benefits of the use of L. lactis in milk fermentation processes including technological and safety features relevant for the dairy industry, probiotic potential, the ability to produce bioactive compounds (e.g., bacteriocins, GABA), and the recent development of such bacteria research methods like whole genome sequencing (WGS).
1. Introduction
Nowadays, consumers not only want to experiment with different flavors and ingredients in dairy products, but also are more careful about their food selections. This is due to the rising number of life-threatening chronic illnesses in Western countries; therefore, consumers also focus their attention on healthy nutrition [1]. In response to consumer demands, dairy companies are always seeking new ideas. This has resulted in the widespread use of probiotics in a variety of dairy products. Numerous companies have also looked into using culture manipulations as a tool for flavor diversification [2,3,4].
Consumer interest in a healthy lifestyle and demand for additional benefits from food apart from a nutritional effect have promoted a functional food market around the world. This growing consumer demand is leading to the increased consumption of dairy foods with functional properties [5]. There is no specific definition for functional foods, though functional food could be described as an ordinary food with components or ingredients that provide a specific health benefit besides a nutritional effect. When a desirable biological, medical, or physiological effect is exerted by microorganisms, such food is called biofunctional food [6]. The term biofunctional concept is typically applied when a beneficial metabolite that results from bacterial metabolic activity in the fermentation process appears in the food product itself. In this way, bacteria can enrich food products with bioactive compounds during fermentation and are not necessarily required to be alive when ingested to produce a beneficial metabolite [7].
The most common method for obtaining microorganisms useful for industrial applications is screening of microorganisms isolated from naturally occurring processes [8]. However, a careful selection of starter microorganisms must be carried out in order to achieve faster, more efficient, controlled, and large-scale fermentation and to ensure the reproducibility of food fermentation at the industrial scale and to have a product with specific properties [9,10]. Desirable technological, nutritional, and sensory qualities are the primary criteria used to select microbial starters because the selection of a starter culture has a significant influence on the final quality of fermented food products [10].
Lactic acid bacteria (LAB) have a long history of application in the food fermentation process, although recently more attention has been directed to their metabolites [11]. LAB are used in the food industry as functional starter cultures that can contribute to food safety and also offer one or more organoleptic, technological, nutritional, or health advantages [12]. LAB include a large amount of bacterial species from which many strains have proven to have multifunctional features such as high fermentative capacity and/or relevant beneficial skills for humans [13]. LAB are also known for production of antimicrobial substances such as organic acids and bacteriocins [14]. Furthermore, LAB can produce some bioactive compounds during fermentation, including enzymes, vitamins, conjugated linoleic acid, exopolysaccharides, and gamma-aminobutyric acid (GABA) [6].
Among all LAB, Lactococcus lactis plays a crucial role in the production of some dairy products including cheese, fermented butter, and other products and is a vital microorganism in the dairy food fermentation due to its role in acidification and flavor development; therefore, selected strains of this species can be considered as main components of starter cultures for dairy fermentation [2,15]. Also, in vitro and in vivo studies on the probiotic potential of L. lactis strains have suggested their use as a probiotic, besides their ability to improve nutritional value of foods [3].
In the past couple of years, several literature reviews concerning L. lactis were published. Several works reviewed probiotic characteristics of L. lactis [16,17]. A review by Li et al. [2] aimed to investigate fermentation characteristics of L. lactis subsp. lactis isolated from naturally fermented dairy products and its potential as starter cultures. Also, a review by Mahony et al. [18] aimed to highlight emerging lactococcal species that may play a role in culture performance and functional attributes of fermented dairy products.
The novelty of this review is the comprehensive and organized presentation of the main benefits of the use of L. lactis in milk fermentation processes including technological and safety features relevant for the dairy industry, probiotic potential, the ability to produce bioactive compounds (e.g., bacteriocins, GABA), and the recent development of such bacteria research methods like whole genome sequencing (WGS).
2. Lactic Acid Bacteria
LAB are a class of anaerobic, Gram-positive, catalase-negative microorganisms that can survive and grow in the presence of oxygen. They have the appearance of cocci or rods and do not form spores; the main byproduct of sugar fermentation is lactic acid [19,20]. The benefits of LAB consumption became apparent in the early 20th century when a scientist proposed that consuming the live microorganisms found in yogurt would lengthen the consumer’s life by improving host health and lowering the number of bacteria in the digestive tract that cause spoilage and toxins [21]. Following their discovery, LAB have been the subject of extensive research, and an impressive amount of data has been gathered about their contribution to the final product’s organoleptic qualities and nutritional value, as well as how they extend the shelf life of fermented foods. This is because LAB produce a wide range of compounds, such as ethanol, hydrogen peroxide, bacteriocins, and organic acids, among others, which work in concert to prevent or eradicate microbial contamination [22,23]. The ability of LAB strains to produce bacteriocins can be seen as a benefit and a functional role in the food industry to enhance food safety and quality [24]. Additionally, LAB produce metabolites, which are technologically attractive and explain how LAB are used in food to produce a wide range of fermented products [25].
LAB can grow in the majority of raw foods and are typically found in nutrient-rich environments [26]. Meat, dairy products, wine, beverages, sourdoughs for baked goods, fruits, vegetables, fish, and sea products are examples of food products that contain LAB. Based on the type of food product, handling techniques, and environment, different microflora predominate in these food products [26,27,28]. Since the majority of LAB’s properties are strain-dependent, the variety of LAB is intriguing overall both at the species and strain levels [25].
Since LAB have been used for centuries in food production and fermentation without posing any health risks, the American Food and Drug Agency (FDA) has classified the majority of LAB as Generally Recognized as Safe (GRAS) at the strain level [19,29,30]. The Qualified Presumption of Safety (QPS) status was also granted by the European Food Safety Authority (EFSA) to the majority of LAB genera at the species level, including Lactococcus, Lactobacillus, Leuconostoc, Pediococcus, and Streptococcus thermophilus [19,20,30]. Oenococcus, which is used in wine; Lactobacillus, which is used in meat, vegetables, dairy, and cereals; and Lactococcus, which is used in dairy products, are some of the most frequently chosen genera for industrial application [31].
2.1. Lactococcus lactis
Gram-positive cocci known as Lactococcus are found solely in pairs or in chains. They are catalase-negative, and facultatively anaerobic L-lactic acid is the main byproduct of the fermentation of glucose during the glycolytic pathway of L. lactis [32]. The lactic acid produced by the anaerobic conditions throughout the glycolysis pathway adds to the sour taste of fermented foods like yogurt [33]. While there are two types of fermentation, homo-lactic acid fermentation and hetero-lactic acid fermentation, Lactococcus is an example of homo-lactic acid fermentation because it produces only lactic acid (pyruvate is created by the glycolysis process using glucose as a carbon source, and lactate dehydrogenase converts it to lactic acid) [33].
L. lactis as one of the best known and characterized species of LAB serves as a model organism for studying LAB [29]. Taxonomically, L. lactis is a mesophilic species that belongs to the Streptococcaceae family. It is further divided into four subspecies: L. lactis subsp. hordniae, L. lactis subsp. lactis (which includes the biovar diacetylactis), L. lactis subsp. tructae, and L. lactis subsp. cremoris [34,35,36]. Known as “dairy” lactococci, L. lactis subsp. lactis and L. lactis subsp. cremoris together constitute the primary LAB components of a dairy starter, a costarter, or flavoring adjunct culture systems used in the production of cheese [31,37]. Recently, Li et al. [36] proposed that L. lactis subsp. cremoris should be elevated to the species level as L. cremoris sp. and L. lactis subsp. tructae should be transferred to L. cremoris as L. cremoris subsp. tructae. This was included in the official list of recognized prokaryotic species [38].
Strains that are isolated from raw milk or even non-dairy environments are referred to as “wild” lactococci. According to studies, wild lactococci isolated from dairy and non-dairy sources can produce particular flavors that are different from those of industrial strains [35,39]. For a very long time, L. lactis has been utilized in milk fermentation, both in well-managed industrial applications and small-scale traditional operations [40]. Technological contribution of lactococci has always been associated with the manufacturing stage; however, because of their proteolytic and amino acid conversion pathways, they can also affect the final texture and flavor of dairy products [41,42].
2.2. L. lactis Application in the Dairy Industry
A significant number of LAB components linked to dairy food fermentation are L. lactis strains. These strains are used in commercial mesophilic starter cultures or small-scale traditional processes like making artisanal cheese, or they are employed in carefully regulated industrial dairy fermentation to produce cheese and ripen it, as well as fermented milk products like buttermilk, butter, and sour cream [18,31,35,40,43,44]. In the dairy industry, mesophilic starter cultures are classified as D-, L-, DL-, or O-cultures according to the application of L. lactis biovar diacetylactis (D-), Leuconostoc (L-), or a combination of both species (DL-) for flavor formation in addition to L. lactis strain(s) that are the major contributors to acid formation. Likewise, O-cultures are those starter cultures that exclusively rely on L. lactis for acidification [18]. It is assessed that through the consumption of fermented dairy products, humans ingest up to 1018 lactococcal cells throughout the year [31]. In dairy starter cultures, the L. lactis primary function is to generate lactic acid at a sufficient rate and to aid in the fermentation process by breaking down milk proteins. These actions have a significant impact on the final dairy product’s microbiological quality and organoleptic characteristics (Table 1) [31].

Table 1.
Properties of L. lactis strains, isolated from dairy sources, useful for dairy food production.
Proteolytic activity, lactose fermentation capabilities, exopolysaccharide (EPS) production, flavor production, and other isolate-dependent traits are among the many traits of L. lactis used as starter cultures for commercial purposes [2]. L. lactis subsp. cremoris and L. lactis subsp. Lactis, also L. lactis subsp. lactis biovar. diacetylactis, are the most common L. lactis subspecies found in dairy environments. L. lactis subsp. lactis is usually differentiated from L. lactis subsp. cremoris by its tolerance to higher temperatures and salt concentrations.
From the technological perspective, two characteristics of L. lactis strains are of significant importance—they can grow at 10 °C and at 40 °C, but not at 45 °C, and can also tolerate high concentrations of NaCl [32]. The strains’ ability to tolerate NaCl allows them to survive the negative effects of high osmotic pressure in the gastrointestinal tract’s high-salt environment and preserve a relative osmotic pressure balance in such circumstances. Because bacterial cells grown in high salt concentrations will experience a loss of turgor pressure that would affect the physiology, enzyme activity, water activity, and cell metabolism, the NaCl tolerance test indicates the degree of osmotolerance exhibited by a given LAB strain. It is known that homofermentative strains like L. lactis are more resistant to NaCl than heterofermentative strains [18]. LAB are described as homofermentative when an end product of fermentation is numerous amounts of lactic acid alone and heterofermentative when lactic acid is produced together with acetic acid, ethanol, and carbon dioxide [19].
The primary criterion for selection of L. lactis subsp. lactis starter cultures is acidification activity, as these cultures are frequently employed in the production of cheese, butter, and other fermented milk products. Slow or medium acid-producing strains are only employed if they have additional desirable qualities. Fast acid-producing strains are most commonly chosen as starter cultures [2]. Additionally, the dairy industry must consider the capacity for acid production during storage because low post-fermentation acidification is a crucial factor for commercial starters used to produce fermented milk because it impacts the quality of the finished product [2,49].
Some Lactococcus strains, such as L. lactis subsp. lactis biovar diacetylactis, exhibit additional fermentation capabilities, such as the fermentation of citrate, which results in the production of diacetyl, a flavor and aroma enhancer. Citrate can be converted by these strains into carbon dioxide and aroma compounds (C4), which enhance the organoleptic properties of fermented foods [2,47]. Diacetyl plays a crucial role in the flavor of many dairy products because, even in small amounts, it gives certain cheeses their distinctive creamy and buttery aroma [47].
2.3. Probiotic Features of L. lactis
Probiotics are defined as live microorganisms, which, when administered in adequate amounts, confer a health benefit on the host [30]. Probiotic-containing food products are also referred to as functional foods (the fastest-growing category of functional food development is probiotic foods) [50,51] and have a number of positive health effects, such as lowering blood pressure and serum cholesterol levels [52,53], improvement in lactose intolerance [54], production of numerous vital bioactive molecules, such as vitamins, amino acids, and gamma-aminobutyric acid, demonstrating health-promoting activity, stimulation of the growth of beneficial microorganisms and reduction in the amount of pathogens, and improving the intestinal microbial balance of the host and lowering the risk of gastro-intestinal diseases [29,55]. Thus, there is a great deal of interest in the isolation of novel probiotic strains that have the potential to enhance the quality of already-existing food products and promote health [56,57]. The optimal daily dosage of probiotic bacteria to achieve the desired effect is between 106 and 109 viable organisms [58,59], although studies have been performed that demonstrate dead cells can also have positive immunological effects [57].
Interesting probiotic potential has also been shown by certain L. lactis strains. Several previous studies have shown that some L. lactis strains that were isolated from raw and fermented milk were resistant to bile salts and acids (pH 2.5–3) and exhibited other properties like antimicrobial activity or growth in cholesterol, classifying them as probiotics [60,61]. The primary component of bile, bile salts, are toxic to living cells because they have the ability to alter the structure of cell membranes [62]. Consequently, in order for probiotic strains to reach the small intestine or colon alive, they must endure exposure to bile [63]. Human bile’s physiological concentration falls between 0.3 and 0.5% [64]. The concentration at which the growth lag time of LAB strains is measured is 0.3%; L. lactis has been reported to be resistant to 0.3–1.0% (w/v) of bile salt [60,65,66].
L. lactis has exhibited resilience in the severe conditions of the gastrointestinal tract, as well as the capacity to remain viable in the intestine and generate lactic acid, which contributes to the preservation of an acidic environment in the gut and prevents the growth of pathogenic bacteria [67]. It has been observed that L. lactis strengthens the tight junctions between intestinal epithelial cells, thereby enhancing the integrity of the gut barrier. This characteristic lowers intestinal permeability and stops harmful compounds from entering the bloodstream [68,69]. Bacteriocins, which inhibit the growth of pathogenic bacteria, are among the antimicrobial substances that L. lactis produces. In order to keep the gut microbiota balanced and healthy, these antimicrobial properties are essential [65]. Detailed information about L. lactis, isolated from dairy products, probiotic characteristics is given in Table 2.

Table 2.
Probiotic features of L. lactis strains isolated from dairy sources.
3. Safety Assessment of L. lactis
Safety evaluation of microorganisms used in the food industry has become crucially important to ensure the health of consumers [77]. LAB have been used for different purposes for centuries, which has allowed them to be classified as GRAS microorganisms by the FDA and QPS by the European Food Safety Authority [78,79]. Lactococcus spp. are generally considered as safe; however, they can carry and also receive virulence genes and become a virulence reservoir in a food system [48,80]. Some studies describe that certain LAB produce toxic compounds and can cause problems such as bacteremia, endocarditis, and peritonitis, especially in individuals with previous pathological conditions [81]. Thus, safety assessment of L. lactis based on virulence, antibiotic resistance, and hemolytic activities is essential in the selection of these bacteria for application in food production [82].
3.1. Virulence Assessment
Virulence assessment of L. lactis is based on the testing for the harboring well-established different Enterococcus spp. virulence genes including gelE (gelatinase), hyl (hyaluronidase), asa1 (aggregation substance), esp (enterococcal surface protein), cylA (cytolisin), efaA (endocarditis antigen), ace (adhesion of collagen), and vanA and vanB (both related to vancomycin resistance) [83]. It is important to verify the presence of these virulence genes in L. lactis, in regard to close relation to Enterococcus spp., which have become important pathogens of nosocomial infections [84]. The gelE gene encodes a protein that is an extracellular protease and has been associated with biofilm formation in Enterococcus spp. [85]. The ability to form biofilms is also related to enterococcal surface protein (esp) and adhesion of collagen (ace) factors [85]. The hyl gene encodes an enzyme called hyaluronidase and the presence of the hyl gene can contribute to virulence and the ability of the bacterium to spread within host tissues [86]. The aggregation substance coded by the asa1 gene is enterococcal surface protein and it often acts as a virulence factor and it transfers antibiotic resistance genes [85,87]. The cell wall adhesin (efa A) is a virulence factor associated with infective endocarditis [86]. Cytolysin (cyl) is known for bactericidal properties toward Gram-negative bacteria and toxic properties (b-hemolysis) [86]. Testing of L. lactis on vancomycin resistance involves special attention because this antibiotic is considered as the drug of choice for C. difficile infection treatment [88].
3.2. Antibiotic Resistance
Antibiotic resistance is an urgent public health concern in the 21st century and is drawing interest all around the world [89]. Antibiotics are widely used in human and veterinary medicine to treat different bacterial infections. However, the antibiotic resistance of bacteria is on the rise. The biggest concern is the spread of antibiotic resistance coding genes in microbial communities. Thus, concerns have been raised about the existence of antibiotic resistance in beneficial bacterial species including LAB that can disseminate the antibiotic resistance through the entire food chain [58]. Gastrointestinal microbiota including LAB are potentially vulnerable to acquired antibiotic resistance [90]. Noteworthily, it is important to consider the genomic location of an antibiotic resistance gene when testing as unlike plasmid-mediated resistance genes, chromosome-mediated resistance genes have a much lower risk of transfer [91].
Safety assessment of L. lactis on the susceptibility to antibiotics is based on the testing of the antibiotics listed in EFSA scientific opinion and commonly used for the treatment of enterococcal infections [92,93]. The cutoff values (μg mL−1) indicated in EFSA include ampicillin (2 μg mL−1), clindamycin (1 μg mL−1), chloramphenicol (8 μg mL−1), erythromycin (1 μg mL−1), gentamicin (32 μg mL−1), kanamycin (64 μg mL−1), streptomycin (32 μg mL−1), tetracycline (4 μg mL−1), and vancomycin (4 μg mL−1). L. lactis isolated from different sources demonstrate different and in certain cases high resistance to antibiotics (Table 3).

Table 3.
Antibiotic resistance of L. lactis isolated from different sources.
3.3. Hemolytic Activity
Hemolytic activity of bacteria is related to the ability to destroy red blood cells and this is a typical property of pathogenic bacteria. Thus, this test provides important information about the tested strain‘s pathogenicity [91]. Only a L. lactis strain with a lack of hemolytic activity is considered as safe and not virulent and the lack of hemolysin ensures that opportunistic virulence will not appear among strains [97]. Evaluation of hemolytic activity is performed in nutrient agar supplemented with 5% defibrinated sheep blood. The hemolytic reaction is evaluated by observing the partial hydrolysis of the red blood cells and the production of a green zone (α-hemolysis), a clear zone around the bacterial colonies (β-hemolysis) or no reaction (γ-hemolysis).
3.4. Biogenic Amines
Biogenic amines (BAs) are low-molecular-weight organic nitrogen compounds formed in food and beverages during enzymatic activities of raw materials or are developed by microbial decarboxylation of amino acids [98,99]. Due to proteolysis, the content of free amino acids increases and decarboxylative activity of bacterial enzymes may favor the formation of BA [100,101,102]. The concentration of BA in fermented foods is influenced by the raw materials and processing hygiene. Fermentation conditions, such as the type of bacteria used, the process’s duration and temperature, and the food matrix, also play a significant role in the formation of these toxic compounds. It is essential to monitor BA content, especially in fermented foods and/or beverages, because higher amounts of these substances signal lower quality and safety of products, due to their potential toxicity to human health [98,103]. Also, the amount of BAs greatly depends on the release of specific amino acids (which are precursors of biogenic amines); for example, the amount of tyrosine affects the higher amount of tyramine, while arginine and ornithine are responsible for a higher putrescine concentration [102,104,105]. Moreover, it is known that LAB are responsible for BA generation in fermented products and Lactococcus spp. are no exception. Despite the broad application of L. lactis subsp. lactis, L. lactis subsp. lactis biovar diacetylactis, and L. lactis subsp. cremoris in the dairy industry, some of these bacteria have been reported to have aminobiogenic activity, both in vitro and in real systems [47,98,106,107].
L. lactis subsp. lactis and L. lactis subsp. cremoris are capable of generating putrescine from agmatine via the agmatine deiminase (AGDI) pathway, which involves the production of ATP and two ammonium ions, and it is a species-level trait. These bacteria strains together with E. faecalis, E. hirae, L. brevis, and L. curvatus are pivotal putrescine producers in dairy foods, especially cheese [99,105,108]. The concentration of putrescine, in different varieties of cheeses produced from cow, sheep, or goat milk and prepared with L. lactis spp. as a starter culture, can reach 1560 mg kg−1; however, there are no EU regulations that set the upper level of BAs in cheese and other foods, except of histamine [109,110,111,112]. The concentration of cadaverine in cheeses is associated with the presence of contaminating bacteria in the raw material; however, a small amount of this BA indicates good quality and hygiene of milk. It is known that cadaverine and putrescine concentrations correlate with ripening time of the cheeses, and in over-ripened cheeses, the amount of detected BA was higher compared with results before the ripening, as well as in products prepared from raw milk [110,113]. Also, the type and part of cheeses have an influence on cadaverine concentration in products. For instance, the content of this BA can vary from 2.57 to 64.22 mg kg−1 in the core, and from 3.70 to 38.17 mg kg−1 in the rind of the cheeses [114,115,116,117]. Histamine and tyramine are the most toxic BAs for humans and also are the most abundant biogenic amines in cheeses prepared from cow and sheep milk [101,114,118]. However, the amount of histamine varies depending on the product. For example, in both semi-hard and hard cheeses, the core has more histamine than the rind [101,114,119]. As previously noted, tyramine is the second most toxic base acid (BA). Other researchers found that this BA was the most prevalent amine, making up 75.4% of the amines in mold cheese, 41.3% in hard cheeses, and 35% in semi-hard cheeses [114,120,121,122]. However, it was reported that L. lactis has an ambiguous effect on tyramine concentration in cheeses. L. lactis, because of the ability to produce nisin, as also used as a starter culture (L. lactis subsp. lactis plus L. lactis subsp. cremoris), can reduce contents of tyramine after ripening [110,118]. However, other researchers published that the latter BA concentration increases during the ripening process of different cheese production, despite the use of starter cultures of loctococci alone or together with an adjunct culture [123,124,125]. Tyramine content in cheeses differs, for instance, in mold-ripened (from 0.97 to 710.5 mg kg−1), semi-hard (from 2.30 to 767.03 mg kg−1), and hard cheeses (from 1.33 to 236.33 mg kg−1) and in all cases, the latter BAs concentration in products’ core was found to be higher compared with the rind [104,114].
It is important to lower putrescine as well as cadaverine concentration in dairy foods, due to their ability to develop an undesirable flavor of cheese [110,126]. Moreover, the latter BAs have an ability to potentiate the toxicity of histamine and tyramine [109,126,127]. Histamine intolerance has become more common in recent years, and even low-histamine foods can cause a wide range of nonspecific gastrointestinal neurological, cardiovascular, respiratory, and skin symptoms. For this reason, it is crucial to reduce the amount of histamine in food [128]. In the European Union, a 50 mg kg−1 histamine concentration in foods is defined as the limit with no adverse effects [112] and it is known that this BA’s concentration is higher in raw milk cheeses than in processed ones [101,114]. Also, 100 to 800 mg kg−1 of tyramine has been described as toxic in foods. However, most EU countries do not establish limited concentrations of tyramine in foods, and 800 mg kg−1 can be considered as the upper legal limit, except for the 200 mg kg−1 limit of the latter BA in cheese applied in The Slovak Republic’s Nutritional Codex [98,127]. Flasarová et al. [104] determined that the higher tyramine concentration in cheese samples prepared with the strains of L. lactis subsp. cremoris CCDM 824 and L. lactis subsp. cremoris CCDM 946 could be explained by tested strains’ ability to decarboxylate tyrosine to form tyramine. Symptoms associated with the adverse effects of the tyramine are gastrointestinal (nausea, cytotoxic in the intestinal epithelium), neurological (headache, palpitation, and migraine), circulatory (sweating, chest pain), respiratory (chest tightness), etc., and these symptoms are called a tyramine reaction or cheese reaction [128]. Other BA contents, for instance, spermidine, spermine, tryptamine, and phenylethylamine, are low in mold-ripened, semi-hard, and hard cheese models during the ripening process [104,110,114,115].
The use of the nisin-producing strain of L. lactis subsp. lactis [110,125], pasteurization [108,110,118,129], which decreases the counts of microorganisms in raw milk, and appropriate ripening time [108,110,111,113,130] could lead to a lower putrescine concentration in the final product. Also, it was reported that there are L. lactis subsp. cremoris strains lacking genes for putrescine biosynthesis and hence do not produce this BA. However, other strains of lactococci have the AGDIc and produce putrescine [105,107]. Other researchers published results, suggesting that in order to lower histamine concentrations in dairy products, L. lactis spp. as a main starter together with other LAB strains as an adjunct culture can be applied [104,110,114,123]. Moreover, environmental conditions of the real food matrix, for instance, dry matter, NaCl content, and pH, have significant influence on BA content in the final product, so it is indispensable to analyze the abilities of microorganisms to produce BA not only in vitro, but also in a real system of foodstuffs. Another important factor for BA control is selection of starter cultures, which are decarboxylase-negative or can degrade these toxic compounds. And finally, a global safety assurance program, from raw milk quality and hygiene, within all processing durations, storage, and retailing of cheeses, is mandatory in order to lower the risk of BA formation in products.
4. Bioactive Compounds Produced by L. lactis
L. lactis has the ability to produce a great variety of bioactive compounds [131], which possess advantageous effects for technological aspects of food quality and safety parameters [132,133,134], as well as human health [68,135,136]. Bioactive compounds produced by L. lactis (exopolysaccharides (EPSs) [44,132]; organic acids, especially lactic acid (LA) [29,137,138]; bacteriocins [139,140,141,142]; bioactive peptides [143,144,145,146]; conjugated linoleic acid (CLA) [147,148]; gamma-aminobutyric acid (GABA) [70,149,150]; vitamins [151,152,153,154]; and enzymes [19,155,156,157]) are sometimes credited as the end byproducts of the fermentation process [44,139,158]. In this section, the advantages and impacts of the above-mentioned compounds produced by L. lactis for better human health and quality of food will be summarized.
4.1. Gamma-Aminobutyric Acid
Food and medicine are among the applications for gamma-aminobutyric acid (GABA), a four-carbon non-protein amino acid that is widely distributed in nature among prokaryotes and eukaryotes [159]. LAB isolates are the most valuable source of naturally produced GABA [55]. It is known that LAB produce GABA during fermentation as a defense mechanism to maintain viability under acidic conditions (below pH 5 in anaerobic conditions) [160,161]. However, GABA production can vary widely among LAB strains [162]. Primarily located in the brain, GABA serves as a crucial inhibitory neurotransmitter [163]. It directly affects the personality and stress management, and has hypotensive, tranquilizing, antidiabetic, and diuretic effects [55,164]. GABA is produced by L. lactis via the enzymatic decarboxylation of glutamic acid [165].
High concentrations of GABA are found in fermented products, especially in fermented dairy products [166]. As L. lactis is the LAB that is most widely used as a primary fermentation starter in the dairy industry for the production of matured cheese, unripened cheeses (cream cheeses and cottage cheese), fermented milk products, sour cream, and fermented butter [165], the ability of L. lactis to produce GABA as value-added property is of great interest. In a study by Mileriene et al. [70], the L. lactis strain, isolated from raw bovine milk, produced 0.372 µmol g−1 of GABA in fermented milk. In another study by Mileriene et al. [70], the same L. lactis strain was used to produce acid whey cheese and GABA concentration reached an average of 7.61 mg 100 g−1 cheese after 14 days of storage. In a study by Inoue et al. [167], Lactobacillus casei and L. lactis YIT 2027 naturally produced GABA in fermented low-fat milk. When this milk was consumed daily for 12 weeks, containing 10–12 mg of GABA, it reduced systolic blood pressure by 17.4 mmHg, diastolic blood pressure by 7.2 mmHg, and mean blood pressure by 10.6 mmHg [161]. Also, in a study conducted by Redruello et al. [165], isolation of six GABA-producing L. lactis subsp. lactis strains from raw camel’s milk collected in Algeria was reported. These strains were used individually as starter cultures for cheese production and generated high GABA concentrations with the mean concentration of 384 mg kg−1.
4.2. Exopolysaccharides (EPSs)
EPSs are defined as extracellular high-molecular-weight polymers that can be of animal and plant origin or excreted by LAB (Lactobacillus, Lactococcus, Bifidobacterium, Leuconostoc, Pediococcus, Streptococcus, Enterococcus, and Weissella sp.) [132,168,169]. EPS are homopolysaccharides (HoPSs) or heteropolysaccharides (HePSs), based on the composition of the main chain, whether it is composed of one or numerous monomers, mechanisms of synthesis, extra- or intra-cellular, respectively [170,171]. Generally, EPS produced by L. lactis plays an important role in food processing, especially in the dairy industry, due to its positive effect on product quality, safety, and sensory parameters. There are several reports that L. lactis strains produce two types of phenotypic products of EPS, mucoid and ropy, which can be neutral or acidic [40,132,172,173,174].
The latter EPSs have a more complex structure because they contain a uronic acid group compared with the neutral type, which has two or more different monosaccharides with the absence of organic acid [133,172,174]. For instance, ropy EPS-producing starter L. lactis ssp. cremoris DPC6532 [134,175], as well as L. lactis subsp. lactis [176,177], remarkably improved the low-fat cheese yield, texture, and cooking properties without negative impact on sensory parameters [178,179,180]. A ropy capsular EPS (LL-1+) contributed to higher gel stiffness, higher water retention, lower particle size, and improved creaminess texture, if compared with a non-ropy EPS (LL-2)-producing strain in low-fat cheese matrices [173,181,182]. In addition to this, rheological parameters (firmness, thickness, and sliminess) of dairy products, such as fermented milk drinks, yogurt, low-fat mozzarella, and semi-fat or low-fat cheddar cheese, depend on the capability of ropy strains of L. lactis subsp. lactis and L. lactis subsp. cremoris to generate nonidentical HePSs, which bestow the texture of the final product [134,181,182,183,184]. However, L. lactis strains have a limited capacity to produce HePS [185,186,187]; nevertheless, the potential to produce these types of EPS depends on the sugar nucleotide biosynthesis path, the genetic potential of the strain, as well as technological parameters (substrate chemical composition, acidity, temperature, and duration of fermentation) [188,189,190,191]. It was reported that the L. lactis subsp. lactis IMAU11823 strain has a potent ability to generate two groups of polysaccharides: i—EPS-1 (simple-structure EPS composed of mannose and glucose (molar ratio of 1.00:7:01)); ii—EPS-2 (non-linear polysaccharide composed of eight sugar residues, of which main sugars were mannose, rhamnose, and glucose (molar ratio of 7.45:1.00:2.34) [191]. Moreover, L. lactis-produced EPSs have strong antioxidant [191,192,193,194], antimicrobial [132,194,195,196], and immunomodulatory activities [170,193,197,198] and no cytotoxicity [170,191,198]. Also, some studies showed that EPS has the ability to reduce cholesterol content in blood, and showed anticancer and anti-diabetic effects; however, these results were observed in vitro or using animal models, leaving out human clinical trials [19,199]. These EPS characteristics could be of high interest not only in dairy but in bakery, meat, and animal feed industries as well as pharmacy and biotechnology development.
4.3. Lactic Acid and Citrate
Fermented products play an important role in the food industry as these foods are profuse sources of prospective advantageous microbes, which possess varieties of bioactive compounds, leading to longer shelf life, better organoleptic characteristics, and improved nutritional value [33,200]. During the fermentation process, sugar is metabolized in different organic acids, such as lactic acid (LA), acetic acid, butyric acid, and propionic acid, depending on the metabolic pathway [33,201]. LA is the main organic acid formed during the metabolic process and contains two isomers, L-lactic acid and D-lactic acid. The latter is a harmful enantiomer and can cause numerous health disorders, for instance, dysbiosis, which can lead to inflammatory bowel disease (IBD), metabolic disorders, autoimmune conditions, and psychiatric and neurological illnesses [202,203,204,205,206]. So, it is important to select an appropriate LAB strain, so that during fermentation, only an L-lactic isomer would be generated [202,203]. It is well documented that Lactococcus is known as the homo-lactic acid fermentation type of LAB, which also produces L-lactic acid [138,207,208,209,210]. Lactose utilization by lactococcus can occur in two ways: via the Leloir or tagatose-6-phosphate pathways [211,212]. Usually, Lactococcus spp. are mainly linked to the tagatose-6-phosphate pathway where lactose-6P is hydrolyzed by a phospho-β-galactosidase into glucose and galactose-6P. Tagatose-6 phosphate end products are triose-3 phosphates, Glyceraldehyde-3Phosphate and Dihydroxyacetone Phosphate, which can subsequently enter the glycolysis pathway [212]. For instance, in L. cremoris LA10, lactose must be utilized through the tagatose-6-phosphate pathway due to its lack of the β-galactosidase-encoding lacZ gene. However, L. garviae ATCC 49156, L. lactis IL 1403, L. lactis SK11, and L. lactis MG1363 utilize lactose and galactose through the Leloir pathway [213]. Moreover, recent studies showed that the L. lactis LA1 genome contains a β-galactosidase-encoding gene, which was located close to the Leloir pathway genes [211]. In the Leloir pathway, lactose is internalized by the product of galactose permease (galP), and then hydrolyzed by a β-galactosidase into glucose and β-galactose. Afterwards, galactose is metabolized into glucose-1P, which is then converted into glucose-6P, an intermediate of glycolysis, by the action of the phosphoglucomutase [212]. According to Rodríguez et al. [211], the gene collections required L. lactis LA1.
Fermentation with L. lactis is important in dairy processing, because of final co-products, such as LA. For instance, isolates of L. lactis subsp. lactis are fast acidifiers and are desirable in cheese and butter production [2,47,214], while L. lactis subsp. cremoris is preferred for less bitterness of the product [2,215,216,217]. Lactococci are able to produce an enzyme called lactase, which catalyzes the hydrolysis of lactose, the sugar found in milk, into its constituent sugars, glucose and galactose. This enzymatic activity is crucial for the fermentation of lactose and the production of LA, which acidifies the milk and is a defining characteristic of dairy products like yogurt [155,218]. Also, there are suggestions that L. lactis strains could be used for LA production from carbons [40,219,220], and used in the food industry as an additive.
Despite the fact that L. lactis mainly generates LA during fermentation, it is known that L. lactis subsp. lactis biovar diacetylactis utilizes citrate and has the ability to prosper at pH values that would interdict the growth of most L. lactis strains [221,222,223]. Moreover, L. lactis subsp. lactis biovar diacetylactis during citrate fermentation is able to produce diacetyl and acetoin, and these compounds furnish desirable flavor characteristics in ripened cheeses [47,123,224].
4.4. Bacteriocins
It is known that Lactococcus spp. generate more than 40 peculiar bacteriocins, antimicrobial proteins synthesized by ribosomes. These bacteriocins mainly belong to two classes: (I) Class I includes post-translationally modified, small (<10 kDa), heat-stable peptides, such as nisin A; (II) Class II involves non-modified, small (<10 kDa), mainly heat-stable peptides, such as lactococcin G [140,141]. Nisin A is generated by L. lactis and contains 34 amino acids. This bacteriocin belongs to a group of cationic peptide antimicrobials collectively called Type A (I) lantibiotics and is known for antimicrobial activity against Gram-positive bacteria, especially foodborne bacteria (Bacillus cereus, Listeria monocytogenes, Clostridium botulinum, and/or Enterococci) [225,226,227,228]. EFSA and FDA approved that nisin is generally regarded as safe (GRAS) to use in the food industry, and it is also nontoxic, odorless, colorless, and tasteless [229,230]. Despite its antibacterial properties, this bacteriocin has a broad application in the food industry due to its resistance to heat treatment [231,232,233,234,235], preservation effect extending the shelf life of the product [228,235,236,237], and effectiveness at low concentrations [226,235,236,237,238], and it can be used for cleaner-label food products [229,239,240]. However, nisin incorporation in food, especially dairy product matrices, has a few limitations. First of all, nisin solubility is strongly dependent on media acidity, and the most significant effect can be achieved at pH 3, so this bacteriocin is not suitable for nonfermented dairy products [237,241,242]. Also, fat content of the raw material and/or product has an influence on antimicrobial activity of nisin. It is known that nisin is a hydrophobic peptide and its distribution and activity strongly depend on fat globule concentration and integrity of the food matrix. For instance, nisin is more effective at inhibiting L. monocytogenes in skim milk compared with whole milk [243,244,245]. Also, the homogenization process has a negative impact on nisin effectiveness, because during this process, fat globules’ surface area increases, leading to stronger nisin adsorption to fat globules, where it is immobilized and loses its activity [237,245,246]. Also, nisin interaction with proteins has an ambiguous effect in the food industry, especially in dairy products. It was reported that at neutral pH, nisin is a cationic peptide; however, caseins and other milk proteins are anionic, proposing that there is a possibility of ionic interactions between nisin and milk proteins, leading to the absorption of nisin to caseins, and lower antimicrobial activity of this bacteriocin [225,237,247,248]. The same tendencies in the other food matrices with higher protein content, especially meat and fish, were observed [244,249,250,251]. On the other hand, the above-mentioned interaction between nisin and casein can be eligible in cheesemaking, guaranteeing a high preservation of nisin in the cheese matrix and reducing the loss of cheese whey, leading to more sustainable production [237,252].
Another L. lactis subsp. lactis-produced bacteriocin is lactococcin G, which belongs to Class II, subclass II-B, which comprises two peptides. The bacteriocin consists of two peptides, LcnG-α and LcnG-β, consisting of 39 amino acid residues and 35 amino acid residues, respectively [24,253,254]. Lactococcin G possesses antimicrobial activity against various LAB as well as some Clostridia strains [254,255,256]. The use of lactococcin G as a food additive is not regulated, but L. lactis that secretes this substance during fermentation is desirable, as the antimicrobial properties of this bacteriocin can help ensure product safety and prolong shelf life.
4.5. Bioactive Peptides
Unique, low-molecular-weight peptide sequences that typically start with two to twenty amino acid residues are known as bioactive peptides, and these substances possess antimicrobial, antihypertensive, and antioxidant activities; blood-lipid-lowering effect; opioid role; antiobesity; ability to bind minerals; and antidiabetic effects. These peptide sequences are inactive within the parent protein but, upon hydrolysis, are liberated from the native polypeptide sequence and become physiologically active [257,258,259].
LAB fermentation of various food matrices, particularly dairy products, is one of the techniques for generating bioactive peptides from precursor protein molecules [259,260]. In general, milk protein and their fractions are valuable sources of bioactive peptides with miscellaneous biological effects, for instance, antithrombotic, antimicrobial, antioxidative, antihypertensive, and immunomodulatory activities [158,258].
Milk fermentation processes executed by LAB are preferable ways to generate peptides due to the GRAS status of these organisms. The proteolytic system of L. lactis has been comprehensively analyzed with respect to the genes and enzymes involved, as well as their regulation [261,262,263,264]. It was reported that during bovine milk fermentation with L. lactis, numerous casein-derived bioactive peptides can be generated, most of which have angiotensin-converting enzyme (ACE) inhibitory activity, leading to an antihypertensive effect [36,260,265]. In addition to this, it was reported that L. lactis NRRL B-50571 and NRRL B-50572 strains in fermented milk produced, not only ACE, but also type 2 diabetes (DPP-IV) and thrombin inhibition enzymes, and their activity was improved after digestion in vitro. So, peptides with multiple enzyme-inhibitory activities can have a significant effect in the management of hypertension, type 2 diabetes, and thrombosis [257,266,267]. Also, in order to increase the quantities and variety of bioactive peptides separated from fermented milk, the proteolytic system of L. lactis can be modulated [257,260,261]. Metal binding peptides or caseinphosphopeptides produced during milk fermentation with L. lactis subsp. cremoris NCFB 712 has the capacity to enlarge the absorption of calcium and other divalent minerals, such as iron. Quantities of released calcium-binding peptides are correlated with proteolytic activity of the strains. L. lactis spp. with such peptides could be used as a starter culture in order to develop higher-functional-value cheese [180,268,269]. Furthermore, different types of dairy matrices (milk, fermented milk, cheese, kefir, yogurt, etc.), milk of different animals (bovine, donkey, goat, camel, etc.), as well as milk and dairy processing have an influence on diversity and bioactivity of peptides [146,257,260,270,271]. For instance, in donkey milk, fermented with L. lactis, production of only ACE inhibitory peptides was observed, while in bovine and human milk treated with the same strain, antimicrobial peptides, against E. coli and S. aureus, were also found [143,146,272,273]. As mentioned before, bioactive peptides, possessing an inhibitory effect of ACE, can be found in goat cheese Festivo fermented with a mixture of Lactococcus sp., Leuconostoc sp., Propionibacterium sp., Lactobacillus sp., and Bifidobacterium sp. strains, and it was reported that bioactivity of these peptides increased during ripening of cheese and activity was forsaken until a certain level of proteolysis was exceeded. Also, antihypertensive peptides β-lactoglobulin f147–148, lactoferrin f288–289, and lactoferrin f319–320 were isolated from mature and vintage cheddar as well as Feta cheese, respectively [274,275,276]. Moreover, anticarcinogenic peptides were found in cheese slurry prepared with L. lactis subsp. lactis as a starter culture [274,277]. Recent studies showed that starter cultures containing L. lactis subsp. lactis, L. lactis subsp. cremoris, L. lactis subsp. lactis biovar diacetylactis, Streptococcus thermophilus, and Lactobacillus delbrueckii subsp. bulgaricus, used for whey of different milk (ewe, cow, and goat) fermentation, generated a specific novel β-casein-derived bioactive polypeptide with an unusual and potent dual antibacterial and anti-gelatinase bioactivity [258,278]. L. lactis, as a biotechnological microorganism, could be used not only for novel and higher-functional-value dairy food development as a starter culture, but also for the production and isolation of bioactive peptides for food supplements.
4.6. Vitamins
Vitamins are essential micronutrients for the smooth functioning of the organism; however, they cannot be generated by humans, so they must be gained with food. The loss of vitamins during food processing can result in inadequate intake, potentially causing public health issues attributed to unbalanced diets [6,279]. Some vitamins, especially water-soluble, which belongs to the B-group (riboflavin, folates, thiamine, and cobalamin), can be synthesized by lactobacilli, bifidobacteria, or lactococci [279,280,281].
It is known that in L. lactis MG1363, a cluster of folate genes, folA, folB, folKE, folP, and folC, are responsible for this vitamin biosynthesis [282]. Folate is generated from the precursors, such as guanosine triphosphate, para-amino benzoic acid, and glutamate, which act as building blocks for the production of various folate derivatives such as tetrahydrofolate, 5-formyl tetrahydrofolate, 5,10-methenyl tetrahydrofolate, 10-formyl tetrahydrofolate, and 5,10-methylene tetrahydrofolate in a number of enzymatic steps. Also, among various LAB, L. lactis is the only organism known to possess a complete biosynthesis route for folate [87,283,284]. L. lactis genes, menF, menA, preA, and mvk, are responsible for vitamin K2 (menaquinone) production. In L. lactis, menaquinones are generated from acetyl-CoA, phosphoenolpyruvate, and D-erythrose-4-phosphate. The precursors are transformed step by step to a hydrophobic polyprenyl diphosphate chain and a hydrophilic naphtoquinone ring, 1,4-dihydroxy-2-napthoate (shikimate and menaquinone pathways) [154]. Also, riboflavin (vitamin B) can be synthesized by the above-mentioned bacteria strains, because it contains riboflavin biosynthetic genes (ribG, ribH, ribB, and ribA) [285].
It is well known that vitamins play different roles in the proper functioning of the organism and that the best source of health benefits is natural food. Folate, or vitamin B9, has a significant part in many metabolic pathways, and shows antioxidant capacity; however, its shortage has been related to poor cognitive function, some types of cancer, neural tube defects, and coronary heart diseases [286,287]. Vitamin B2 is also an essential component of basal cellular metabolism and is the precursor of the enzymes flavin mononucleotide and flavin adenine dinucleotide that are electron acceptors and involved in many oxidation–reduction reactions [288]. A lack of riboflavin results in problems with metabolization, which can cause fatigue, skin conditions, hyperemia, and angular stomatitis. On the other hand, a severe vitamin B deficiency can cause anemia and cataract development [289]. In addition to vitamins B2 and B9, menaquinone also has a positive effect on kin health and bone metabolism, promotes proper brain function, and prevents heart-related diseases by regulating calcium homeostasis [6,290].
One of the strategies to improve folate daily intake could be inclusion of fermented milk in the diet. Milk fermentation with L. lactis subsp. lactis can be used for folate-fortified food development to mitigate folate insufficiency effectively [281,283,284,288]. Fermented milks could be considered one of the best product matrices for folate fortification due to folate-binding proteins of milk increasing folate stability, improving the bioavailability of both 5-methyltetrahydrofolate and folic acid [281,291,292].The same approach could be applied in order to increase daily intake of riboflavin. For instance, the use of starter cultures of L. lactis spp. for cow milk fermentation significantly increased the riboflavin concentration in milk, and final products such as cheese could be achieved [279,285,293,294,295]. In addition to this, goat milk is known to contain about the same amount of riboflavin and folate as cow’s milk, but most of these nutrients are lost during the cheesemaking process. Because of this, adding vitamin-producing L. lactis isolated from a similar kind of product to cheeses could boost folate and riboflavin content and provide the health benefits linked to consuming these vitamins with food [279,288,292,296]. In addition, apart from water-soluble vitamin production, L. lactis can synthesize fat-soluble vitamin K2, which principally is of bacterial origin. It is known that cheese prepared with L. lactis is the main source of vitamin K2 in the Western diet and the content of this vitamin can reach up to 110 μg 100 g−1 [154,297,298]. The concentration of menaquinone found in L. lactis is strain-dependent, and fermentation (medium, temperature, aerobic vs. anaerobic conditions) and product processing conditions play a significant role, and when these conditions are optimized, it is possible to increase vitamin K2 concentration in fermented milk up to 50% [152,154,299]. Also, menaquinone can be biosynthesized by L. lactis in industrially fermented cheese, buttermilk, sour cream, cottage cheese, and kefir [6,299,300].
In order to avoid the use of synthetic folate, as excessive usage could be linked to colorectal cancer, fermented milks could be a good source of natural folate. However, the concentration of this substance depends on the composition and processing conditions of raw milk, strain and species of starter bacteria, size of inoculum, incubation time, and storage conditions [87,284,301]. Moreover, Sybesma et al. [302] reported that direct mutagenesis followed by selection and metabolic engineering could modify L. lactis (strain NZ9000), resulting in simultaneous overproduction of both folate and riboflavin. For instance, this method can be used for controlling secondary metabolism, such as folate biosynthesis by overexpressing folKE and folC. Such a pathway leads to higher production of folate, which remains intracellular. These findings are explained by the fact that increased potency of folate synthetase leads to higher retention of this vitamin in the cell due to an increased enzymatic capacity to elongate the glutamyl tail of the extra folate produced by the overexpression of folKE [282]. Moreover, L. lactis subsp. cremoris NZ9000, which generates riboflavin, can be used in biotechnology processes for vitamin production [285,294,303,304,305,306,307,308]. Riboflavin (vitamin B2) is an essential component of basal cellular metabolism and is the precursor of the enzymes flavin mononucleotide and flavin adenine dinucleotide that are electron acceptors and involved in many oxidation–reduction reactions [288]. The genes mvk, preA, menF, and menA are important contributors to menaquinone levels as single overexpression of these genes double and more than triple the total menaquinone content in the culture. Combined overexpression of mvk, preA, and menA increased menaquinone levels to a higher level than obtained individually. When the overproducing strains were applied for milk fermentations, vitamin K2 content was effectively increased three-fold compared to the wild type [154].
4.7. Conjugated Linoleic Acid (CLA)
Conjugated linoleic acid (CLA) has many positive effects on health, for instance, decreasing cancer incidence and atherosclerosis severity. Nevertheless, dietary CLA intake is relatively low in order to gain the desired physiological effects. The recommended intake of CLA is approximately 3 g day−1, and through its consumption, many health effects are manifested in humans. The CLA bacterial production in foods is a technological challenge and the ability of probiotics to produce CLA is currently under research [309,310,311,312]. In the literature, there is ambiguous information about the ability of lactococci to produce CLA. For instance, L. lactis subsp. cremoris MRS 47 increased CLA and polyunsaturated fatty acid content while simultaneously decreasing saturated fatty acids in fermented whole milk. These findings can be explained by the L. lactis subsp. cremoris MRS 47 ability to produce CLA in whole milk during fermentation, through the lipolysis of milk fat [309,313,314,315]. It is known that during fermentation in dairy products, CLA precursor fatty acids, for instance, vaccenic acid, linoleic acid, and ricinoleic acid, as well as oils containing a high level of these fatty acids, can be turned into CLA [312]. In addition to this, Salam et al. [314] found that it is possible to increase CLA content to 21.6 mg 100 mL−1 in milk, fermented with probiotic L. lactis subsp. Lactis using sesame oil processed by lipases as the lipid source. Also, it is important to control the microbial growth phase, pH, temperature, and incubation time and select an appropriate bacterial strain, in order to obtain the amount and type of CLA wanted [312]. However, according to Renes et al. [315], strains producing CLA do not generate GABA, and vice versa.
4.8. Enzymes
Microbial enzymes have a broad spectrum of catalytic activities and high yields, and their rapid growth on inexpensive media and the stability of their enzyme products make bacteria the preferred enzyme source in the food industry, especially dairy [316,317]. It is known that enzymes such as esterase, lactase, lipases, proteases, and catalase are used in dairy and food technology [19]. The combination of L. lactis subsp. lactis biovar. Diacetylactis and L. lactis subsp. cremoris strains is very popular for fermented milks, while L. lactis subsp. lactis is used in cheesemaking, and the mix of Lactococcus strains is applied in cheddar, most Dutch cheese, Camembert, and farmhouse-made goat milk cheese preparation [19,318]. During cheese ripening, after the acidification period, the Lactococcus level decreases due to early self-degradation of the peptidoglycan by specific hydrolases. Such changes have an influence on further destruction of the cell and the release of the intracellular content outside the cell, for instance, enzymes, lipases, esterases, peptidases, and DNAases, which have an influence on cheese ripening time acceleration [318,319]. Lactococcus’ proteolytic enzyme can hydrolyze caseins and peptides, and there are enzymes able to degrade amino acids, for instance, transaminases transfer amino acids into α-cetoacids, which can be further converted into aldehydes, alcohols, hydroxyacids, all aroma compounds, or aroma precursors [106,153,318,320]. Moreover, it is known that L. lactis subsp. lactis possesses an arginine dehydrolase, which enables the release of ammonia from arginine, and L. lactis subsp. lactis biovar. diacetylactis is also able to produce diacetyl from citrate [47,321,322]. Due to enzymatic activity of Lactoccocus, all the above-mentioned compounds are very important in dairy product processing, because these substances have a significant influence on nutty, creamy, and buttery aroma that develops in cheeses, creams, and butters [15,19,156,322]. Lipolysis is hydrolysis of triglycerides by the enzyme lipase, resulting in formation of free fatty acids, which may be precursors of aromatic components such as methyl ketones, secondary alcohols, esters, and lactones, which have a significant influence on the cheese flavor development. However, Lactoccocus, as well as other LAB species, is considered as not possessing or showing weak lipase activity [15,157,318,323]. Also, L. lactis isolated from dairy products does not show any esterase activity outside the cell [15,93,318]; however, environmental L. lactis strains have opposite traits [157]. Lactate dehydrogenase is another Lactococcus-utilized enzyme, which is responsible for carbon source conversion to L-lactate from pyruvate. Due to metabolic engineering efforts in L. lactis, and the use of lactate dehydrogenase, α-acetolactate in place of lactate, where the former is a reduced carboxylated form of diacetyl, was generated. These findings suggested that modified L. lactis during fermentation could produce important flavor compounds for dairy products, such as diacetyl, acetaldehyde, and acetoin [44,106,156,324].
In conclusion, the above-mentioned bioactive compounds generated by Lactococcus strains have many advantageous characteristics not only for human health but also in the dairy industry (Figure 1). The application of bioactive compounds could lead to innovative (bio) technology expansion and/or new product formulation development in a sustainable manner. This contemporary approach could lead to higher food security as well as improve people’s nutrition by valorizing raw materials and creating functional products and/or foods, as well as food additives or pharmaceuticals.
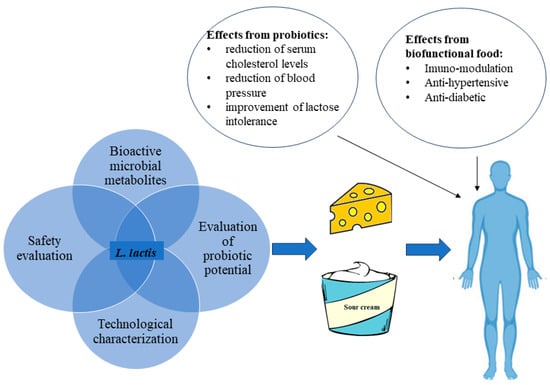
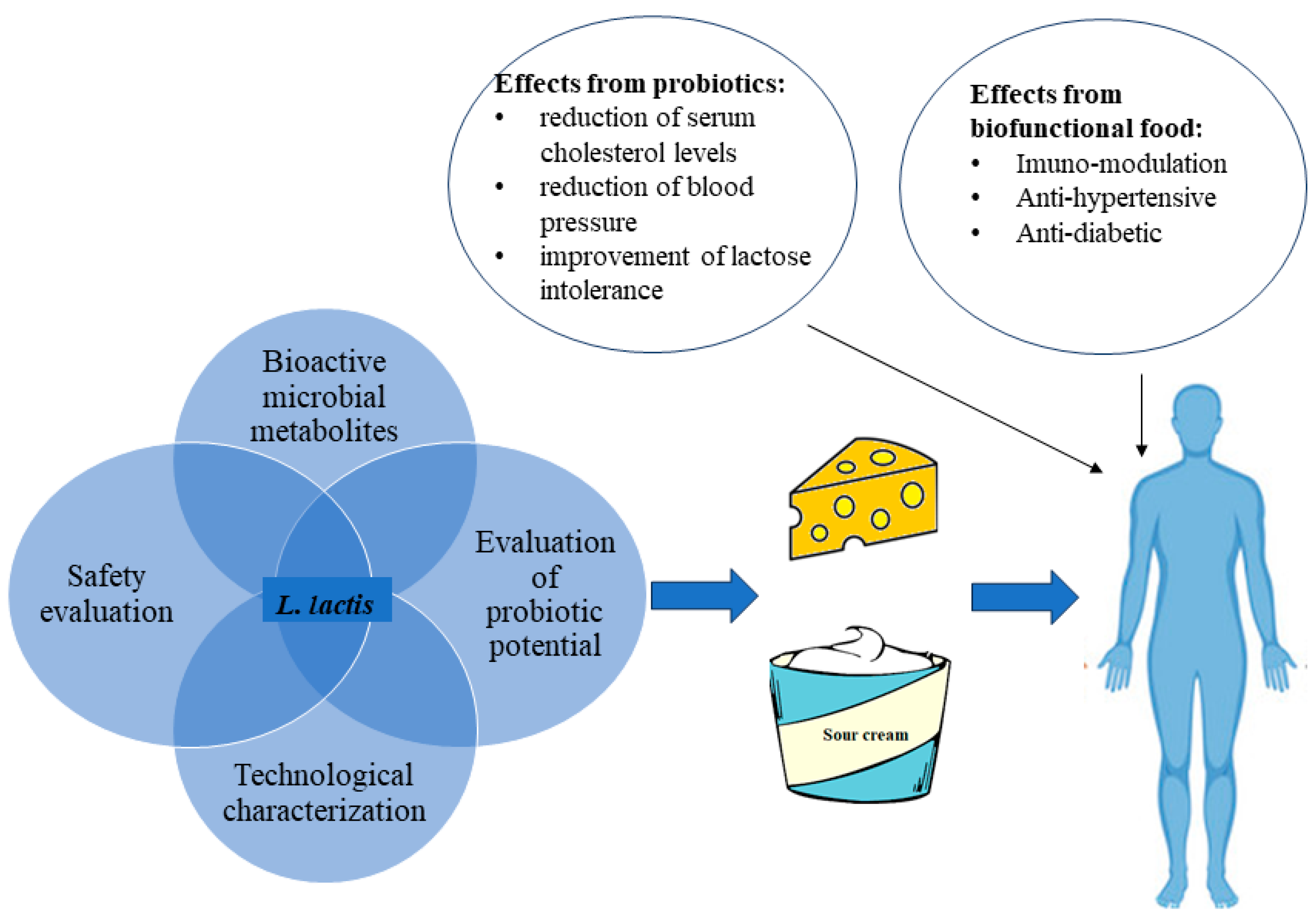
Figure 1.
Some beneficial effects resulting from the consumption of biofunctional fermented dairy foods or probiotic dairy foods containing L. lactis.
5. Bioinformatic Tool Application for L. lactis Evaluation
The field of genomics has experienced a transformative revolution with the introduction of high-throughput sequencing (HTS) technologies, which allow for the rapid analysis of vast quantities of genetic material. Over the past decade, whole genome sequencing (WGS) has become an essential tool in routine laboratory practices, enabling high-speed and high-resolution investigations, as well as surveillance of a wide range of microorganisms, including Lactococcus species [325,326]. DNA sequencing technologies, such as next-generation sequencing (NGS), and third-generation sequencing techniques like PacBio or Nanopore sequencing, are employed to identify and characterize Lactococcus species within the sample. By utilizing sequencing data and bioinformatic tools, it is possible to accurately determine the specific type or species of Lactococcus by comparing the genetic information to reference sequences [83,326].
5.1. Prediction of Potential Antimicrobial Resistance and the Detection of Antimicrobial Resistance Genes (ARGs)
A significant concern revolves around the possibility that probiotic bacteria could inherently possess antibiotic resistance genes (ARGs) or harbor transferable antibiotic resistance genes that should not be disseminated to other organisms. Consequently, when contemplating their commercial use, and in light of the presence and potential transfer of these genes to humans or animals, meticulous monitoring becomes imperative. This monitoring involves analyzing antibiotic resistance genes using whole genome sequences as a data analysis tool. Advancements in bioinformatics have significantly transformed the ability to explore antibiotic resistance by studying and characterizing ARG and providing insights into the genetic basis of resistance within L. lactis [327,328,329,330,331]. By comparing the sequenced genomes to comprehensive databases of AMR genes, we can precisely identify the presence of known resistance genes. This helps in characterizing the potential resistance profile of L. lactis strains. To predict antibiotic resistance/virulence related to AMR/virulence genes, the ABRicate pipeline can be used [332], which conducts mass screening of contigs against multiple databases, including the Comprehensive Antibiotic Resistance Database (CARD) [333], ResFinder [334], Antibiotic Resistance Gene-ANNOTation (ARG-ANNOT) [335], and MEGARes [336]. ABRicate generates a tab-separated output file containing essential outputs such as gene names, proportion of the gene in the query sequence, information about alignment gaps (potentially indicating pseudogenes), gene coverage percentage, exact nucleotide match percentage, sequence database details, and other important information (Table 4) [332].

Table 4.
Example of ABRicate pipeline output with description.
CARD stands as a widely recognized and inclusive repository encompassing an extensive array of meticulously curated antibiotic resistance genes and their corresponding mutations. Its scope extends across a diverse spectrum of bacterial species, as well as L. lactis. The database boasts a repository of high-quality, thoroughly annotated data, solidifying its status as a dependable resource for the identification and delineation of AMR genes. Moreover, the integration of the Resistance Gene Identifier (RGI) tool [337] within CARD amplifies its utility by furnishing users with a user-friendly and potent platform for AMR detection. RGI identifies open reading frames (ORFs) through the use of Prodigal [338], detects homologs using DIAMOND [339], and assesses matches against CARD’s database, applying model cutoff values for prediction [340,341].
5.2. Metabolic Pathway Prediction
Clarifying the probiotic effects of Lactococcus or any other probiotic microorganism involves a multidisciplinary approach that combines microbiological experiments, in vitro assays, and bioinformatic analyses. Databases like KEGG [342], MetaCyc [343], and EggNOG [344] help annotate the functional potential of microbial communities, including the probiotic strain, by identifying metabolic pathways, functional genes, and enzymes that might play a role in probiotic effects [68].
L. lactis primarily metabolizes carbohydrates through glycolysis, and using the KEGG database can provide detailed information on the enzymes and reactions involved in glycolysis and its reverse process, gluconeogenesis, and also about the biosynthesis and metabolism of amino acids, which are crucial for protein synthesis and various cellular processes [106,345]. Using different bioinformatic pipelines can provide information related to utilizing cofactors and vitamins of L. lactis, which are essential for various enzymatic reactions. Tools like BLAST [346] and InterProScan [347] can help in the prediction of the functions of genes and proteins related to cofactor and vitamin utilization based on sequence homology and functional annotation. Using comparative genomics can predict cofactor and vitamin utilization pathways in L. lactis with related species to identify unique features or differences.
One more of the quick and valuable computational mining tools associated with probiotic functions is iProbiotics [348], which utilizes k-mer composition and the support vector machine (SVM) algorithm to identify potential strain-specific markers. The tool incorporates three SVM models trained on different datasets. Upon uploading bacterial genome sequences in FASTA format, iProbiotics generates probabilities for various classifications, including the likelihood that the microbe is a probiotic, a probiotic Lactococcus, a probiotic Bifidobacterium, or another probiotic strain. Additionally, iProbiotics can explore gene expression patterns within the probiotic’s datasets.
KBase and Pathway Tools are specialized software platforms for predicting metabolic pathways and reactions in L. lactis, which can be used for metabolic modeling and pathway analyses. Kbase [349], short for the DOE Systems Biology Knowledgebase, is a comprehensive platform that includes a variety of tools for predicting metabolic pathways in L. lactis based on genome annotations. These tools use complex algorithms to infer potential metabolic reactions and pathways. KBase allows for the creation and analysis of metabolic models for L. lactis, which can be used to simulate and predict cellular metabolism under different conditions, making it possible to study how the organism utilizes various nutrients and produces metabolites. Pathway Tools is a versatile software package designed for the prediction, visualization, and analysis of metabolic pathways and cellular functions in a wide range of microorganisms. This tool provides capabilities for the exploration of metabolic networks, genome-scale modeling, and pathway analyses. Pathway Tools uses a combination of genome annotation data, enzyme databases, and reaction databases to infer potential metabolic reactions and pathways. In the context of metabolic modeling, an Integrated Flux Balance Analysis (FBA) is a crucial technique. FBA is a constraint-based modeling approach seamlessly integrated into the Pathway Tools platform. This integrated FBA capability allows the investigation and forecasting of metabolic fluxes within L. lactis across a range of growth conditions. It facilitates simulating the bacterium’s growth and metabolic behavior [350,351,352].
5.3. Bacteriocin and Other Bioactive Compound Prediction and Identification
Bacteriocins, antimicrobial peptides produced by bacteria, including L. lactis, are essential for inhibiting the growth of other microorganisms. Identifying bacteriocin genes in L. lactis is a multifaceted process that requires a combination of computational and experimental approaches.
Identifying bacteriocin genes in L. lactis using bioinformatics tools involves a systematic approach to predict, annotate, and validate these antimicrobial peptide-encoding genes. Several bioinformatics tools are designed specifically for bacteriocin prediction. Tools such as BAGEL (Bacteriocin Genome Mining Tool) [353] and BaPreS (Bacteriocin PredictionTool) [354] are commonly employed for the identification of bacteriocin gene clusters.
BAGEL is a publicly available online tool that scans the genomic sequences for genes encoding bacteriocins, as well as associated genes involved in bacteriocin production and transportation. BAGEL classifies predicted bacteriocins into different classes or groups based on structural and functional features and helps with understanding the diversity of bacteriocins within a given genome. BAGEL can integrate external databases and known bacteriocin information to enhance prediction accuracy and functional annotation of the identified bacteriocins [353].
BaPreS is a software package that utilizes a high-performing machine learning model and an easy-to-use graphical interface to generate essential features for precise protein sequence prediction. BaPreS is flexible for evaluating multiple sequences and integrating new data, whether related to bacteriocins, to enhance prediction accuracy. The software is designed to automatically extract optimal features and can predict bacteriocin sequences, and its performance is compared to the sequence-matching-based tool BLASTP [354].
BACTIBASE is also an available online tool that curates experimentally validated and annotated bacteriocins and provides comprehensive information, including sequences, classifications, and functional data. BACTIBASE may also provide structural data for some bacteriocins. This information can be useful for predicting the 3D structures of bacteriocins and helps with understanding their potential modes of action [355].
The widely used tool for protein sequence similarity searches, BLASTP, can also be utilized to identify putative bacteriocin sequences through homology to known bacteriocins [356]. BLASTP [356,357] is not a specialized bacteriocin prediction tool, but it is a versatile and commonly used approach to identify potential bacteriocins. While BLASTP is a valuable tool for identifying potential bacteriocin sequences based on sequence similarity, it has limitations, particularly when it comes to identifying sequences that are highly dissimilar to known bacteriocins. These limitations are not unique to BLASTP but apply to similarity-based approaches in general. Bacteriocins can undergo various post-translational modifications, which can change their structure and function, and these modifications may not be captured by sequence similarity searches. Purposely addressing these limitations can employ combinations of some approaches, for example, HMMER [358], PSI-BLAST [359], a structural analysis, and functional annotation. HMMER is a tool that can be used to search for hidden Markov models (HMMs) of known bacteriocins in Lactococcus genomes. After creating custom HMMs from known bacteriocin sequences, HMMER is used to search for homologous genes. For the prediction of bacteriocins using HMMER, a profile HMM representing bacteriocin families or domains needs to be created or obtained. Profile HMMs can be built based on known bacteriocin sequences or specific protein domains associated with bacteriocins or can use existing profiles from databases, e.g., Pfam or many of the databases that participate in Interpro. The effectiveness of HMMER-based bacteriocin prediction depends on the quality of the profile HMM and the specificity of the database you are searching against. Using profile HMMs specific to bacteriocin domains or families is crucial for accurate prediction [360,361].
The functional annotation is a crucial step in bacteriocin prediction and characterization, as it involves assessing the functional properties of potential bacteriocins. Many aspects of functional annotation for bacteriocins can be determined or at least predicted using bioinformatic tools. The gene cluster analysis can be used to identify and analyze gene clusters associated with bacteriocin production. The gene cluster analysis is an important part of predicting bacteriocins because many bacteriocins are produced as part of gene clusters that include not only the structural genes encoding the bacteriocin itself but also genes responsible for transport and regulation. Analyzing gene clusters provides data regarding the genetic context and functional properties of bacteriocins. Some bacteriocins share structural features even if their primary sequences are dissimilar [362,363,364,365]. Structural analysis tools, e.g., SWISS-MODEL [366] and I-TASSER [367], can help identify these common structural elements. The combination of the selection of suitable bioinformatic tools can help to comprehensively analyze the structural features of bacteriocins of L. lactis.
AntiSMASH (Antibiotics & Secondary Metabolite Analysis Shell) is another comprehensive bioinformatics tool specifically designed to identify and analyze secondary metabolite gene clusters. While AntiSMASH was initially developed to identify a wide range of secondary metabolites, it can also be used to predict the presence of bioactive compounds, including those involved in the biosynthesis of bacteriocins produced by Lactococcus strains. AntiSMASH identifies gene clusters and predicts specific modules, and algorithms of bacteriocins based on gene cluster characteristics, structural motifs, and known bacteriocin features [368,369].
5.4. Genetic Element Annotation and Identification
For the additional annotation of specific bacterial genetic features, PlasmidFinder [370], PlasFlow [371], or plasmidSPAdes [372] can be employed to identify plasmids within the genome. A genomic analysis goes beyond plasmids and encompasses the identification of other important genetic elements. Tools like IslandViewer [373], PHASTER, and Phigaro are used for annotating prophages [374], while CRISPR/Cas Finder [375] is a valuable tool in annotating CRISPRs.
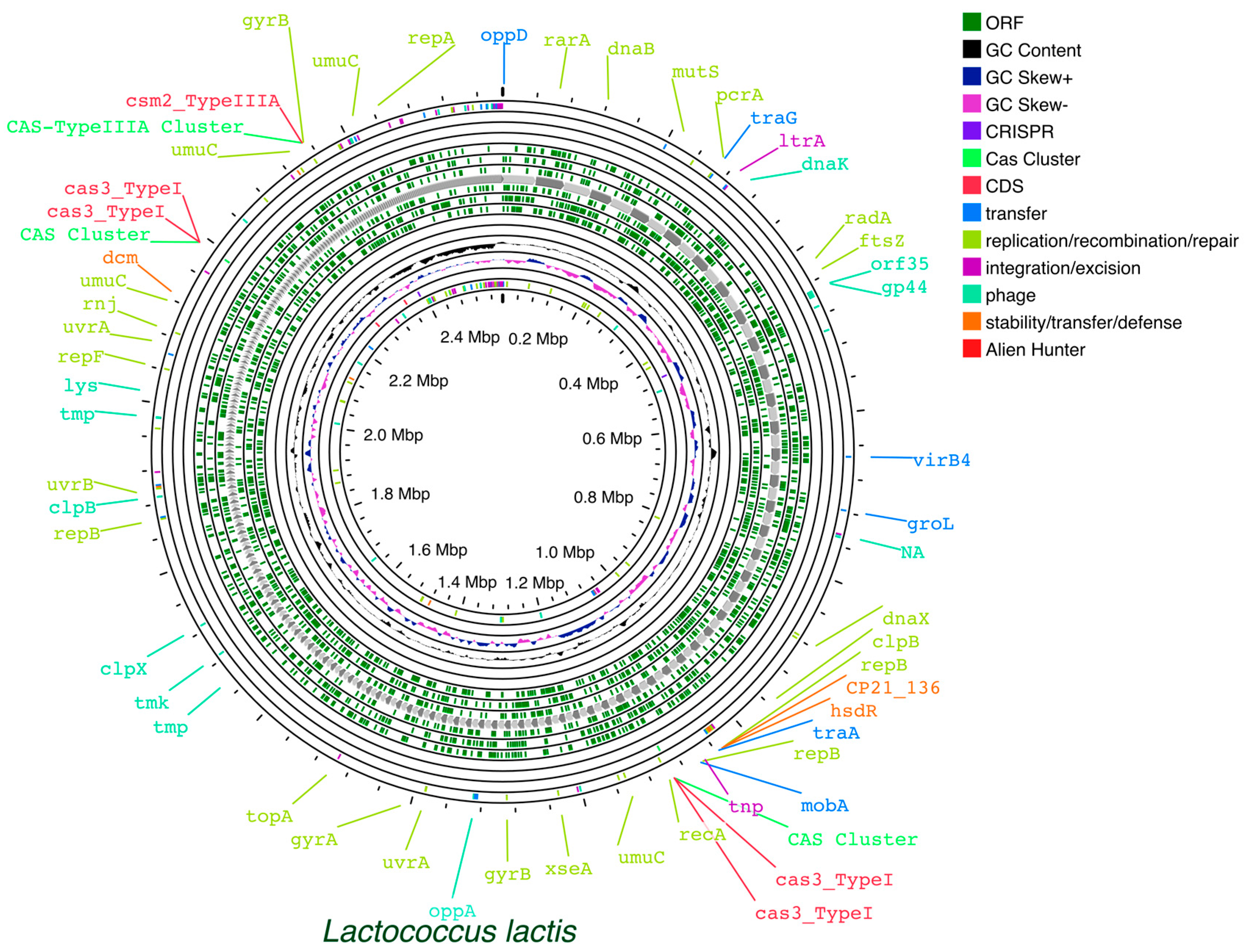
To facilitate the comprehensive annotation of genetic elements in L. lactis, a valuable tool is Proksee. This tool can assist in predicting and identifying specific genetic elements [376]. This user-friendly web server facilitates the assembly and annotation of bacterial genomes and enables the visualization of predictions from various third-party tools on a single graphical map. The integration of a variety of tools, e.g., FastANI [377], mobileOG-db [378], CRISPR/Cas Finder, pLannotate [379], and CGView Builder [380], and their associated visualization capabilities facilitates the identification of genome elements of interest. Maps generated using Proksee (Figure 2) allow rapid identification and evaluation of specific regions, resulting in the creation of visually appealing and informative maps that can be downloaded in SVG format.
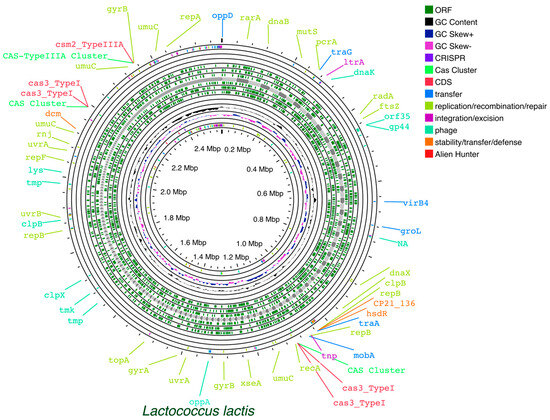
Figure 2.
Example of Proksee generated map of L. lactis, facilitating rapid identification and evaluation of specific regions.
The FastANI tool integrated into the Proksee platform is designed for fast and efficient whole genome Average Nucleotide Identity (ANI) calculations. FastANI provides efficient genomic comparisons, taxonomy verification, and quality control and gives data about the relatedness and diversity of Lactococcus species. The use of ANI calculations is helpful in characterizing L. lactis strains and confirming their taxonomic identity.
6. Conclusions
In conclusion, L. lactis, a versatile bacterium in dairy fermentation, reveals its potential and offers both technological and health-promoting advantages to the dairy industry and consumers.
The application of L. lactis in dairy fermentation provides numerous benefits in terms of flavor development, acidification, and the production of bioactive compounds like GABA, bacteriocins, and others. As consumers increasingly seek healthy and functional dairy food options, L. lactis has gained attention in the dairy industry due to its probiotic potential apart from the already known and applied technological properties, which can enhance the nutritional value of dairy products and promote health and makes L. lactis a valuable choice for the dairy industry.
However, safety assessment of these bacteria remains critical in strain selection for dairy product development. The antibiotic resistance of L. lactis is a significant concern, emphasizing the need for prudent evaluation of antibiotic susceptibility in various strains. Strategies such as use of specific starter cultures and proper ripening should be considered to mitigate and monitor formation of biogenic amines in dairy products linked to Lactococcus spp. to ensure the quality and safety due to their potential health risks. The application of bioinformatic tools has enabled the prediction of metabolic pathways of L. lactis and provides comprehensive insights of L. lactis functions based on its genetic features.
The future research and industry development of L. lactis in dairy fermentation are likely to be directed at disclosure of new opportunities of this versatile bacterium, which enables the development of a broad range of innovative and health-promoting dairy products, reflecting challenges caused by changing consumer preferences and regulatory developments.
Author Contributions
Conceptualization, K.K., P.Z., A.N., J.A., and M.M.; software, J.A.; collected the papers and performed the systematic selection of the papers, K.K., P.Z., A.N., and J.A.; visualization, K.K. and J.A.; writing—original draft preparation, K.K., P.Z., A.N., J.A., and M.M.; writing—review and editing, K.K. and M.M.; supervision, M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Petrescu, D.C.; Vermeir, I.; Petrescu-Mag, R.M. Consumer Understanding of Food Quality, Healthiness, and Environmental Impact: A Cross-National Perspective. Int. J. Environ. Res. Public Health 2020, 17, 169. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ren, M.; Duo, L.; Li, J.; Wang, S.; Sun, Y.; Li, M.; Ren, W.; Hou, Q.; Yu, J.; et al. Fermentation Characteristics of Lactococcus lactis subsp. lactis Isolated From Naturally Fermented Dairy Products and Screening of Potential Starter Isolates. Front. Microbiol. 2020, 11, 1794. [Google Scholar] [CrossRef] [PubMed]
- Yerlikaya, O. Probiotic Potential and Biochemical and Technological Properties of Lactococcus lactis ssp. lactis Strains Isolated from Raw Milk and Kefir Grains. J. Dairy Sci. 2019, 102, 124–134. [Google Scholar] [CrossRef]
- Yadav, M.; Shukla, P. Efficient Engineered Probiotics Using Synthetic Biology Approaches: A Review. Biotechnol. Appl. Biochem. 2020, 67, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.T.; Lu, P.; Parrella, J.A.; Leggette, H.R. Consumer Acceptance toward Functional Foods: A Scoping Review. Int. J. Environ. Res. Public Health 2022, 19, 1217. [Google Scholar] [CrossRef] [PubMed]
- Linares, D.M.; Gómez, C.; Renes, E.; Fresno, J.M.; Tornadijo, M.E.; Ross, R.P.; Stanton, C. Lactic Acid Bacteria and Bifidobacteria with Potential to Design Natural Biofunctional Health-Promoting Dairy Foods. Front. Microbiol. 2017, 8, 846. [Google Scholar] [CrossRef]
- Farnworth, E.R.; Champagne, C.P. Chapter 20—Production of Probiotic Cultures and Their Incorporation into Foods. In Probiotics, Prebiotics, and Synbiotics; Watson, R.R., Preedy, V.R., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 303–318. ISBN 978-0-12-802189-7. [Google Scholar]
- Olicón-Hernández, D.R.; Guerra-Sánchez, G.; Porta, C.J.; Santoyo-Tepole, F.; Hernández-Cortez, C.; Tapia-García, E.Y.; Chávez-Camarillo, G.M. Fundaments and Concepts on Screening of Microorganisms for Biotechnological Applications. Mini Review. Curr Microbiol 2022, 79, 373. [Google Scholar] [CrossRef]
- Carnevali, P.; Ciati, R.; Leporati, A.; Paese, M. Liquid Sourdough Fermentation: Industrial Application Perspectives. Food Microbiol. 2007, 24, 150–154. [Google Scholar] [CrossRef]
- Manini, F.; Casiraghi, M.C.; Poutanen, K.; Brasca, M.; Erba, D.; Plumed-Ferrer, C. Characterization of Lactic Acid Bacteria Isolated from Wheat Bran Sourdough. LWT-Food Sci. Technol. 2016, 66, 275–283. [Google Scholar] [CrossRef]
- de Souza, E.L.; de Oliveira, K.Á.; de Oliveira, M.E. Influence of Lactic Acid Bacteria Metabolites on Physical and Chemical Food Properties. Curr. Opin. Food Sci. 2023, 49, 100981. [Google Scholar] [CrossRef]
- Ribeiro, S.C.; O’Connor, P.M.; Ross, R.P.; Stanton, C.; Silva, C.C.G. An Anti-Listerial Lactococcus lactis Strain Isolated from Azorean Pico Cheese Produces Lacticin 481. Int. Dairy J. 2016, 63, 18–28. [Google Scholar] [CrossRef]
- Arena, M.P.; Capozzi, V.; Russo, P.; Drider, D.; Spano, G.; Fiocco, D. Immunobiosis and Probiosis: Antimicrobial Activity of Lactic Acid Bacteria with a Focus on Their Antiviral and Antifungal Properties. Appl. Microbiol. Biotechnol. 2018, 102, 9949–9958. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Nero, L.A.; Todorov, S.D. Safety Profiles of Beneficial Lactic Acid Bacteria Isolated from Dairy Systems. Braz. J. Microbiol. 2020, 51, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Bojanic Rasovic, M.; Mayrhofer, S.; Martinovic, A.; Dürr, K.; Domig, K.J. Lactococci of Local Origin as Potential Starter Cultures for Traditional Montenegrin Cheese Production. Food Technol. Biotechnol. 2017, 55, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Xie, X.; Du, T.; Jiang, X.; Miao, W.; Wang, T. Lactococcus lactis, a Bacterium with Probiotic Functions and Pathogenicity. World J. Microbiol. Biotechnol. 2023, 39, 325. [Google Scholar] [CrossRef] [PubMed]
- Cano-Lozano, J.A.; Villamil Diaz, L.M.; Melo Bolivar, J.F.; Hume, M.E.; Ruiz Pardo, R.Y. Probiotics in Tilapia (Oreochromis Niloticus) Culture: Potential Probiotic Lactococcus lactis Culture Conditions. J. Biosci. Bioeng. 2022, 133, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Bottacini, F.; van Sinderen, D. Towards the Diversification of Lactococcal Starter and Non-Starter Species in Mesophilic Dairy Culture Systems. Microb. Biotechnol. 2023, 16, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Castellone, V.; Bancalari, E.; Rubert, J.; Gatti, M.; Neviani, E.; Bottari, B. Eating Fermented: Health Benefits of LAB-Fermented Foods. Foods 2021, 10, 2639. [Google Scholar] [CrossRef]
- Mokoena, M.P.; Omatola, C.A.; Olaniran, A.O. Applications of Lactic Acid Bacteria and Their Bacteriocins against Food Spoilage Microorganisms and Foodborne Pathogens. Molecules 2021, 26, 7055. [Google Scholar] [CrossRef]
- Burgain, J.; Scher, J.; Francius, G.; Borges, F.; Corgneau, M.; Revol-Junelles, A.M.; Cailliez-Grimal, C.; Gaiani, C. Lactic Acid Bacteria in Dairy Food: Surface Characterization and Interactions with Food Matrix Components. Adv. Colloid Interface Sci. 2014, 213, 21–35. [Google Scholar] [CrossRef]
- Grosu-Tudor, S.-S.; Stancu, M.-M.; Pelinescu, D.; Zamfir, M. Characterization of Some Bacteriocins Produced by Lactic Acid Bacteria Isolated from Fermented Foods. World J. Microbiol. Biotechnol. 2014, 30, 2459–2469. [Google Scholar] [CrossRef] [PubMed]
- Casaburi, A.; Di Martino, V.; Ferranti, P.; Picariello, L.; Villani, F. Technological Properties and Bacteriocins Production by Lactobacillus curvatus 54M16 and Its Use as Starter Culture for Fermented Sausage Manufacture. Food Control 2016, 59, 31–45. [Google Scholar] [CrossRef]
- Lahiri, D.; Nag, M.; Dutta, B.; Sarkar, T.; Pati, S.; Basu, D.; Abdul Kari, Z.; Wei, L.S.; Smaoui, S.; Wen Goh, K.; et al. Bacteriocin: A Natural Approach for Food Safety and Food Security. Front. Bioeng. Biotechnol. 2022, 10, 1005918. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, E.; LeBlanc, J.G.; Pérez, G. Technological Properties of Lactic Acid Bacteria Isolated from Raw Cereal Material. LWT 2016, 70, 185–191. [Google Scholar] [CrossRef]
- Lahiri, D.; Nag, M.; Sarkar, T.; Ray, R.R.; Shariati, M.A.; Rebezov, M.; Bangar, S.P.; Lorenzo, J.M.; Domínguez, R. Lactic Acid Bacteria (LAB): Autochthonous and Probiotic Microbes for Meat Preservation and Fortification. Foods 2022, 11, 2792. [Google Scholar] [CrossRef] [PubMed]
- Sadiq, S.; Imran, M.; Hassan, M.N.; Iqbal, M.; Zafar, Y.; Hafeez, F.Y. Potential of Bacteriocinogenic Lactococcus lactis subsp. lactis Inhabiting Low pH Vegetables to Produce Nisin Variants. LWT-Food Sci. Technol. 2014, 59, 204–210. [Google Scholar] [CrossRef]
- Ünlü, G.; Nielsen, B.; Ionita, C. Production of Antilisterial Bacteriocins from Lactic Acid Bacteria in Dairy-Based Media: A Comparative Study. Probiotics Antimicro. Prot. 2015, 7, 259–274. [Google Scholar] [CrossRef]
- Tavares, L.M.; de Jesus, L.C.L.; da Silva, T.F.; Barroso, F.A.L.; Batista, V.L.; Coelho-Rocha, N.D.; Azevedo, V.; Drumond, M.M.; Mancha-Agresti, P. Novel Strategies for Efficient Production and Delivery of Live Biotherapeutics and Biotechnological Uses of Lactococcus lactis: The Lactic Acid Bacterium Model. Front. Bioeng. Biotechnol. 2020, 8, 517166. [Google Scholar] [CrossRef]
- Martín, R.; Langella, P. Emerging Health Concepts in the Probiotics Field: Streamlining the Definitions. Front. Microbiol. 2019, 10, 1047. [Google Scholar] [CrossRef]
- Cavanagh, D.; Fitzgerald, G.F.; McAuliffe, O. From Field to Fermentation: The Origins of Lactococcus lactis and Its Domestication to the Dairy Environment. Food Microbiol. 2015, 47, 45–61. [Google Scholar] [CrossRef]
- Nicolescu, C.M.; Bumbac, M.; Buruleanu, C.L.; Popescu, E.C.; Stanescu, S.G.; Georgescu, A.A.; Toma, S.M. Biopolymers Produced by Lactic Acid Bacteria: Characterization and Food Application. Polymers 2023, 15, 1539. [Google Scholar] [CrossRef] [PubMed]
- Abdul Hakim, B.N.; Xuan, N.J.; Oslan, S.N.H. A Comprehensive Review of Bioactive Compounds from Lactic Acid Bacteria: Potential Functions as Functional Food in Dietetics and the Food Industry. Foods 2023, 12, 2850. [Google Scholar] [CrossRef] [PubMed]
- Passerini, D.; Beltramo, C.; Coddeville, M.; Quentin, Y.; Ritzenthaler, P.; Daveran-Mingot, M.-L.; Bourgeois, P.L. Genes but Not Genomes Reveal Bacterial Domestication of Lactococcus lactis. PLoS ONE 2010, 5, e15306. [Google Scholar] [CrossRef] [PubMed]
- Tidona, F.; Meucci, A.; Povolo, M.; Pelizzola, V.; Zago, M.; Contarini, G.; Carminati, D.; Giraffa, G. Applicability of Lactococcus hircilactis and Lactococcus laudensis as Dairy Cultures. Int. J. Food Microbiol. 2018, 271, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Li, T.T.; Tian, W.L.; Gu, C.T. Elevation of Lactococcus lactis subsp. cremoris to the Species Level as Lactococcus cremoris sp. nov. and Transfer of Lactococcus lactis subsp. tructae to Lactococcus cremoris as Lactococcus cremoris subsp. tructae comb. nov. Int. J. Syst. Evol. Microbiol. 2019, 71, 004727. [Google Scholar] [CrossRef] [PubMed]
- Samelis, J.; Giannou, E.; Pappa, E.C.; Bogović-Matijašić, B.; Lianou, A.; Parapouli, M.; Drainas, C. Behavior of Artificial Listerial Contamination in Model Greek Graviera Cheeses Manufactured with the Indigenous Nisin A-Producing Strain Lactococcus lactis subsp. cremoris M104 as Costarter Culture. J. Food Saf. 2017, 37, e12326. [Google Scholar] [CrossRef]
- Oren, A.; Garrity, G.M. Notification That New Names of Prokaryotes, New Combinations and New Taxonomic Opinions Have Appeared in Volume 71, Part 3 of the IJSEM. Int. J. Syst. Evol. Microbiol. 2021, 71, 004812. [Google Scholar] [CrossRef]
- Bozoudi, D.; Kotzamanidis, C.; Hatzikamari, M.; Tzanetakis, N.; Menexes, G.; Litopoulou-Tzanetaki, E. A Comparison for Acid Production, Proteolysis, Autolysis and Inhibitory Properties of Lactic Acid Bacteria from Fresh and Mature Feta PDO Greek Cheese, Made at Three Different Mountainous Areas. Int. J. Food Microbiol. 2015, 200, 87–96. [Google Scholar] [CrossRef]
- Kleerebezemab, M.; Hols, P.; Hugenholtz, J. Lactic Acid Bacteria as a Cell Factory: Rerouting of Carbon Metabolism in Lactococcus lactis by Metabolic Engineering. Enzym. Microb. Technol. 2000, 26, 840–848. [Google Scholar] [CrossRef]
- Ruggirello, M.; Cocolin, L.; Dolci, P. Fate of Lactococcus lactis Starter Cultures during Late Ripening in Cheese Models. Food Microbiol. 2016, 59, 112–118. [Google Scholar] [CrossRef]
- Rattanachaikunsopon, P.; Phumkhachorn, P. Lactic Acid Bacteria: Their Antimicrobial Compounds and Their Uses in Food Production. Ann. Biol. Res. 2010, 1, 218–228. [Google Scholar]
- Alegría, A.; Delgado, S.; Roces, C.; López, B.; Mayo, B. Bacteriocins Produced by Wild Lactococcus lactis Strains Isolated from Traditional, Starter-Free Cheeses Made of Raw Milk. Int. J. Food Microbiol. 2010, 143, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Song, A.A.-L.; In, L.L.A.; Lim, S.H.E.; Rahim, R.A. A Review on Lactococcus lactis: From Food to Factory. Microb. Cell Fact. 2017, 16, 55. [Google Scholar] [CrossRef] [PubMed]
- Mileriene, J.; Serniene, L.; Kondrotiene, K.; Lauciene, L.; Kasetiene, N.; Sekmokiene, D.; Andruleviciute, V.; Malakauskas, M. Quality and Nutritional Characteristics of Traditional Curd Cheese Enriched with Thermo-Coagulated Acid Whey Protein and Indigenous Lactococcus lactis Strain. Int. J. Food Sci. Technol. 2021, 56, 2853–2863. [Google Scholar] [CrossRef]
- Mileriene, J.; Serniene, L.; Kondrotiene, K.; Santarmaki, V.; Kourkoutas, Y.; Vasiliauskaite, A.; Lauciene, L.; Malakauskas, M. Indigenous Lactococcus lactis with Probiotic Properties: Evaluation of Wet, Thermally- and Freeze-Dried Raisins as Supports for Cell Immobilization, Viability and Aromatic Profile in Fresh Curd Cheese. Foods 2022, 11, 1311. [Google Scholar] [CrossRef] [PubMed]
- Fusieger, A.; Martins, M.C.F.; de Freitas, R.; Nero, L.A.; de Carvalho, A.F. Technological Properties of Lactococcus lactis subsp. lactis bv. diacetylactis Obtained from Dairy and Non-Dairy Niches. Braz. J. Microbiol. 2020, 51, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Perin, L.M.; Miranda, R.O.; Todorov, S.D.; de Melo Franco, B.D.G.; Nero, L.A. Virulence, Antibiotic Resistance and Biogenic Amines of Bacteriocinogenic Lactococci and Enterococci Isolated from Goat Milk. Int. J. Food Microbiol. 2014, 185, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Dan, T.; Chen, Y.; Chen, X.; Sun, C.; Wang, X.; Wang, J.; Zhang, H. Isolation and Characterisation of a Lactobacillus delbrueckii subsp. bulgaricus Mutant with Low H+-ATP Ase Activity. Int. J. Dairy Technol. 2015, 68, 527–532. [Google Scholar] [CrossRef]
- Tripathi, M.K.; Giri, S.K. Probiotic Functional Foods: Survival of Probiotics during Processing and Storage. J. Funct. Foods 2014, 9, 225–241. [Google Scholar] [CrossRef]
- Shori, A.B. The Potential Applications of Probiotics on Dairy and Non-Dairy Foods Focusing on Viability during Storage. Biocatal. Agric. Biotechnol. 2015, 4, 423–431. [Google Scholar] [CrossRef]
- Turgut, T.; Cakmakci, S. Probiotic Strawberry Yogurts: Microbiological, Chemical and Sensory Properties. Probiotics Antimicro. Prot. 2018, 10, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Panghal, A.; Janghu, S.; Virkar, K.; Gat, Y.; Kumar, V.; Chhikara, N. Potential Non-Dairy Probiotic Products—A Healthy Approach. Food Biosci. 2018, 21, 80–89. [Google Scholar] [CrossRef]
- Angmo, K.; Kumari, A.; Savitri; Bhalla, T.C. Probiotic Characterization of Lactic Acid Bacteria Isolated from Fermented Foods and Beverage of Ladakh. LWT-Food Sci. Technol. 2016, 66, 428–435. [Google Scholar] [CrossRef]
- Sokovic Bajic, S.; Djokic, J.; Dinic, M.; Veljovic, K.; Golic, N.; Mihajlovic, S.; Tolinacki, M. GABA-Producing Natural Dairy Isolate From Artisanal Zlatar Cheese Attenuates Gut Inflammation and Strengthens Gut Epithelial Barrier in Vitro. Front. Microbiol. 2019, 10, 527. [Google Scholar] [CrossRef] [PubMed]
- Abushelaibi, A.; Al-Mahadin, S.; El-Tarabily, K.; Shah, N.P.; Ayyash, M. Characterization of Potential Probiotic Lactic Acid Bacteria Isolated from Camel Milk. LWT-Food Sci. Technol. 2017, 79, 316–325. [Google Scholar] [CrossRef]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.-A.G.; Tsakalidou, E.; Nychas, G.-J.E.; Panagou, E.Z.; Tassou, C.C. Selection of Potential Probiotic Lactic Acid Bacteria from Fermented Olives by in Vitro Tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic Resistance among Commercially Available Probiotics. Food Res. Int. 2014, 57, 176–195. [Google Scholar] [CrossRef]
- Vasudha, S.; Mishra, H.N. Non Dairy Probiotic Beverages. Int. Food Res. J. 2013, 20, 7. [Google Scholar]
- Kondrotiene, K.; Lauciene, L.; Andruleviciute, V.; Kasetiene, N.; Serniene, L.; Sekmokiene, D.; Malakauskas, M. Safety Assessment and Preliminary In Vitro Evaluation of Probiotic Potential of Lactococcus lactis Strains Naturally Present in Raw and Fermented Milk. Curr. Microbiol. 2020, 77, 3013–3023. [Google Scholar] [CrossRef]
- Akbar, A.; Sadiq, M.B.; Ali, I.; Anwar, M.; Muhammad, N.; Muhammad, J.; Shafee, M.; Ullah, S.; Gul, Z.; Qasim, S.; et al. Lactococcus lactis subsp. lactis Isolated from Fermented Milk Products and Its Antimicrobial Potential. CyTA-J. Food 2019, 17, 214–220. [Google Scholar] [CrossRef]
- Hermanns, G.; Funck, G.D.; Schmidt, J.T.; Pereira, J.Q.; Brandelli, A.; Richards, N.S.P.d.S. Evaluation of Probiotic Characteristics of Lactic Acid Bacteria Isolated from Artisan Cheese. J. Food Saf. 2014, 34, 380–387. [Google Scholar] [CrossRef]
- Das, P.; Khowala, S.; Biswas, S. In Vitro Probiotic Characterization of Lactobacillus casei Isolated from Marine Samples. LWT 2016, 73, 383–390. [Google Scholar] [CrossRef]
- Zielińska, D.; Rzepkowska, A.; Radawska, A.; Zieliński, K. In Vitro Screening of Selected Probiotic Properties of Lactobacillus Strains Isolated from Traditional Fermented Cabbage and Cucumber. Curr. Microbiol. 2015, 70, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Jawan, R.; Abbasiliasi, S.; Mustafa, S.; Kapri, M.R.; Halim, M.; Ariff, A.B. In Vitro Evaluation of Potential Probiotic Strain Lactococcus lactis Gh1 and Its Bacteriocin-Like Inhibitory Substances for Potential Use in the Food Industry. Probiotics Antimicro. Prot. 2021, 13, 422–440. [Google Scholar] [CrossRef] [PubMed]
- Vinderola, C.G.; Reinheimer, J.A. Lactic Acid Starter and Probiotic Bacteria: A Comparative “in Vitro” Study of Probiotic Characteristics and Biological Barrier Resistance. Food Res. Int. 2003, 36, 895–904. [Google Scholar] [CrossRef]
- Jung, M.Y.; Lee, C.; Seo, M.-J.; Roh, S.W.; Lee, S.H. Characterization of a Potential Probiotic Bacterium Lactococcus raffinolactis WiKim0068 Isolated from Fermented Vegetable Using Genomic and in Vitro Analyses. BMC Microbiol. 2020, 20, 136. [Google Scholar] [CrossRef]
- Sałański, P.; Kowalczyk, M.; Bardowski, J.K.; Szczepankowska, A.K. Health-Promoting Nature of Lactococcus lactis IBB109 and Lactococcus lactis IBB417 Strains Exhibiting Proliferation Inhibition and Stimulation of Interleukin-18 Expression in Colorectal Cancer Cells. Front. Microbiol. 2022, 13, 822912. [Google Scholar] [CrossRef]
- Tenea, G.N.; Suárez, J. Probiotic Potential and Technological Properties of Bacteriocinogenic Lactococcus lactis subsp. lactis UTNGt28 from a Native Amazonian Fruit as a Yogurt Starter Culture. Microorganisms 2020, 14, 733. [Google Scholar] [CrossRef]
- Mileriene, J.; Serniene, L.; Kasparaviciene, B.; Lauciene, L.; Kasetiene, N.; Zakariene, G.; Kersiene, M.; Leskauskaite, D.; Viskelis, J.; Kourkoutas, Y.; et al. Exploring the Potential of Sustainable Acid Whey Cheese Supplemented with Apple Pomace and GABA-Producing Indigenous Lactococcus lactis Strain. Microorganisms 2023, 11, 436. [Google Scholar] [CrossRef]
- Hurtado-Romero, A.; Del Toro-Barbosa, M.; Gradilla-Hernández, M.S.; Garcia-Amezquita, L.E.; García-Cayuela, T. Probiotic Properties, Prebiotic Fermentability, and GABA-Producing Capacity of Microorganisms Isolated from Mexican Milk Kefir Grains: A Clustering Evaluation for Functional Dairy Food Applications. Foods 2021, 10, 2275. [Google Scholar] [CrossRef]
- Jaskulski, I.B.; Uecker, J.; Bordini, F.; Moura, F.; Gonçalves, T.; Chaves, N.G.; Camargo, F.; Grecco, F.B.; Fiorentini, Â.M.; da Silva, W.P.; et al. In Vivo Action of Lactococcus lactis subsp. lactis Isolate (R7) with Probiotic Potential in the Stabilization of Cancer Cells in the Colorectal Epithelium. Process Biochem. 2020, 91, 165–171. [Google Scholar] [CrossRef]
- Ramalho, J.B.; Soares, M.B.; Spiazzi, C.C.; Bicca, D.F.; Soares, V.M.; Pereira, J.G.; da Silva, W.P.; Sehn, C.P.; Cibin, F.W.S. In Vitro Probiotic and Antioxidant Potential of Lactococcus lactis subsp. cremoris LL95 and Its Effect in Mice Behaviour. Nutrients 2019, 11, 901. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Singh, N.; Singh, S.; Khare, S.K.; Nain, P.K.S.; Nain, L. Potent γ-Amino Butyric Acid Producing Psychobiotic Lactococcus lactis LP-68 from Non-Rhizospheric Soil of Syzygium Cumini (Black Plum). Arch. Microbiol. 2021, 204, 82. [Google Scholar] [CrossRef] [PubMed]
- da Silva, L.A.; Lopes Neto, J.H.P.; Cardarelli, H.R. Safety and Probiotic Functionality of Isolated Goat Milk Lactic Acid Bacteria. Ann. Microbiol. 2019, 69, 1497–1505. [Google Scholar] [CrossRef]
- Kazancıgil, E.; Demirci, T.; Öztürk-Negiş, H.İ.; Akın, N. Isolation, Technological Characterization and in Vitro Probiotic Evaluation of Lactococcus Strains from Traditional Turkish Skin Bag Tulum Cheeses. Ann. Microbiol. 2019, 69, 1275–1287. [Google Scholar] [CrossRef]
- Colautti, A.; Arnoldi, M.; Comi, G.; Iacumin, L. Antibiotic Resistance and Virulence Factors in Lactobacilli: Something to Carefully Consider. Food Microbiol. 2022, 103, 103934. [Google Scholar] [CrossRef]
- EFSA, E.F.S. Introduction of a Qualified Presumption of Safety (QPS) Approach for Assessment of Selected Microorganisms Referred to EFSA—Opinion of the Scientific Committee. EFSA J. 2007, 5, 587. [Google Scholar] [CrossRef]
- EFSA, E.F.S. EFSA Statement on the Requirements for Whole Genome Sequence Analysis of Microorganisms Intentionally Used in the Food Chain. EFSA J. 2021, 19, e06506. [Google Scholar]
- Herreros, M.A.; Sandoval, H.; González, L.; Castro, J.M.; Fresno, J.M.; Tornadijo, M.E. Antimicrobial Activity and Antibiotic Resistance of Lactic Acid Bacteria Isolated from Armada Cheese (a Spanish Goats’ Milk Cheese). Food Microbiol. 2005, 22, 455–459. [Google Scholar] [CrossRef]
- Colautti, A.; Orecchia, E.; Comi, G.; Iacumin, L. Lactobacilli, a Weapon to Counteract Pathogens through the Inhibition of Their Virulence Factors. J. Bacteriol. 2022, 204, e0027222. [Google Scholar] [CrossRef]
- Pieniz, S.; de Moura, T.M.; Cassenego, A.P.V.; Andreazza, R.; Frazzon, A.P.G.; Camargo, F.A.d.O.; Brandelli, A. Evaluation of Resistance Genes and Virulence Factors in a Food Isolated Enterococcus durans with Potential Probiotic Effect. Food Control 2015, 51, 49–54. [Google Scholar] [CrossRef]
- Sylvere, N.; Mustopa, A.Z.; Budiarti, S.; Meilina, L.; Hertati, A.; Handayani, I. Whole-Genome Sequence Analysis and Probiotic Characteristics of Lactococcus lactis subsp. lactis Strain Lac3 Isolated from Traditional Fermented Buffalo Milk (Dadih). J. Genet. Eng. Biotechnol. 2023, 21, 49. [Google Scholar] [CrossRef] [PubMed]
- Brinkwirth, S.; Ayobami, O.; Eckmanns, T.; Markwart, R. Hospital-Acquired Infections Caused by Enterococci: A Systematic Review and Meta-Analysis, WHO European Region, 1 January 2010 to 4 February 2020. Eurosurveillance 2021, 26, 2001628. [Google Scholar] [CrossRef]
- Șchiopu, P.; Toc, D.A.; Colosi, I.A.; Costache, C.; Ruospo, G.; Berar, G.; Gălbău Ștefan, G.; Ghilea, A.C.; Botan, A.; Pană, A.-G.; et al. An Overview of the Factors Involved in Biofilm Production by the Enterococcus Genus. Int. J. Mol. Sci. 2023, 24, 11577. [Google Scholar] [CrossRef] [PubMed]
- Chajęcka-Wierzchowska, W.; Zadernowska, A.; Łaniewska-Trokenheim, Ł. Virulence Factors of Enterococcus spp. Presented in Food. LWT 2017, 75, 670–676. [Google Scholar] [CrossRef]
- Czarnowska-Kujawska, M.; Paszczyk, B. Changes in the Folate Content and Fatty Acid Profile in Fermented Milk Produced with Different Starter Cultures during Storage. Molecules 2021, 26, 6063. [Google Scholar] [CrossRef] [PubMed]
- Eubank, T.A.; Gonzales-Luna, A.J.; Hurdle, J.G.; Garey, K.W. Genetic Mechanisms of Vancomycin Resistance in Clostridioides Difficile: A Systematic Review. Antibiotics 2022, 11, 258. [Google Scholar] [CrossRef] [PubMed]
- Cavera, V.L.; Arthur, T.D.; Kashtanov, D.; Chikindas, M.L. Bacteriocins and Their Position in the next Wave of Conventional Antibiotics. Int. J. Antimicrob. Agents 2015, 46, 494–501. [Google Scholar] [CrossRef]
- Zycka-Krzesinska, J.; Boguslawska, J.; Aleksandrzak-Piekarczyk, T.; Jopek, J.; Bardowski, J.K. Identification and Characterization of Tetracycline Resistance in Lactococcus lactis Isolated from Polish Raw Milk and Fermented Artisanal Products. Int. J. Food Microbiol. 2015, 211, 134–141. [Google Scholar] [CrossRef]
- Miquel, S.; Beaumont, M.; Martín, R.; Langella, P.; Braesco, V.; Thomas, M. A Proposed Framework for an Appropriate Evaluation Scheme for Microorganisms as Novel Foods with a Health Claim in Europe. Microb. Cell Fact. 2015, 14, 48. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP). Guidance on the Assessment of Bacterial Susceptibility to Antimicrobials of Human and Veterinary Importance. EFSA J. 2012, 10, 2740.
- Kim, T.; Mondal, S.C.; Jeong, C.; Kim, S.; Ban, O.; Jung, Y.H.; Yang, J.; Kim, S. Safety Evaluation of Lactococcus lactis IDCC 2301 Isolated from Homemade Cheese. Food Sci. Nutr. 2021, 10, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.; Paek, J.J.; Lee, Y. Antimicrobial Resistance of Seventy Lactic Acid Bacteria Isolated from Commercial Probiotics in Korea. J. Microbiol. Biotechnol. 2023, 33, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Araújo, C.; Muñoz-Atienza, E.; Ramírez, M.; Poeta, P.; Igrejas, G.; Hernández, P.E.; Herranz, C.; Cintas, L.M. Safety Assessment, Genetic Relatedness and Bacteriocin Activity of Potential Probiotic Lactococcus lactis Strains from Rainbow Trout (Oncorhynchus Mykiss, Walbaum) and Rearing Environment. Eur. Food Res. Technol. 2015, 241, 647–662. [Google Scholar] [CrossRef]
- Ammor, M.S.; Flórez, A.B.; van Hoek, A.H.A.M.; de Los Reyes-Gavilán, C.G.; Aarts, H.J.M.; Margolles, A.; Mayo, B. Molecular Characterization of Intrinsic and Acquired Antibiotic Resistance in Lactic Acid Bacteria and Bifidobacteria. J. Mol. Microbiol. Biotechnol. 2008, 14, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Casarotti, S.N.; Carneiro, B.M.; Todorov, S.D.; Nero, L.A.; Rahal, P.; Penna, A.L.B. In Vitro Assessment of Safety and Probiotic Potential Characteristics of Lactobacillus Strains Isolated from Water Buffalo Mozzarella Cheese. Ann. Microbiol. 2017, 67, 289–301. [Google Scholar] [CrossRef]
- Doeun, D.; Davaatseren, M.; Chung, M.-S. Biogenic Amines in Foods. Food Sci. Biotechnol. 2017, 26, 1463–1474. [Google Scholar] [CrossRef]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic Amine Production by Lactic Acid Bacteria: A Review. Foods 2019, 8, 17. [Google Scholar] [CrossRef]
- Kononiuk, A.D.; Karwowska, M. Influence of Freeze-Dried Acid Whey Addition on Biogenic Amines Formation in a Beef and Deer Dry Fermented Sausages without Added Nitrite. Asian-Australas J. Anim. Sci. 2020, 33, 332–338. [Google Scholar] [CrossRef]
- Moniente, M.; García-Gonzalo, D.; Llamas-Arriba, M.G.; Garate, J.; Ontañón, I.; Jaureguibeitia, A.; Virto, R.; Pagán, R.; Botello-Morte, L. The Significance of Cheese Sampling in the Determination of Histamine Concentration: Distribution Pattern of Histamine in Ripened Cheeses. LWT 2022, 171, 114099. [Google Scholar] [CrossRef]
- Costantini, A.; Vaudano, E.; Pulcini, L.; Carafa, T.; Garcia-Moruno, E. An Overview on Biogenic Amines in Wine. Beverages 2019, 5, 19. [Google Scholar] [CrossRef]
- Gao, X.; Li, C.; He, R.; Zhang, Y.; Wang, B.; Zhang, Z.-H.; Ho, C.-T. Research Advances on Biogenic Amines in Traditional Fermented Foods: Emphasis on Formation Mechanism, Detection and Control Methods. Food Chem. 2023, 405, 134911. [Google Scholar] [CrossRef]
- Flasarová, R.; Pachlová, V.; Buňková, L.; Menšíková, A.; Georgová, N.; Dráb, V.; Buňka, F. Biogenic Amine Production by Lactococcus lactis subsp. cremoris Strains in the Model System of Dutch-Type Cheese. Food Chem. 2016, 194, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Ladero, V.; Cañedo, E.; Pérez, M.; Martín, M.C.; Fernández, M.; Alvarez, M.A. Multiplex qPCR for the Detection and Quantification of Putrescine-Producing Lactic Acid Bacteria in Dairy Products. Food Control 2012, 27, 307–313. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef]
- Ladero, V.; Rattray, F.P.; Mayo, B.; Martín, M.C.; Fernández, M.; Alvarez, M.A. Sequencing and Transcriptional Analysis of the Biosynthesis Gene Cluster of Putrescine-Producing Lactococcus lactis. Appl. Environ. Microbiol. 2011, 77, 6409–6418. [Google Scholar] [CrossRef]
- del Rio, B.; Linares, D.M.; Ladero, V.; Redruello, B.; Fernández, M.; Martin, M.C.; Alvarez, M.A. Putrescine Production via the Agmatine Deiminase Pathway Increases the Growth of Lactococcus lactis and Causes the Alkalinization of the Culture Medium. Appl. Microbiol. Biotechnol. 2015, 99, 897–905. [Google Scholar] [CrossRef]
- del Rio, B.; Redruello, B.; Linares, D.M.; Ladero, V.; Ruas-Madiedo, P.; Fernandez, M.; Martin, M.C.; Alvarez, M.A. The Biogenic Amines Putrescine and Cadaverine Show in Vitro Cytotoxicity at Concentrations That Can Be Found in Foods. Sci. Rep. 2019, 9, 120. [Google Scholar] [CrossRef]
- Garbowska, M.; Pluta, A.; Berthold-Pluta, A. Impact of Nisin-Producing Strains of Lactococcus lactis on the Contents of Bioactive Dipeptides, Free Amino Acids, and Biogenic Amines in Dutch-Type Cheese Models. Materials 2020, 13, 1835. [Google Scholar] [CrossRef]
- Spizzirri, U.G.; Restuccia, D.; Curcio, M.; Parisi, O.I.; Iemma, F.; Picci, N. Determination of Biogenic Amines in Different Cheese Samples by LC with Evaporative Light Scattering Detector. J. Food Compos. Anal. 2013, 29, 43–51. [Google Scholar] [CrossRef]
- EFSA Scientific Opinion on Risk Based Control of Biogenic Amine Formation in Fermented foods-European Food Safety Authority Panel on Biological Hazards (BIOHAZ). EFSA J. 2011, 9, 2393–2486. [CrossRef]
- Moret, S.; Bortolomeazzi, R.; Feruglio, M.; Lercker, G. Determination of Biogenic Amines in Italian Cheeses [Amine Biogeniche Nei Formaggi Italiani]. Sci. E Tec. Latt.-Casearia 1992, 43, 187–198. [Google Scholar]
- Zdolec, N.; Bogdanović, T.; Severin, K.; Dobranić, V.; Kazazić, S.; Grbavac, J.; Pleadin, J.; Petričević, S.; Kiš, M. Biogenic Amine Content in Retailed Cheese Varieties Produced with Commercial Bacterial or Mold Cultures. Processes 2022, 10, 10. [Google Scholar] [CrossRef]
- Kandasamy, S.; Yoo, J.; Yun, J.; Kang, H.B.; Seol, K.-H.; Ham, J.-S. Quantitative Analysis of Biogenic Amines in Different Cheese Varieties Obtained from the Korean Domestic and Retail Markets. Metabolites 2021, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Berthoud, H.; Wechsler, D.; Irmler, S. Production of Putrescine and Cadaverine by Paucilactobacillus Wasatchensis. Front. Microbiol. 2022, 13, 842403. [Google Scholar] [CrossRef] [PubMed]
- Buňka, F.; Zálešáková, L.; Flasarová, R.; Pachlová, V.; Budinský, P.; Buňková, L. Biogenic Amines Content in Selected Commercial Fermented Products of Animal Origin. J. Microbiol. Biotechnol. Food Sci. 2012, 2, 209–218. [Google Scholar]
- Novella-Rodríguez, S.; Veciana-Nogués, M.T.; Roig-Sagués, A.X.; Trujillo-Mesa, A.J.; Vidal-Carou, M.C. Evaluation of Biogenic Amines and Microbial Counts throughout the Ripening of Goat Cheeses from Pasteurized and Raw Milk. J. Dairy Res. 2004, 71, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Marijan, A.; Džaja, P.; Bogdanović, T.; Škoko, I.; Cvetnić, Ž.; Dobranić, V.; Zdolec, N.; Šatrović, E.; Severin, K. Influence of Ripening Time on the Amount of Certain Biogenic Amines in Rind and Core of Cow Milk Livno Cheese. Mljekarstvo Časopis Za Unaprjeđenje Proizv. I Prerade Mlijeka 2014, 64, 159–169. [Google Scholar]
- Bogdanović, T.; Petričević, S.; Brkljača, M.; Listeš, I.; Pleadin, J. Biogenic Amines in Selected Foods of Animal Origin Obtained from the Croatian Retail Market. Food Addit. Contam. Part A 2020, 37, 815–830. [Google Scholar] [CrossRef]
- Shalaby, M.A.; Kassem, M.A.; Morsy, O.; Mohamed, N.M. Anti-Tyramine Potential of Lactobacillus rhamnosus (LGG®) in Cheese Samples Collected from Alexandria, Egypt. Food Biotechnol. 2020, 34, 243–261. [Google Scholar] [CrossRef]
- Schirone, M.; Esposito, L.; D’Onofrio, F.; Visciano, P.; Martuscelli, M.; Mastrocola, D.; Paparella, A. Biogenic Amines in Meat and Meat Products: A Review of the Science and Future Perspectives. Foods 2022, 11, 788. [Google Scholar] [CrossRef]
- Decadt, H.; Weckx, S.; De Vuyst, L. The Rotation of Primary Starter Culture Mixtures Results in Batch-to-Batch Variations during Gouda Cheese Production. Front. Microbiol. 2023, 14, 1128394. [Google Scholar] [CrossRef] [PubMed]
- Ladero, V.; Martín, M.C.; Redruello, B.; Mayo, B.; Flórez, A.B.; Fernández, M.; Alvarez, M.A. Genetic and Functional Analysis of Biogenic Amine Production Capacity among Starter and Non-Starter Lactic Acid Bacteria Isolated from Artisanal Cheeses. Eur. Food Res. Technol. 2015, 241, 377–383. [Google Scholar] [CrossRef]
- Perin, L.M.; Dal Bello, B.; Belviso, S.; Zeppa, G.; de Carvalho, A.F.; Cocolin, L.; Nero, L.A. Microbiota of Minas Cheese as Influenced by the Nisin Producer Lactococcus lactis subsp. lactis GLc05. Int. J. Food Microbiol. 2015, 214, 159–167. [Google Scholar] [CrossRef]
- Alvarez, M.A.; Moreno-Arribas, M.V. The Problem of Biogenic Amines in Fermented Foods and the Use of Potential Biogenic Amine-Degrading Microorganisms as a Solution. Trends Food Sci. Technol. 2014, 39, 146–155. [Google Scholar] [CrossRef]
- Omer, A.K.; Mohammed, R.R.; Ameen, P.S.M.; Abas, Z.A.; Ekici, K. Presence of Biogenic Amines in Food and Their Public Health Implications: A Review. J. Food Prot. 2021, 84, 1539–1548. [Google Scholar] [CrossRef]
- Dala-Paula, B.M.; Custódio, F.B.; Gloria, M.B. Health Concerns Associated with Biogenic Amines in Food and Interaction with Amine Oxidase Drugs. Curr. Opin. Food Sci. 2023, 54, 101090. [Google Scholar] [CrossRef]
- Fernández, M.; Linares, D.M.; Del Río, B.; Ladero, V.; Alvarez, M.A. HPLC Quantification of Biogenic Amines in Cheeses: Correlation with PCR-Detection of Tyramine-Producing Microorganisms. J. Dairy Res. 2007, 74, 276–282. [Google Scholar] [CrossRef]
- Barone, C.; Barebera, M.; Barone, M.; Parisi, S.; Zaccheo, A.; Barone, C.; Barbera, M.; Barone, M.; Parisi, S.; Zaccheo, A. Biogenic Amines in Cheeses: Types and Typical Amounts. In Chemical Evolution of Nitrogen-based Compounds in Mozzarella Cheeses; Springer: Cham, Switzerland, 2018; pp. 1–18. [Google Scholar] [CrossRef]
- Perez, R.H.; Ancuelo, A.E.; Perez, R.H.; Ancuelo, A.E. Diverse Bioactive Molecules from the Genus Lactobacillus. In Lactobacillus—A Multifunctional Genus; IntechOpen: London, UK, 2022; ISBN 978-1-80355-445-7. [Google Scholar]
- Nehal, F.; Sahnoun, M.; Smaoui, S.; Jaouadi, B.; Bejar, S.; Mohammed, S. Characterization, High Production and Antimicrobial Activity of Exopolysaccharides from Lactococcus lactis F-Mou. Microb. Pathog. 2019, 132, 10–19. [Google Scholar] [CrossRef]
- Tiwari, O.N.; Sasmal, S.; Kataria, A.K.; Devi, I. Application of Microbial Extracellular Carbohydrate Polymeric Substances in Food and Allied Industries. 3 Biotech 2020, 10, 221. [Google Scholar] [CrossRef]
- Costa, N.E.; Hannon, J.A.; Guinee, T.P.; Auty, M.A.E.; McSweeney, P.L.H.; Beresford, T.P. Effect of Exopolysaccharide Produced by Isogenic Strains of Lactococcus lactis on Half-Fat Cheddar Cheese. J. Dairy Sci. 2010, 93, 3469–3486. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, Y.; Ye, L.; Wang, C. The Anti-Cancer Effects and Mechanisms of Lactic Acid Bacteria Exopolysaccharides in Vitro: A Review. Carbohydr. Polym. 2021, 253, 117308. [Google Scholar] [CrossRef] [PubMed]
- Muscariello, L.; De Siena, B.; Marasco, R. Lactobacillus Cell Surface Proteins Involved in Interaction with Mucus and Extracellular Matrix Components. Curr. Microbiol. 2020, 77, 3831–3841. [Google Scholar] [CrossRef] [PubMed]
- Nugroho, A.D.W.; Kleerebezem, M.; Bachmann, H. A Novel Method for Long-Term Analysis of Lactic Acid and Ammonium Production in Non-Growing Lactococcus lactis Reveals Pre-Culture and Strain Dependence. Front. Bioeng. Biotechnol. 2020, 8, 580090. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.N.; Awang Adeni, D.S.; Suhaili, N.; Bujang, K. Optimisation of Pre-Harvest Sago Frond Sap for the Production of l-Lactic Acid Using Lactococcus lactis IO-1. Biocatal. Agric. Biotechnol. 2022, 43, 102435. [Google Scholar] [CrossRef]
- Azhar, N.S.; Zin, N.H.M.; Hamid, T.H.T.A. Lactococcus Lactis Strain A5 Producing Nisin-like Bacteriocin Active against Gram Positive and Negative Bacteria. Trop. Life Sci. Res. 2017, 28, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Acedo, J.Z.; Chiorean, S.; Vederas, J.C.; van Belkum, M.J. The Expanding Structural Variety among Bacteriocins from Gram-Positive Bacteria. FEMS Microbiol. Rev. 2018, 42, 805–828. [Google Scholar] [CrossRef] [PubMed]
- Takala, T.M.; Mokhtari, S.; Ahonen, S.L.; Wan, X.; Saris, P.E.J. Wild-Type Lactococcus lactis Producing Bacteriocin-like Prophage Lysins. Front. Microbiol. 2023, 14, 1219723. [Google Scholar] [CrossRef]
- Chandran, C.; Tham, H.Y.; Rahim, R.A.; Lim, S.H.E.; Yusoff, K.; Song, A.A.-L. Lactococcus lactis Secreting Phage Lysins as a Potential Antimicrobial against Multi-Drug Resistant Staphylococcus aureus. PeerJ 2022, 10, e12648. [Google Scholar] [CrossRef]
- Nebbia, S.; Lamberti, C.; Lo Bianco, G.; Cirrincione, S.; Laroute, V.; Cocaign-Bousquet, M.; Cavallarin, L.; Giuffrida, M.G.; Pessione, E. Antimicrobial Potential of Food Lactic Acid Bacteria: Bioactive Peptide Decrypting from Caseins and Bacteriocin Production. Microorganisms 2020, 9, 65. [Google Scholar] [CrossRef]
- Kurbanova, M.; Voroshilin, R.; Kozlova, O.; Atuchin, V. Effect of Lactobacteria on Bioactive Peptides and Their Sequence Identification in Mature Cheese. Microorganisms 2022, 10, 2068. [Google Scholar] [CrossRef] [PubMed]
- Kocak, A.; Sanli, T.; Anli, E.A.; Hayaloglu, A.A. Role of Using Adjunct Cultures in Release of Bioactive Peptides in White-Brined Goat-Milk Cheese. LWT 2020, 123, 109127. [Google Scholar] [CrossRef]
- Cirrincione, S.; Luganini, A.; Lamberti, C.; Manfredi, M.; Cavallarin, L.; Giuffrida, M.G.; Pessione, E. Donkey Milk Fermentation by Lactococcus lactis subsp. cremoris and Lactobacillus rhamnosus Affects the Antiviral and Antibacterial Milk Properties. Molecules 2021, 26, 5100. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Liu, R.h. Increase of Conjugated Linoleic Acid Content in Milk by Fermentation with Lactic Acid Bacteria. J. Food Sci. 2002, 67, 1731–1737. [Google Scholar] [CrossRef]
- Puniya, A.K.; Chaitanya, S.; Tyagi, A.K.; De, S.; Singh, K. Conjugated Linoleic Acid Producing Potential of Lactobacilli Isolated from the Rumen of Cattle. J. Ind. Microbiol. Biotechnol. 2008, 35, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Renes, E.; Linares, D.M.; González, L.; Fresno, J.M.; Tornadijo, M.E.; Stanton, C. Production of Conjugated Linoleic Acid and Gamma-Aminobutyric Acid by Autochthonous Lactic Acid Bacteria and Detection of the Genes Involved. J. Funct. Foods 2017, 34, 340–346. [Google Scholar] [CrossRef]
- Laroute, V.; Mazzoli, R.; Loubière, P.; Pessione, E.; Cocaign-Bousquet, M. Environmental Conditions Affecting GABA Production in Lactococcus lactis NCDO 2118. Microorganisms 2021, 9, 122. [Google Scholar] [CrossRef]
- Khalili, M.; Rad, A.H.; Khosroushahi, A.Y.; Khosravi, H.; Jafarzadeh, S. Application of Probiotics in Folate Bio-Fortification of Yoghurt. Probiotics Antimicrob. Proteins 2020, 12, 756–763. [Google Scholar] [CrossRef]
- Liu, Y.; van Bennekom, E.O.; Zhang, Y.; Abee, T.; Smid, E.J. Long-Chain Vitamin K2 Production in Lactococcus lactis Is Influenced by Temperature, Carbon Source, Aeration and Mode of Energy Metabolism. Microb. Cell Factories 2019, 18, 129. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, S.; Hao, G.; Yu, H.; Tian, H.; Zhao, G. Role of Lactic Acid Bacteria on the Yogurt Flavour: A Review. Int. J. Food Prop. 2017, 20, S316–S330. [Google Scholar] [CrossRef]
- Bøe, C.A.; Holo, H. Engineering Lactococcus lactis for Increased Vitamin K2 Production. Front. Bioeng. Biotechnol. 2020, 8, 191. [Google Scholar] [CrossRef] [PubMed]
- Mir Khan, U.; Selamoglu, Z. Use of Enzymes in Dairy Industry: A Review of Current Progress. Arch. Razi Inst. 2020, 75, 131–136. [Google Scholar] [CrossRef]
- Hernandez-Valdes, J.A.; Solopova, A.; Kuipers, O.P. Development of Lactococcus lactis Biosensors for Detection of Diacetyl. Front. Microbiol. 2020, 11, 1032. [Google Scholar] [CrossRef] [PubMed]
- Román Naranjo, D.; Callanan, M.; Thierry, A.; McAuliffe, O. Superior Esterolytic Activity in Environmental Lactococcus lactis Strains Is Linked to the Presence of the SGNH Hydrolase Family of Esterases. JDS Commun. 2020, 1, 25–28. [Google Scholar] [CrossRef]
- Park, Y.W.; Nam, M.S. Bioactive Peptides in Milk and Dairy Products: A Review. Korean J. Food Sci. Anim. Resour. 2015, 35, 831. [Google Scholar] [CrossRef]
- Ham, S.; Bhatia, S.K.; Gurav, R.; Choi, Y.-K.; Jeon, J.-M.; Yoon, J.-J.; Choi, K.-Y.; Ahn, J.; Kim, H.T.; Yang, Y.-H. Gamma Aminobutyric Acid (GABA) Production in Escherichia coli with Pyridoxal Kinase (pdxY) Based Regeneration System. Enzym. Microb. Technol. 2022, 155, 109994. [Google Scholar] [CrossRef] [PubMed]
- Santos-Espinosa, A.; Beltrán-Barrientos, L.M.; Reyes-Díaz, R.; Mazorra-Manzano, M.Á.; Hernández-Mendoza, A.; González-Aguilar, G.A.; Sáyago-Ayerdi, S.G.; Vallejo-Cordoba, B.; González-Córdova, A.F. Gamma-Aminobutyric Acid (GABA) Production in Milk Fermented by Specific Wild Lactic Acid Bacteria Strains Isolated from Artisanal Mexican Cheeses. Ann. Microbiol. 2020, 70, 12. [Google Scholar] [CrossRef]
- Pouliot-Mathieu, K.; Gardner-Fortier, C.; Lemieux, S.; St-Gelais, D.; Champagne, C.P.; Vuillemard, J.-C. Effect of Cheese Containing Gamma-Aminobutyric Acid-Producing Lactic Acid Bacteria on Blood Pressure in Men. PharmaNutrition 2013, 1, 141–148. [Google Scholar] [CrossRef]
- Hudec, J.; Kobida, Ľ.; Čanigová, M.; Lacko-Bartošová, M.; Ložek, O.; Chlebo, P.; Mrázová, J.; Ducsay, L.; Bystrická, J. Production of γ-Aminobutyric Acid by Microorganisms from Different Food Sources. J. Sci. Food Agric. 2015, 95, 1190–1198. [Google Scholar] [CrossRef]
- Sarasa, S.B.; Mahendran, R.; Muthusamy, G.; Thankappan, B.; Selta, D.R.F.; Angayarkanni, J. A Brief Review on the Non-Protein Amino Acid, Gamma-Amino Butyric Acid (GABA): Its Production and Role in Microbes. Curr. Microbiol. 2020, 77, 534–544. [Google Scholar] [CrossRef]
- Dhakal, R.; Bajpai, V.K.; Baek, K.-H. Production of Gaba (γ-Aminobutyric Acid) by Microorganisms: A Review. Braz. J. Microbiol. 2012, 43, 1230–1241. [Google Scholar] [CrossRef] [PubMed]
- Redruello, B.; Saidi, Y.; Sampedro, L.; Ladero, V.; del Rio, B.; Alvarez, M.A. GABA-Producing Lactococcus lactis Strains Isolated from Camel’s Milk as Starters for the Production of GABA-Enriched Cheese. Foods 2021, 10, 633. [Google Scholar] [CrossRef] [PubMed]
- Diana, M.; Quílez, J.; Rafecas, M. Gamma-Aminobutyric Acid as a Bioactive Compound in Foods: A Review. J. Funct. Foods 2014, 10, 407–420. [Google Scholar] [CrossRef]
- Inoue, K.; Shirai, T.; Ochiai, H.; Kasao, M.; Hayakawa, K.; Kimura, M.; Sansawa, H. Blood-Pressure-Lowering Effect of a Novel Fermented Milk Containing γ-Aminobutyric Acid (GABA) in Mild Hypertensives. Eur. J. Clin. Nutr. 2003, 57, 490–495. [Google Scholar] [CrossRef]
- Ruas-Madiedo, P.; de los Reyes-Gavilán, C.G. Invited Review: Methods for the Screening, Isolation, and Characterization of Exopolysaccharides Produced by Lactic Acid Bacteria. J. Dairy Sci. 2005, 88, 843–856. [Google Scholar] [CrossRef]
- Jiang, Y.; Yang, Z. A Functional and Genetic Overview of Exopolysaccharides Produced by Lactobacillus plantarum. J. Funct. Foods 2018, 47, 229–240. [Google Scholar] [CrossRef]
- Jeong, H.; Kim, S.; Hwang, U.-S.; Choi, H.; Park, Y.-S. Immunostimulatory Activity of Lactococcus lactis subsp. lactis CAB701 Isolated from Jeju Cabbage. Microorganisms 2023, 11, 1718. [Google Scholar] [CrossRef]
- Lynch, K.M.; Coffey, A.; Arendt, E.K. Exopolysaccharide Producing Lactic Acid Bacteria: Their Techno-Functional Role and Potential Application in Gluten-Free Bread Products. Food Res. Int. 2018, 110, 52–61. [Google Scholar] [CrossRef]
- Knoshaug, E.P.; Ahlgren, J.A.; Trempy, J.E. Exopolysaccharide Expression in Lactococcus lactis subsp. cremoris Ropy352: Evidence for Novel Gene Organization. Appl. Environ. Microbiol. 2007, 73, 897–905. [Google Scholar] [CrossRef]
- Surber, G.; Schäper, C.; Wefers, D.; Rohm, H.; Jaros, D. Exopolysaccharides from Lactococcus lactis Affect Manufacture, Texture and Sensory Properties of Concentrated Acid Milk Gel Suspensions (Fresh Cheese). Int. Dairy J. 2021, 112, 104854. [Google Scholar] [CrossRef]
- Zhou, Y.; Cui, Y.; Qu, X. Exopolysaccharides of Lactic Acid Bacteria: Structure, Bioactivity and Associations: A Review. Carbohydr. Polym. 2019, 207, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Kristo, E.; Miao, Z.; Corredig, M. The Role of Exopolysaccharide Produced by Lactococcus lactis subsp. cremoris in Structure Formation and Recovery of Acid Milk Gels. Int. Dairy J. 2011, 21, 656–662. [Google Scholar] [CrossRef]
- Poulsen, V.K.; Derkx, P.; Oregaard, G. High-Throughput Screening for Texturing Lactococcus Strains. FEMS Microbiol. Lett. 2019, 366, fnz001. [Google Scholar] [CrossRef] [PubMed]
- Allam, M.G.; Darwish, A.M.; Ayad, E.H.; Shokery, E.S.; Darwish, S.M. Lactococcus Species for Conventional Karish Cheese Conservation. LWT-Food Sci. Technol. 2017, 79, 625–631. [Google Scholar] [CrossRef]
- Mende, S.; Rohm, H.; Jaros, D. Influence of Exopolysaccharides on the Structure, Texture, Stability and Sensory Properties of Yoghurt and Related Products. Int. Dairy J. 2016, 52, 57–71. [Google Scholar] [CrossRef]
- Mailam, M.; Abd El-Razek, H.; A Shaaban, H. Effect of Exopolysaccharides-Producing Starter Culture on the Flavor Profile and Characteristics of Low Fat Ras Cheese. Pak. J. Biol Sci. 2020, 23, 691–700. [Google Scholar] [CrossRef]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Lactic Acid Bacteria in Raw-Milk Cheeses: From Starter Cultures to Probiotic Functions. Foods 2022, 11, 2276. [Google Scholar] [CrossRef]
- Werning, M.L.; Hernández-Alcántara, A.M.; Ruiz, M.J.; Soto, L.P.; Dueñas, M.T.; López, P.; Frizzo, L.S. Biological Functions of Exopolysaccharides from Lactic Acid Bacteria and Their Potential Benefits for Humans and Farmed Animals. Foods 2022, 11, 1284. [Google Scholar] [CrossRef]
- De Vuyst, L.; Degeest, B. Heteropolysaccharides from Lactic Acid Bacteria. FEMS Microbiol. Rev. 1999, 23, 153–177. [Google Scholar] [CrossRef]
- Jurášková, D.; Ribeiro, S.C.; Silva, C.C.G. Exopolysaccharides Produced by Lactic Acid Bacteria: From Biosynthesis to Health-Promoting Properties. Foods 2022, 11, 156. [Google Scholar] [CrossRef]
- Broadbent, J.R.; Barnes, M.; Brennand, C.; Strickland, M.; Houck, K.; Johnson, M.E.; Steele, J.L. Contribution of Lactococcus lactis Cell Envelope Proteinase Specificity to Peptide Accumulation and Bitterness in Reduced-Fat Cheddar Cheese. Appl. Environ. Microbiol. 2002, 68, 1778–1785. [Google Scholar] [CrossRef] [PubMed]
- Mozzi, F.; Vaningelgem, F.; Hébert, E.M.; Van der Meulen, R.; Foulquié Moreno, M.R.; Font de Valdez, G.; De Vuyst, L. Diversity of Heteropolysaccharide-Producing Lactic Acid Bacterium Strains and Their Biopolymers. Appl. Environ. Microbiol. 2006, 72, 4431–4435. [Google Scholar] [CrossRef] [PubMed]
- Gruter, M.; Leeflang, B.R.; Kuiper, J.; Kamerling, J.P.; Vliegenthart, J.F. Structure of the Exopolysaccharide Produced by Lactococcus lactis subspecies cremoris H414 Grown in a Defined Medium or Skimmed Milk. Carbohydr. Res. 1992, 231, 273–291. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van Kranenburg, R.; Vos, H.R.; van Swam, I.I.; Kleerebezem, M.; de Vos, W.M. Functional Analysis of Glycosyltransferase Genes from Lactococcus lactis and Other Gram-Positive Cocci: Complementation, Expression, and Diversity. J. Bacteriol. 1999, 181, 6347–6353. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Leroy, F. Functional Role of Yeasts, Lactic Acid Bacteria and Acetic Acid Bacteria in Cocoa Fermentation Processes. FEMS Microbiol. Rev. 2020, 44, 432–453. [Google Scholar] [CrossRef] [PubMed]
- Prete, R.; Alam, M.K.; Perpetuini, G.; Perla, C.; Pittia, P.; Corsetti, A. Lactic Acid Bacteria Exopolysaccharides Producers: A Sustainable Tool for Functional Foods. Foods 2021, 10, 1653. [Google Scholar] [CrossRef]
- Torino, M.; Font de Valdez, G.; Mozzi, F. Biopolymers from Lactic Acid Bacteria. Novel Applications in Foods and Beverages. Front. Microbiol. 2015, 6, 834. [Google Scholar] [CrossRef]
- Li, M.; Li, W.; Li, D.; Tian, J.; Xiao, L.; Kwok, L.-Y.; Li, W.; Sun, Z. Structure Characterization, Antioxidant Capacity, Rheological Characteristics and Expression of Biosynthetic Genes of Exopolysaccharides Produced by Lactococcus lactis subsp. lactis IMAU11823. Food Chem. 2022, 384, 132566. [Google Scholar] [CrossRef]
- Lo, T.C.T.; Kang, M.W.; Wang, B.C.; Chang, C.A. Glycosyl Linkage Characteristics and Classifications of Exo-Polysaccharides of Some Regionally Different Strains of Lentinula Edodes by Amplified Fragment Length Polymorphism Assay and Cluster Analysis. Anal. Chim. Acta 2007, 592, 146–153. [Google Scholar] [CrossRef]
- Guo, Y.; Pan, D.; Sun, Y.; Xin, L.; Li, H.; Zeng, X. Antioxidant Activity of Phosphorylated Exopolysaccharide Produced by Lactococcus lactis subsp. lactis. Carbohydr. Polym. 2013, 97, 849–854. [Google Scholar] [CrossRef]
- Nguyen, A.T.-B.; Picart-Palmade, L.; Nigen, M.; Jimenez, L.; Ait-Abderahim, H.; Marchesseau, S. Effect of Mono or Co-Culture of EPS-Producing Streptococcus thermophilus Strains on the Formation of Acid Milk Gel and the Appearance of Texture Defects. Int. Dairy J. 2019, 98, 17–24. [Google Scholar] [CrossRef]
- Feng, J.; Cai, Z.; Chen, Y.; Zhu, H.; Chang, X.; Wang, X.; Liu, Z.; Zhang, J.; Nie, G. Effects of an Exopolysaccharide from Lactococcus lactis Z-2 on Innate Immune Response, Antioxidant Activity, and Disease Resistance against Aeromonas Hydrophila in Cyprinus Carpio L. Fish Shellfish Immunol. 2020, 98, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Dowdell, P.; Chankhamhaengdecha, S.; Panbangred, W.; Janvilisri, T.; Aroonnual, A. Probiotic Activity of Enterococcus Faecium and Lactococcus lactis Isolated from Thai Fermented Sausages and Their Protective Effect Against Clostridium Difficile. Probiotics Antimicro. Prot. 2020, 12, 641–648. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, Y.; Suzuki, S.; Amako, M.; Kitamura, S.; Toda, T. Effect of Orally Administered Exopolysaccharides Produced by Lactococcus lactis subsp. cremoris FC on a Mouse Model of Dermatitis Induced by Repeated Exposure to 2,4,6-Trinitro-1-Chlorobenzene. J. Funct. Foods 2017, 35, 43–50. [Google Scholar] [CrossRef]
- Cho, Y.; Han, H.T.; Kim, T.; Sohn, M.; Park, Y.-S. Immunostimulatory Activity of Lactococcus lactis LM1185 Isolated from Hydrangea Macrophylla. Food Sci. Biotechnol. 2023, 32, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Holst, O.A. Molinaro in Microbial Glycobiology; Academic Press: San Diego, CA, USA, 2010. [Google Scholar]
- Şanlier, N.; Gökcen, B.B.; Sezgin, A.C. Health Benefits of Fermented Foods. Crit. Rev. Food Sci. Nutr. 2019, 59, 506–527. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Vestergaard, M.; Jensen, T.G.; Shen, J.; Dufva, M.; Solem, C.; Jensen, P.R. Finding the Needle in the Haystack—The Use of Microfluidic Droplet Technology to Identify Vitamin-Secreting Lactic Acid Bacteria. mBio 2017, 8, 10–1128. [Google Scholar] [CrossRef]
- Pohanka, M. D-Lactic Acid as a Metabolite: Toxicology, Diagnosis, and Detection. Biomed. Res. Int. 2020, 2020, 3419034. [Google Scholar] [CrossRef]
- Remund, B.; Yilmaz, B.; Sokollik, C. D-Lactate: Implications for Gastrointestinal Diseases. Children 2023, 10, 945. [Google Scholar] [CrossRef]
- Weiss, G.A.; Hennet, T. Mechanisms and Consequences of Intestinal Dysbiosis. Cell. Mol. Life Sci. 2017, 74, 2959–2977. [Google Scholar] [CrossRef]
- Mishima, Y.; Sartor, R.B. Manipulating Resident Microbiota to Enhance Regulatory Immune Function to Treat Inflammatory Bowel Diseases. J. Gastroenterol. 2020, 55, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Kalliomäki, M.; Carmen Collado, M.; Salminen, S.; Isolauri, E. Early Differences in Fecal Microbiota Composition in Children May Predict Overweight2. Am. J. Clin. Nutr. 2008, 87, 534–538. [Google Scholar] [CrossRef] [PubMed]
- Maślak, E.; Złoch, M.; Arendowski, A.; Sugajski, M.; Janczura, I.; Rudnicka, J.; Walczak-Skierska, J.; Buszewska-Forajta, M.; Rafińska, K.; Pomastowski, P.; et al. Isolation and Identification of Lactococcus lactis and Weissella Cibaria Strains from Fermented Beetroot and an Investigation of Their Properties as Potential Starter Cultures and Probiotics. Foods 2022, 11, 2257. [Google Scholar] [CrossRef] [PubMed]
- Hatti-Kaul, R.; Chen, L.; Dishisha, T.; Enshasy, H.E. Lactic Acid Bacteria: From Starter Cultures to Producers of Chemicals. FEMS Microbiol. Lett. 2018, 365, fny213. [Google Scholar] [CrossRef] [PubMed]
- Gandini, C.; Tarraran, L.; Kalemasi, D.; Pessione, E.; Mazzoli, R. Recombinant Lactococcus lactis for Efficient Conversion of Cellodextrins into L-Lactic Acid. Biotechnol. Bioeng. 2017, 114, 2807–2817. [Google Scholar] [CrossRef]
- Bintsis, T. Lactic Acid Bacteria as Starter Cultures: An Update in Their Metabolism and Genetics. AIMS Microbiol. 2018, 4, 665. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.; Vázquez, L.; Flórez, A.B.; Mayo, B. Phenotype Testing, Genome Analysis, and Metabolic Interactions of Three Lactic Acid Bacteria Strains Existing as a Consortium in a Naturally Fermented Milk. Front. Microbiol. 2022, 13, 1000683. [Google Scholar] [CrossRef]
- Iskandar, C.F.; Cailliez-Grimal, C.; Borges, F.; Revol-Junelles, A.-M. Review of Lactose and Galactose Metabolism in Lactic Acid Bacteria Dedicated to Expert Genomic Annotation. Trends Food Sci. Technol. 2019, 88, 121–132. [Google Scholar] [CrossRef]
- Fortina, M.G.; Ricci, G.; Borgo, F. A Study of Lactose Metabolism in Lactococcus garvieae Reveals a Genetic Marker for Distinguishing between Dairy and Fish Biotypes. J. Food Prot. 2009, 72, 1248–1254. [Google Scholar] [CrossRef]
- Bukvicki, D.; Siroli, L.; D’Alessandro, M.; Cosentino, S.; Fliss, I.; Said, L.B.; Hassan, H.; Lanciotti, R.; Patrignani, F. Unravelling the Potential of Lactococcus lactis Strains to Be Used in Cheesemaking Production as Biocontrol Agents. Foods 2020, 9, 1815. [Google Scholar] [CrossRef]
- Poudel, R.; Thunell, R.K.; Oberg, C.J.; Overbeck, S.; Lefevre, M.; Oberg, T.S.; McMahon, D.J. Comparison of Growth and Survival of Single Strains of Lactococcus lactis and Lactococcus cremoris during Cheddar Cheese Manufacture. J. Dairy Sci. 2022, 105, 2069–2081. [Google Scholar] [CrossRef] [PubMed]
- Taïbi, A.; Dabour, N.; Lamoureux, M.; Roy, D.; LaPointe, G. Comparative Transcriptome Analysis of Lactococcus lactis subsp. cremoris Strains under Conditions Simulating Cheddar Cheese Manufacture. Int. J. Food Microbiol. 2011, 146, 263–275. [Google Scholar] [CrossRef] [PubMed]
- Vedamuthu, E.R.; Sandine, W.E.; Elliker, P.R. Flavor and Texture in Cheddar Cheese. I. Role of Mixed Strain Lactic Starter Cultures. J. Dairy Sci. 1966, 49, 144–150. [Google Scholar] [CrossRef]
- Li, A.; Zheng, J.; Han, X.; Yang, S.; Cheng, S.; Zhao, J.; Zhou, W.; Lu, Y. Advances in Low-Lactose/Lactose-Free Dairy Products and Their Production. Foods 2023, 12, 2553. [Google Scholar] [CrossRef]
- Serna Cock, L.; Rodriguez De Stouvenel, A. Lactic Acid Production by a Strain of Lactococcus lactis subs lactis Isolated from Sugar Cane Plants. Electron. J. Biotechnol. 2006, 9, 40–45. [Google Scholar] [CrossRef]
- Petrov, K.; Urshev, Z.; Petrova, P. L(+)-Lactic Acid Production from Starch by a Novel Amylolytic Lactococcus lactis subsp. lactis B84. Food Microbiol. 2008, 25, 550–557. [Google Scholar] [CrossRef]
- Sánchez, C.; Neves, A.R.; Cavalheiro, J.; dos Santos, M.M.; García-Quintáns, N.; López, P.; Santos, H. Contribution of Citrate Metabolism to the Growth of Lactococcus lactis CRL264 at Low pH. Appl. Environ. Microbiol. 2008, 74, 1136–1144. [Google Scholar] [CrossRef]
- Magni, C.; de Mendoza, D.; Konings, W.N.; Lolkema, J.S. Mechanism of Citrate Metabolism in Lactococcus lactis: Resistance against Lactate Toxicity at Low pH. J. Bacteriol. 1999, 181, 1451–1457. [Google Scholar] [CrossRef]
- Laëtitia, G.; Pascal, D.; Yann, D. The Citrate Metabolism in Homo- and Heterofermentative LAB: A Selective Means of Becoming Dominant over Other Microorganisms in Complex Ecosystems. Food Nutr. Sci. 2014, 5, 953–969. [Google Scholar] [CrossRef]
- Monnet, C.; Aymes, F.; Corrieu, G. Diacetyl and α-Acetolactate Overproduction by Lactococcus lactis subsp. lactis biovar diacetylactis Mutants That Are Deficient in α-Acetolactate Decarboxylase and Have a Low Lactate Dehydrogenase Activity. Appl. Environ. Microbiol. 2000, 66, 5518–5520. [Google Scholar] [CrossRef]
- Shin, J.M.; Gwak, J.W.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Biomedical Applications of Nisin. J. Appl. Microbiol. 2016, 120, 1449–1465. [Google Scholar] [CrossRef] [PubMed]
- Kajwadkar, R.; Shin, J.M.; Lin, G.-H.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. High-Purity Nisin Alone or in Combination with Sodium Hypochlorite Is Effective against Planktonic and Biofilm Populations of Enterococcus faecalis. J. Endod. 2017, 43, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Setiarto, R.H.B.; Anshory, L.; Wardana, A.A. Biosynthesis of Nisin, Antimicrobial Mechanism and Its Applications as a Food Preservation: A Review. IOP Conf. Ser. Earth Environ. Sci. 2023, 1169, 012105. [Google Scholar] [CrossRef]
- Hassan, H.; St-Gelais, D.; Gomaa, A.; Fliss, I. Impact of Nisin and Nisin-Producing Lactococcus lactis ssp. lactis on Clostridium tyrobutyricum and Bacterial Ecosystem of Cheese Matrices. Foods 2021, 10, 898. [Google Scholar] [CrossRef]
- Senan, S.; El-aal, H.A.; Dave, R.; Hassan, A. Production and Stability of Nisin in Whey Protein Concentrate. LWT-Food Sci. Technol. 2016, 71, 125–129. [Google Scholar] [CrossRef]
- EFSA Panel on Food Additives and Nutrient Sources added to Food (ANS); Younes, M.; Aggett, P.; Aguilar, F.; Crebelli, R.; Dusemund, B.; Filipič, M.; Frutos, M.J.; Galtier, P.; Gundert-Remy, U.; et al. Safety of Nisin (E 234) as a Food Additive in the Light of New Toxicological Data and the Proposed Extension of Use. EFSA J. 2017, 15, e05063. [Google Scholar] [CrossRef]
- Novickij, V.; Stanevičienė, R.; Staigvila, G.; Gruškienė, R.; Sereikaitė, J.; Girkontaitė, I.; Novickij, J.; Servienė, E. Effects of Pulsed Electric Fields and Mild Thermal Treatment on Antimicrobial Efficacy of Nisin-Loaded Pectin Nanoparticles for Food Preservation. LWT 2020, 120, 108915. [Google Scholar] [CrossRef]
- Gharsallaoui, A.; Oulahal, N.; Joly, C.; Degraeve, P. Nisin as a Food Preservative: Part 1: Physicochemical Properties, Antimicrobial Activity, and Main Uses. Crit. Rev. Food Sci. Nutr. 2016, 56, 1262–1274. [Google Scholar] [CrossRef]
- Belfiore, C.; Castellano, P.; Vignolo, G. Reduction of Escherichia Coli Population Following Treatment with Bacteriocins from Lactic Acid Bacteria and Chelators. Food Microbiol. 2007, 24, 223–229. [Google Scholar] [CrossRef]
- Delves-Broughton, J. 16-Use of the Natural Food Preservatives, Nisin and Natamycin, to Reduce Detrimental Thermal Impact on Product Quality. In In-Pack Processed Foods; Richardson, P., Ed.; Woodhead Publishing Series in Food Science, Technology and Nutrition; Woodhead Publishing: Thorston, UK, 2008; pp. 319–337. ISBN 978-1-84569-246-9. [Google Scholar]
- Junior, B.R.D.C.L.; Tribst, A.A.L. Use of Nisin and Bioprotective Lactic Cultures to Extend the Shelf Life of Sheep and Goat Cheese Whey. Food Biosci. 2022, 50, 102096. [Google Scholar] [CrossRef]
- Popa, E.E.; Miteluț, A.C.; Râpă, M.; Popescu, P.A.; Drăghici, M.C.; Geicu-Cristea, M.; Popa, M.E. Antimicrobial Active Packaging Containing Nisin for Preservation of Products of Animal Origin: An Overview. Foods 2022, 11, 3820. [Google Scholar] [CrossRef] [PubMed]
- Ibarra-Sánchez, L.A.; El-Haddad, N.; Mahmoud, D.; Miller, M.J.; Karam, L. Invited Review: Advances in Nisin Use for Preservation of Dairy Products. J. Dairy Sci. 2020, 103, 2041–2052. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, N.; Otani, T.; Inai, T. Nisin, a Food Preservative Produced by Lactococcus lactis, Affects the Localization Pattern of Intermediate Filament Protein in HaCaT Cells. Anat. Sci. Int. 2019, 94, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Smaoui, S.; Echegaray, N.; Kumar, M.; Chaari, M.; D’Amore, T.; Shariati, M.A.; Rebezov, M.; Lorenzo, J.M. Beyond Conventional Meat Preservation: Saddling the Control of Bacteriocin and Lactic Acid Bacteria for Clean Label and Functional Meat Products. Appl. Biochem. Biotechnol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Santiesteban-López, N.A.; Gómez-Salazar, J.A.; Santos, E.M.; Campagnol, P.C.B.; Teixeira, A.; Lorenzo, J.M.; Sosa-Morales, M.E.; Domínguez, R. Natural Antimicrobials: A Clean Label Strategy to Improve the Shelf Life and Safety of Reformulated Meat Products. Foods 2022, 11, 2613. [Google Scholar] [CrossRef] [PubMed]
- Abbasiliasi, S.; Shun Tan, J.; Ibrahim, T.A.T.; Bashokouh, F.; Ramanan Ramakrishnan, N.; Mustafa, S.; Ariff, A.B. Fermentation Factors Influencing the Production of Bacteriocins by Lactic Acid Bacteria: A Review. RSC Adv. 2017, 7, 29395–29420. [Google Scholar] [CrossRef]
- Modugno, C.; Loupiac, C.; Bernard, A.; Jossier, A.; Neiers, F.; Perrier-Cornet, J.-M.; Simonin, H. Effect of High Pressure on the Antimicrobial Activity and Secondary Structure of the Bacteriocin Nisin. Innov. Food Sci. Emerg. Technol. 2018, 47, 9–15. [Google Scholar] [CrossRef]
- Jung, D.-S.; Bodyfelt, F.W.; Daeschel, M.A. Influence of Fat and Emulsifiers on the Efficacy of Nisin in Inhibiting Listeria monocytogenes in Fluid Milk. J. Dairy Sci. 1992, 75, 387–393. [Google Scholar] [CrossRef]
- Chen, R.; Skeens, J.W.; Wiedmann, M.; Guariglia-Oropeza, V. The Efficacy of Nisin against Listeria monocytogenes on Cold-Smoked Salmon at Natural Contamination Levels Is Concentration-Dependent and Varies by Serotype. Front. Microbiol. 2022, 13, 930400. [Google Scholar] [CrossRef]
- Soltani, S.; Hammami, R.; Cotter, P.D.; Rebuffat, S.; Said, L.B.; Gaudreau, H.; Bédard, F.; Biron, E.; Drider, D.; Fliss, I. Bacteriocins as a New Generation of Antimicrobials: Toxicity Aspects and Regulations. FEMS Microbiol. Rev. 2021, 45, fuaa039. [Google Scholar] [CrossRef]
- Zapico, P.; de Paz, M.; Medina, M.; Nuñez, M. The Effect of Homogenization of Whole Milk, Skim Milk and Milk Fat on Nisin Activity against Listeria Innocua. Int. J. Food Microbiol. 1999, 46, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ramos, A.; Madi-Moussa, D.; Coucheney, F.; Drider, D. Current Knowledge of the Mode of Action and Immunity Mechanisms of LAB-Bacteriocins. Microorganisms 2021, 9, 2107. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Hao, L.; Li, J.; Yi, J.; Kang, Q.; Liu, X.; Lu, L.; Lu, J. An Innovative Method to Enhance Protease Tolerance of Nisin in Endogenous Proteases. J. Dairy Sci. 2020, 103, 3038–3044. [Google Scholar] [CrossRef] [PubMed]
- Aasen, I.M.; Markussen, S.; Møretrø, T.; Katla, T.; Axelsson, L.; Naterstad, K. Interactions of the Bacteriocins Sakacin P and Nisin with Food Constituents. Int. J. Food Microbiol. 2003, 87, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zang, M.; Wang, S.; Zhao, B.; Bai, J.; Xu, C.; Shi, Y.; Qiao, X. Nisin: From a Structural and Meat Preservation Perspective. Food Microbiol. 2023, 111, 104207. [Google Scholar] [CrossRef]
- Ucar, Y.; Ozogul, Y.; Durmuş, M.; Ozogul, F. The Effects of Nisin on the Growth of Foodborne Pathogens and Biogenic Amine Formation: In Vivo and in Vitro Studies. Food Biosci. 2021, 43, 101266. [Google Scholar] [CrossRef]
- Felicio, B.A.; Pinto, M.S.; Oliveira, F.S.; Lempk, M.W.; Pires, A.C.S.; Lelis, C.A. Effects of Nisin on Staphylococcus aureus Count and Physicochemical Properties of Minas Frescal Cheese. J. Dairy Sci. 2015, 98, 4364–4369. [Google Scholar] [CrossRef]
- Oppegård, C.; Rogne, P.; Kristiansen, P.E.; Nissen-Meyer, J. Structure Analysis of the Two-Peptide Bacteriocin Lactococcin G by Introducing d-Amino Acid Residues. Microbiology 2010, 156, 1883–1889. [Google Scholar] [CrossRef]
- Moll, G.; Hildeng-Hauge, H.; Nissen-Meyer, J.; Nes, I.F.; Konings, W.N.; Driessen, A.J. Mechanistic Properties of the Two-Component Bacteriocin Lactococcin G. J. Bacteriol. 1998, 180, 96–99. [Google Scholar] [CrossRef]
- Moll, G.; Ubbink-Kok, T.; Hildeng-Hauge, H.; Nissen-Meyer, J.; Nes, I.F.; Konings, W.N.; Driessen, A.J. Lactococcin G Is a Potassium Ion-Conducting, Two-Component Bacteriocin. J. Bacteriol. 1996, 178, 600–605. [Google Scholar] [CrossRef]
- Zendo, T.; Koga, S.; Shigeri, Y.; Nakayama, J.; Sonomoto, K. Lactococcin Q, a Novel Two-Peptide Bacteriocin Produced by Lactococcus lactis QU 4. Appl. Environ. Microbiol. 2006, 72, 3383–3389. [Google Scholar] [CrossRef] [PubMed]
- Rendón-Rosales, M.Á.; Torres-Llanez, M.J.; Mazorra-Manzano, M.A.; González-Córdova, A.F.; Hernández-Mendoza, A.; Vallejo-Cordoba, B. In Vitro and in Silico Evaluation of Multifunctional Properties of Bioactive Synthetic Peptides Identified in Milk Fermented with Lactococcus lactis NRRL B-50571 and NRRL B-50572. LWT 2022, 154, 112581. [Google Scholar] [CrossRef]
- Guha, S.; Sharma, H.; Deshwal, G.K.; Rao, P.S. A Comprehensive Review on Bioactive Peptides Derived from Milk and Milk Products of Minor Dairy Species. Food Prod. Process. Nutr. 2021, 3, 2. [Google Scholar] [CrossRef]
- Raveschot, C.; Cudennec, B.; Coutte, F.; Flahaut, C.; Fremont, M.; Drider, D.; Dhulster, P. Production of Bioactive Peptides by Lactobacillus Species: From Gene to Application. Front. Microbiol. 2018, 9, 2354. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, D.; Masoodi, F.A.; Wani, S.M.; Mir, S.A. Bioactive Peptides from Fermented Foods and Their Relevance in COVID-19 Mitigation. Food Prod. Process. Nutr. 2023, 5, 53. [Google Scholar] [CrossRef]
- Huang, C.; Kok, J. Editing of the Proteolytic System of Lactococcus lactis Increases Its Bioactive Potential. Appl. Environ. Microbiol. 2020, 86, e01319-20. [Google Scholar] [CrossRef] [PubMed]
- Forler, B.; Horstmann, G.; Schäfer, J.; Michel, C.; Weiss, A.; Stressler, T.; Fischer, L.; Hinrichs, J.; Schmidt, H. Effects of Protein, Calcium, and pH on Gene Transcription, Cell-Envelope Peptidase Activity of Lactococcus lactis Strains, and the Formation of Bitter Peptides. Foods 2021, 10, 1588. [Google Scholar] [CrossRef]
- Guédon, E.; Renault, P.; Ehrlich, S.D.; Delorme, C. Transcriptional Pattern of Genes Coding for the Proteolytic System of Lactococcus lactis and Evidence for Coordinated Regulation of Key Enzymes by Peptide Supply. J. Bacteriol. 2001, 183, 3614–3622. [Google Scholar] [CrossRef]
- den Hengst, C.D.; van Hijum, S.A.F.T.; Geurts, J.M.W.; Nauta, A.; Kok, J.; Kuipers, O.P. The Lactococcus lactis CodY Regulon: Identification of a conserved Cis-Regulatory Element*[Boxs]. J. Biol. Chem. 2005, 280, 34332–34342. [Google Scholar] [CrossRef]
- Bounouala, F.Z.; Roudj, S.; Karam, N.-E.; Recio, I.; Miralles, B. Casein Hydrolysates by Lactobacillus Brevis and Lactococcus Lactis Proteases: Peptide Profile Discriminates Strain-Dependent Enzyme Specificity. J. Agric. Food Chem. 2017, 65, 9324–9332. [Google Scholar] [CrossRef]
- Han, R.; Maycock, J.; Murray, B.S.; Boesch, C. Identification of Angiotensin Converting Enzyme and Dipeptidyl Peptidase-IV Inhibitory Peptides Derived from Oilseed Proteins Using Two Integrated Bioinformatic Approaches. Food Res. Int. 2019, 115, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Feng, L.; Wang, Z.; Qiao, M.; Shahidi, F.; Lu, W.; Du, M. Sequence Analysis and Molecular Docking of Antithrombotic Peptides from Casein Hydrolysate by Trypsin Digestion. J. Funct. Foods 2017, 32, 313–323. [Google Scholar] [CrossRef]
- Figueroa-Hernández, C.; Cruz-Guerrero, A.; Rodríguez-Serrano, G.; Gómez-Ruiz, L.; García-Garibay, M.; Jiménez-Guzmán, J. Calcium and Iron Binding Peptides Production by Lactococcus lactis subsp. cremoris NCFB 712. Rev. Mex. Ing. Química 2012, 11, 259–267. [Google Scholar]
- Dimitrov, Z. Characterization of Bioactive Peptides with Calcium-Binding Activity Released by Specially Designed Cheese Starter. Biotechnol. Biotechnol. Equip. 2009, 23, 927–930. [Google Scholar] [CrossRef]
- Raw Milk Kefir: Microbiota, Bioactive Peptides, and Immune Modulation. Food Funct. 2023, 14, 1648–1661. [CrossRef] [PubMed]
- Gobbetti, M.; Ferranti, P.; Smacchi, E.; Goffredi, F.; Addeo, F. Production of Angiotensin-I-Converting-Enzyme-Inhibitory Peptides in Fermented Milks Started by Lactobacillus delbrueckii subsp. bulgaricus SS1 and Lactococcus lactis subsp. cremoris FT4. Appl. Environ. Microbiol. 2000, 66, 3898–3904. [Google Scholar] [CrossRef] [PubMed]
- Gagnaire, V.; Jardin, J.; Jan, G.; Lortal, S. Invited Review: Proteomics of Milk and Bacteria Used in Fermented Dairy Products: From Qualitative to Quantitative Advances. J. Dairy Sci. 2009, 92, 811–825. [Google Scholar] [CrossRef]
- Sedaghati, M.; Ezzatpanah, H.; Boojar, M.M.A.; Ebrahimi, M.T.; Kobarfard, F. Isolation and Identification of Some Antibacterial Peptides in the Plasmin-Digest of β-Casein. LWT-Food Sci. Technol. 2016, 68, 217–225. [Google Scholar] [CrossRef]
- Choi, J.; Sabikhi, L.; Hassan, A.; Anand, S. Bioactive Peptides in Dairy Products. Int. J. Dairy Technol. 2012, 65, 1–12. [Google Scholar] [CrossRef]
- Rafiq, S.; Gulzar, N.; Sameen, A.; Huma, N.; Hayat, I.; Ijaz, R. Functional Role of Bioactive Peptides with Special Reference to Cheeses. Int. J. Dairy Technol. 2021, 74, 1–16. [Google Scholar] [CrossRef]
- Rafiq, S.; Huma, N.; Pasha, I.; Shahid, M.; Xiao, H. Angiotensin-converting Enzyme-inhibitory and Antithrombotic Activities of Soluble Peptide Extracts from Buffalo and Cow Milk Cheddar Cheeses. Int. J. Dairy Technol. 2017, 70, 380–388. [Google Scholar] [CrossRef]
- Kim, H.D.; Lee, H.J.; Shin, Z.I.; Nam, H.S.; Woo, H.J. Anticancer Effects of Hydrophobic Peptides Derived from a Cheese Slurry. Food Sci. Biotechnol. 1995, 4, 268–272. [Google Scholar]
- Santos, M.I.; Lima, A.; Mota, J.; Rebelo, P.; Ferreira, R.B.; Pedroso, L.; Ferreira, M.A.; Sousa, I. Extended Cheese Whey Fermentation Produces a Novel Casein-Derived Antibacterial Polypeptide That Also Inhibits Gelatinases MMP-2 and MMP-9. Int. J. Mol. Sci. 2021, 22, 11130. [Google Scholar] [CrossRef] [PubMed]
- González-González, F.; Delgado, S.; Ruiz, L.; Margolles, A.; Ruas-Madiedo, P. Functional Bacterial Cultures for Dairy Applications: Towards Improving Safety, Quality, Nutritional and Health Benefit Aspects. J. Appl. Microbiol. 2022, 133, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Acevedo-Rocha, C.G.; Gronenberg, L.S.; Mack, M.; Commichau, F.M.; Genee, H.J. Microbial Cell Factories for the Sustainable Manufacturing of B Vitamins. Curr. Opin. Biotechnol. 2019, 56, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.; Wang, S.; Guan, J.; Shi, J.; Evivie, S.E.; Yan, F.; Li, N.; Chen, J.; Li, B.; Huo, G. Milk Fermented with Lactococcus lactis KLDS4.0325 Alleviates Folate Status in Deficient Mice. Food Funct. 2020, 11, 4571–4581. [Google Scholar] [CrossRef] [PubMed]
- Sybesma, W.; Starrenburg, M.; Kleerebezem, M.; Mierau, I.; de Vos, W.M.; Hugenholtz, J. Increased Production of Folate by Metabolic Engineering of Lactococcus lactis. Appl. Environ. Microbiol. 2003, 69, 3069–3076. [Google Scholar] [CrossRef] [PubMed]
- Gangadharan, D.; Nampoothiri, K.M. Folate Production Using Lactococcus Lactis Ssp Cremoris with Implications for Fortification of Skim Milk and Fruit Juices. LWT-Food Sci. Technol. 2011, 44, 1859–1864. [Google Scholar] [CrossRef]
- Divya, J.B.; Nampoothiri, K.M. Folate Fortification of Skim Milk by a Probiotic Lactococcus lactis CM28 and Evaluation of Its Stability in Fermented Milk on Cold Storage. J. Food Sci. Technol. 2015, 52, 3513–3519. [Google Scholar] [CrossRef][Green Version]
- Burgess, C.M.; Slotboom, D.J.; Geertsma, E.R.; Duurkens, R.H.; Poolman, B.; van Sinderen, D. The Riboflavin Transporter RibU in Lactococcus lactis: Molecular Characterization of Gene Expression and the Transport Mechanism. J. Bacteriol. 2006, 188, 2752–2760. [Google Scholar] [CrossRef]
- Shulpekova, Y.; Nechaev, V.; Kardasheva, S.; Sedova, A.; Kurbatova, A.; Bueverova, E.; Kopylov, A.; Malsagova, K.; Dlamini, J.C.; Ivashkin, V. The Concept of Folic Acid in Health and Disease. Molecules 2021, 26, 3731. [Google Scholar] [CrossRef]
- Albu, C.-C.; Bencze, M.-A.; Dragomirescu, A.-O.; Suciu, I.; Tănase, M.; Albu, Ş.-D.; Russu, E.-A.; Ionescu, E. Folic Acid and Its Role in Oral Health: A Narrative Review. Processes 2023, 11, 1994. [Google Scholar] [CrossRef]
- Pacheco Da Silva, F.F.; Biscola, V.; LeBlanc, J.G.; Gombossy de Melo Franco, B.D. Effect of Indigenous Lactic Acid Bacteria Isolated from Goat Milk and Cheeses on Folate and Riboflavin Content of Fermented Goat Milk. LWT-Food Sci. Technol. 2016, 71, 155–161. [Google Scholar] [CrossRef]
- Mahabadi, N.; Bhusal, A.; Banks, S.W. Riboflavin Deficiency. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Hariri, E.; Kassis, N.; Iskandar, J.-P.; Schurgers, L.J.; Saad, A.; Abdelfattah, O.; Bansal, A.; Isogai, T.; Harb, S.C.; Kapadia, S. Vitamin K2-a Neglected Player in Cardiovascular Health: A Narrative Review. Open Heart 2021, 8, e001715. [Google Scholar] [CrossRef]
- Saubade, F.; Hemery, Y.M.; Guyot, J.-P.; Humblot, C. Lactic Acid Fermentation as a Tool for Increasing the Folate Content of Foods. Crit. Rev. Food Sci. Nutr. 2017, 57, 3894–3910. [Google Scholar] [CrossRef]
- Mahara, F.A.; Nuraida, L.; Lioe, H. Fermentation of Milk Using Folate-Producing Lactic Acid Bacteria to Increase Natural Folate Content: A Review. J. Apple Biotechnol. Rep. 2019, 6, 129–136. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Laiño, J.E.; del Valle, M.J.; Vannini, V.; van Sinderen, D.; Taranto, M.P.; de Valdez, G.F.; de Giori, G.S.; Sesma, F. B-Group Vitamin Production by Lactic Acid Bacteria—Current Knowledge and Potential Applications. J. Appl. Microbiol. 2011, 111, 1297–1309. [Google Scholar] [CrossRef]
- Averianova, L.A.; Balabanova, L.A.; Son, O.M.; Podvolotskaya, A.B.; Tekutyeva, L.A. Production of Vitamin B2 (Riboflavin) by Microorganisms: An Overview. Front. Bioeng. Biotechnol. 2020, 8, 1172. [Google Scholar] [CrossRef]
- Gu, Q.; Li, P.; Gu, Q.; Li, P. Biosynthesis of Vitamins by Probiotic Bacteria. In Probiotics and Prebiotics in Human Nutrition and Health; IntechOpen: London, UK, 2016; ISBN 978-953-51-2476-4. [Google Scholar]
- Nayik, G.A.; Jagdale, Y.D.; Gaikwad, S.A.; Devkatte, A.N.; Dar, A.H.; Ansari, M.J. Nutritional Profile, Processing and Potential Products: A Comparative Review of Goat Milk. Dairy 2022, 3, 44. [Google Scholar] [CrossRef]
- Manoury, E.; Jourdon, K.; Boyaval, P.; Fourcassié, P. Quantitative Measurement of Vitamin K2 (Menaquinones) in Various Fermented Dairy Products Using a Reliable High-Performance Liquid Chromatography Method. J. Dairy Sci. 2013, 96, 1335–1346. [Google Scholar] [CrossRef]
- Vermeer, C.; Raes, J.; Van’t Hoofd, C.; Knapen, M.H.; Xanthoulea, S. Menaquinone Content of Cheese. Nutrients 2018, 10, 446. [Google Scholar] [CrossRef]
- Walther, B.; Guggisberg, D.; Schmidt, R.S.; Portmann, R.; Risse, M.-C.; Badertscher, R.; Chollet, M. Quantitative Analysis of Menaquinones (Vitamin K2) in Various Types of Cheese from Switzerland. Int. Dairy J. 2021, 112, 104853. [Google Scholar] [CrossRef]
- Walther, B.; Karl, J.P.; Booth, S.L.; Boyaval, P. Menaquinones, Bacteria, and the Food Supply: The Relevance of Dairy and Fermented Food Products to Vitamin K Requirements. Adv. Nutr. 2013, 4, 463–473. [Google Scholar] [CrossRef]
- Homayouni Rad, A.; Yari Khosroushahi, A.; Khalili, M.; Jafarzadeh, S. Folate Bio-Fortification of Yoghurt and Fermented Milk: A Review. Dairy Sci. Technol. 2016, 96, 427–441. [Google Scholar] [CrossRef]
- Sybesma, W.; Burgess, C.; Starrenburg, M.; van Sinderen, D.; Hugenholtz, J. Multivitamin Production in Lactococcus lactis Using Metabolic Engineering. Metab. Eng. 2004, 6, 109–115. [Google Scholar] [CrossRef]
- Thakur, K.; Tomar, S.K.; De, S. Lactic Acid Bacteria as a Cell Factory for Riboflavin Production. Microb. Biotechnol. 2016, 9, 441–451. [Google Scholar] [CrossRef]
- Green, J.M.; Matthews, R.G. Folate Biosynthesis, Reduction, and Polyglutamylation and the Interconversion of Folate Derivatives. EcoSal Plus 2007, 2, 10-1128. [Google Scholar] [CrossRef]
- Klaus, S.M.; Wegkamp, A.; Sybesma, W.; Hugenholtz, J.; Gregory, J.F.; Hanson, A.D. A Nudix Enzyme Removes Pyrophosphate from Dihydroneopterin Triphosphate in the Folate Synthesis Pathway of Bacteria and Plants. J. Biol. Chem. 2005, 280, 5274–5280. [Google Scholar] [CrossRef]
- Strandler, H.S.; Patring, J.; Jägerstad, M.; Jastrebova, J. Challenges in the Determination of Unsubstituted Food Folates: Impact of Stabilities and Conversions on Analytical Results. J. Agric. Food Chem. 2015, 63, 2367–2377. [Google Scholar] [CrossRef]
- Patel, A.; Shah, N.; Prajapati, J.B. Biosynthesis of Vitamins and Enzymes in Fermented Foods by Lactic Acid Bacteria and Related Genera—A Promising Approach. Croat. J. Food Sci. Technol. 2013, 5, 85–91. [Google Scholar]
- Brooijmans, R.; Smit, B.; Santos, F.; van Riel, J.; de Vos, W.M.; Hugenholtz, J. Heme and Menaquinone Induced Electron Transport in Lactic Acid Bacteria. Microb. Cell Factories 2009, 8, 28. [Google Scholar] [CrossRef]
- Vieira, C.P.; Cabral, C.C.; da Costa Lima, B.R.C.; Paschoalin, V.M.F.; Leandro, K.C.; Conte-Junior, C.A. Lactococcus lactis ssp. cremoris MRS47, a Potential Probiotic Strain Isolated from Kefir Grains, Increases Cis-9, Trans-11-CLA and PUFA Contents in Fermented Milk. J. Funct. Foods 2017, 31, 172–178. [Google Scholar] [CrossRef]
- den Hartigh, L.J. Conjugated Linoleic Acid Effects on Cancer, Obesity, and Atherosclerosis: A Review of Pre-Clinical and Human Trials with Current Perspectives. Nutrients 2019, 11, 370. [Google Scholar] [CrossRef]
- Dachev, M.; Bryndová, J.; Jakubek, M.; Moučka, Z.; Urban, M. The Effects of Conjugated Linoleic Acids on Cancer. Processes 2021, 9, 454. [Google Scholar] [CrossRef]
- Nasrollahzadeh, A.; Tavani, S.M.; Arjeh, E.; Jafari, S.M. Production of Conjugated Linoleic Acid by Lactic Acid Bacteria; Important Factors and Optimum Conditions. Food Chem. X 2023, 20, 100942. [Google Scholar] [CrossRef]
- Gong, M.; Hu, Y.; Wei, W.; Jin, Q.; Wang, X. Production of Conjugated Fatty Acids: A Review of Recent Advances. Biotechnol. Adv. 2019, 37, 107454. [Google Scholar] [CrossRef]
- Abd El-Salam, M.H.; el-Shafei, K.; Sharaf, O.M.; Effat, B.A.; Asem, F.M.; el-Aasar, M. Screening of Some Potentially Probiotic Lactic Acid Bacteria for Their Ability to Synthesis Conjugated Linoleic Acid. Int. J. Dairy Technol. 2010, 63, 62–69. [Google Scholar] [CrossRef]
- Renes, E.; Ladero, V.; Tornadijo, M.E.; Fresno, J.M. Production of Sheep Milk Cheese with High γ-Aminobutyric Acid and Ornithine Concentration and with Reduced Biogenic Amines Level Using Autochthonous Lactic Acid Bacteria Strains. Food Microbiol. 2019, 78, 1–10. [Google Scholar] [CrossRef]
- Konkit, M.; Kim, W. Activities of Amylase, Proteinase, and Lipase Enzymes from Lactococcus chungangensis and Its Application in Dairy Products. J. Dairy Sci. 2016, 99, 4999–5007. [Google Scholar] [CrossRef]
- Farkye, N.Y. Cheese Technology. Int. J. Dairy Technol. 2004, 57, 91–98. [Google Scholar] [CrossRef]
- Demarigny, Y. LACTOCOCCUS|Lactococcus lactis subspecies lactis and cremoris. In Encyclopedia of Food Microbiology, 2nd ed.; Batt, C.A., Tortorello, M.L., Eds.; Academic Press: Oxford, UK, 2014; pp. 442–446. ISBN 978-0-12-384733-1. [Google Scholar]
- Rani, S.; Jagtap, S. Acceleration of Swiss Cheese Ripening by Microbial Lipase without Affecting Its Quality Characteristics. J. Food Sci. Technol. 2019, 56, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Yvon, M.; Thirouin, S.; Rijnen, L.; Fromentier, D.; Gripon, J.C. An Aminotransferase from Lactococcus lactis Initiates Conversion of Amino Acids to Cheese Flavor Compounds. Appl. Environ. Microbiol. 1997, 63, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, C.; Irmler, S.; Bisig, W.; Guggisberg, D.; Roetschi, A.; Portmann, R.; Wechsler, D.; Fröhlich-Wyder, M.-T. The Effect of Starters with a Functional Arginine Deiminase Pathway on Cheese Ripening and Quality. Int. Dairy J. 2018, 85, 191–200. [Google Scholar] [CrossRef]
- van Mastrigt, O.; Mager, E.E.; Jamin, C.; Abee, T.; Smid, E.J. Citrate, Low pH and Amino Acid Limitation Induce Citrate Utilization in Lactococcus lactis biovar diacetylactis. Microb. Biotechnol. 2018, 11, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Medina, R.B.; Katz, M.B.; González, S.; Oliver, G. Determination of Esterolytic and Lipolytic Activities of Lactic Acid Bacteria. Public Health Microbiol. Methods Protoc. 2004, 268, 465–470. [Google Scholar]
- Harper, A.R.; Dobson, R.C.J.; Morris, V.K.; Moggré, G. Fermentation of Plant-based Dairy Alternatives by Lactic Acid Bacteria. Microb. Biotechnol. 2022, 15, 1404–1421. [Google Scholar] [CrossRef]
- Mardis, E.R. The Impact of Next-Generation Sequencing Technology on Genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar] [CrossRef]
- Gautam, S.S.; KC, R.; Leong, K.W.; Aogáin, M.M.; O’Toole, R.F. A Step-by-Step Beginner. J. Biol. Methods 2019, 6, e110. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Bortolaia, V.; Tate, H.; Tyson, G.H.; Aarestrup, F.M.; McDermott, P.F. Using Genomics to Track Global Antimicrobial Resistance. Front. Public Health 2019, 7, 242. [Google Scholar] [CrossRef]
- Lal Gupta, C.; Kumar Tiwari, R.; Cytryn, E. Platforms for Elucidating Antibiotic Resistance in Single Genomes and Complex Metagenomes. Environ. Int. 2020, 138, 105667. [Google Scholar] [CrossRef]
- Seoane, A.; Bou, G. Bioinformatics Approaches to the Study of Antimicrobial Resistance. Rev. Esp. Quim. 2021, 34, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Tóth, A.G.; Csabai, I.; Judge, M.F.; Maróti, G.; Becsei, Á.; Spisák, S.; Solymosi, N. Mobile Antimicrobial Resistance Genes in Probiotics. Antibiotics 2021, 10, 1287. [Google Scholar] [CrossRef] [PubMed]
- Fatahi-Bafghi, M.; Naseri, S.; Alizehi, A. Genome Analysis of Probiotic Bacteria for Antibiotic Resistance Genes. Antonie Van Leeuwenhoek 2022, 115, 375–389. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. ABRicate. 2023. Available online: https://bio.tools/ABRicate (accessed on 12 November 2023).
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool To Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Bonin, N.; Doster, E.; Worley, H.; Pinnell, L.J.; Bravo, J.E.; Ferm, P.; Marini, S.; Prosperi, M.; Noyes, N.; Morley, P.S.; et al. MEGARes and AMR++, v3.0: An Updated Comprehensive Database of Antimicrobial Resistance Determinants and an Improved Software Pipeline for Classification Using High-Throughput Sequencing. Nucleic Acids Res. 2022, 51, D744–D752. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2022, 51, D690–D699. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic Gene Recognition and Translation Initiation Site Identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Tenea, G.N. Metabiotics Signature through Genome Sequencing and In Vitro Inhibitory Assessment of a Novel Lactococcus lactis Strain UTNCys6-1 Isolated from Amazonian Camu-Camu Fruits. Int. J. Mol. Sci. 2023, 24, 6127. [Google Scholar] [CrossRef] [PubMed]
- Wissel, E.F.; Talbot, B.M.; Toyosato, N.A.B.; Petit, R.A.; Hertzberg, V.; Dunlop, A.; Read, T.D. hAMRoaster: A Tool for Comparing Performance of AMR Gene Detection Software. bioRxiv 2023. bioRxiv:2022.01.13.476279. [Google Scholar]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for Taxonomy-Based Analysis of Pathways and Genomes. Nucleic Acids Res. 2022, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Altman, T.; Billington, R.; Dreher, K.; Foerster, H.; Fulcher, C.A.; Holland, T.A.; Keseler, I.M.; Kothari, A.; Kubo, A.; et al. The MetaCyc Database of Metabolic Pathways and Enzymes and the BioCyc Collection of Pathway/Genome Databases. Nucleic Acids Res. 2014, 42, D459–D471. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, X.; Hong, R.; Liu, F. Combined Proteomics and Transcriptomics Analysis of Lactococcus lactis under Different Culture Conditions. J. Dairy Sci. 2021, 104, 2564–2580. [Google Scholar] [CrossRef]
- Camacho, C.; Boratyn, G.M.; Joukov, V.; Vera Alvarez, R.; Madden, T.L. ElasticBLAST: Accelerating Sequence Search via Cloud Computing. BMC Bioinform. 2023, 24, 117. [Google Scholar] [CrossRef]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Sun, Y.; Li, H.; Zheng, L.; Li, J.; Hong, Y.; Liang, P.; Kwok, L.-Y.; Zuo, Y.; Zhang, W.; Zhang, H. iProbiotics: A Machine Learning Platform for Rapid Identification of Probiotic Properties from Whole-Genome Primary Sequences. Brief. Bioinform. 2022, 23, bbab477. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Hartleb, D.; Fritzemeier, C.J.; Lercher, M.J. Automated High-Quality Reconstruction of Metabolic Networks from High-Throughput Data. bioRxiv 2018. [Google Scholar] [CrossRef]
- Henry, C.S.; DeJongh, M.; Best, A.A.; Frybarger, P.M.; Linsay, B.; Stevens, R.L. High-Throughput Generation, Optimization and Analysis of Genome-Scale Metabolic Models. Nat. Biotechnol. 2010, 28, 977–982. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, E.; Schulz, C.; Almaas, E. Automated Generation of Genome-Scale Metabolic Draft Reconstructions Based on KEGG. BMC Bioinform. 2018, 19, 467. [Google Scholar] [CrossRef] [PubMed]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Akhter, S.; Miller, J.H. BaPreS: A Software Tool for Predicting Bacteriocins Using an Optimal Set of Features. BMC Bioinform. 2023, 24, 313. [Google Scholar] [CrossRef] [PubMed]
- Hammami, R.; Zouhir, A.; Le Lay, C.; Ben Hamida, J.; Fliss, I. BACTIBASE Second Release: A Database and Tool Platform for Bacteriocin Characterization. BMC Microbiol. 2010, 10, 22. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A More Efficient Report with Usability Improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Eddy, S.R. HMMER User’s Guide. 2023. Available online: http://eddylab.org/software/hmmer/Userguide.pdf (accessed on 12 November 2023).
- Bhagwat, M.; Aravind, L. PSI-BLAST Tutorial. In Comparative Genomics: Volumes 1 and 2; Humana Press: Totowa, NJ, USA, 2007. [Google Scholar]
- Yoon, B.-J. Hidden Markov Models and Their Applications in Biological Sequence Analysis. Curr. Genom. 2009, 10, 402–415. [Google Scholar] [CrossRef]
- Walsh, C.J.; Guinane, C.M.; O’Toole, P.W.; Cotter, P.D. A Profile Hidden Markov Model to Investigate the Distribution and Frequency of LanB-Encoding Lantibiotic Modification Genes in the Human Oral and Gut Microbiome. PeerJ 2017, 5, e3254. [Google Scholar] [CrossRef]
- Golneshin, A.; Gor, M.-C.; Williamson, N.; Vezina, B.; Van, T.T.H.; May, B.K.; Smith, A.T. Discovery and Characterisation of Circular Bacteriocin Plantacyclin B21AG from Lactiplantibacillus plantarum B21. Heliyon 2020, 6, e04715. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, L.L.; Hoffmann, C.; Lacorte, G.A.; de Melo Franco, B.D.G.; Todorov, S.D. Genomic and Functional Characterization of Bacteriocinogenic Lactic Acid Bacteria Isolated from Boza, a Traditional Cereal-Based Beverage. Sci. Rep. 2022, 12, 1460. [Google Scholar] [CrossRef]
- Vezina, B.; Rehm, B.H.A.; Smith, A.T. Bioinformatic Prospecting and Phylogenetic Analysis Reveals 94 Undescribed Circular Bacteriocins and Key Motifs. BMC Microbiol. 2020, 20, 77. [Google Scholar] [CrossRef] [PubMed]
- Ladjouzi, R.; Dussert, E.; Teiar, R.; Belguesmia, Y.; Drider, D. A Review on Enterocin DD14, the Leaderless Two-Peptide Bacteriocin with Multiple Biological Functions and Unusual Transport Pathway. Antibiotics 2023, 12, 1188. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein Structure and Function Prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Krawczyk, P.S.; Lipinski, L.; Dziembowski, A. PlasFlow: Predicting Plasmid Sequences in Metagenomic Data Using Genome Signatures. Nucleic Acids Res. 2018, 46, e35. [Google Scholar] [CrossRef]
- Antipov, D.; Hartwick, N.; Shen, M.; Raiko, M.; Lapidus, A.; Pevzner, P.A. plasmidSPAdes: Assembling Plasmids from Whole Genome Sequencing Data. Bioinformatics 2016, 32, 3380–3387. [Google Scholar] [CrossRef] [PubMed]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Simon Fraser University Research Computing Group; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S. IslandViewer 4: Expanded Prediction of Genomic Islands for Larger-Scale Datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an Update of CRISRFinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Jain, C.; Rodriguez, R.L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-Db: A Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022, 88, e00991-22. [Google Scholar] [CrossRef]
- McGuffie, M.J.; Barrick, J.E. pLannotate: Engineered Plasmid Annotation. Nucleic Acids Res. 2021, 49, W516–W522. [Google Scholar] [CrossRef]
- Stothard, P.; Grant, J.R.; Van Domselaar, G. Visualizing and Comparing Circular Genomes Using the CGView Family of Tools. Brief. Bioinform. 2017, 20, 1576–1582. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).