Improving the Synthesis of Odd-Chain Fatty Acids in the Oleaginous Yeast Yarrowia lipolytica
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Media
2.2. Fermentation Process
2.2.1. Preparation of Stock Cell Culture
2.2.2. Culture Media Composition
2.2.3. Fermentation Conditions
2.2.4. Optimization of the Concentrations of Fatty Acid Precursors
2.3. Lipid Extraction and Fatty Acid Analysis
2.3.1. Lipid Extraction from Y. lipolytica Biomass
2.3.2. Preparation and Analysis of Fatty Acid Methyl Esters
2.4. Statistical Analysis
3. Results and Discussion
3.1. YlOLE1-Overexpressing Strain JMY9178
3.2. Impact of Fatty Acid Precursors on Yeast Cell Growth and OCFA Synthesis
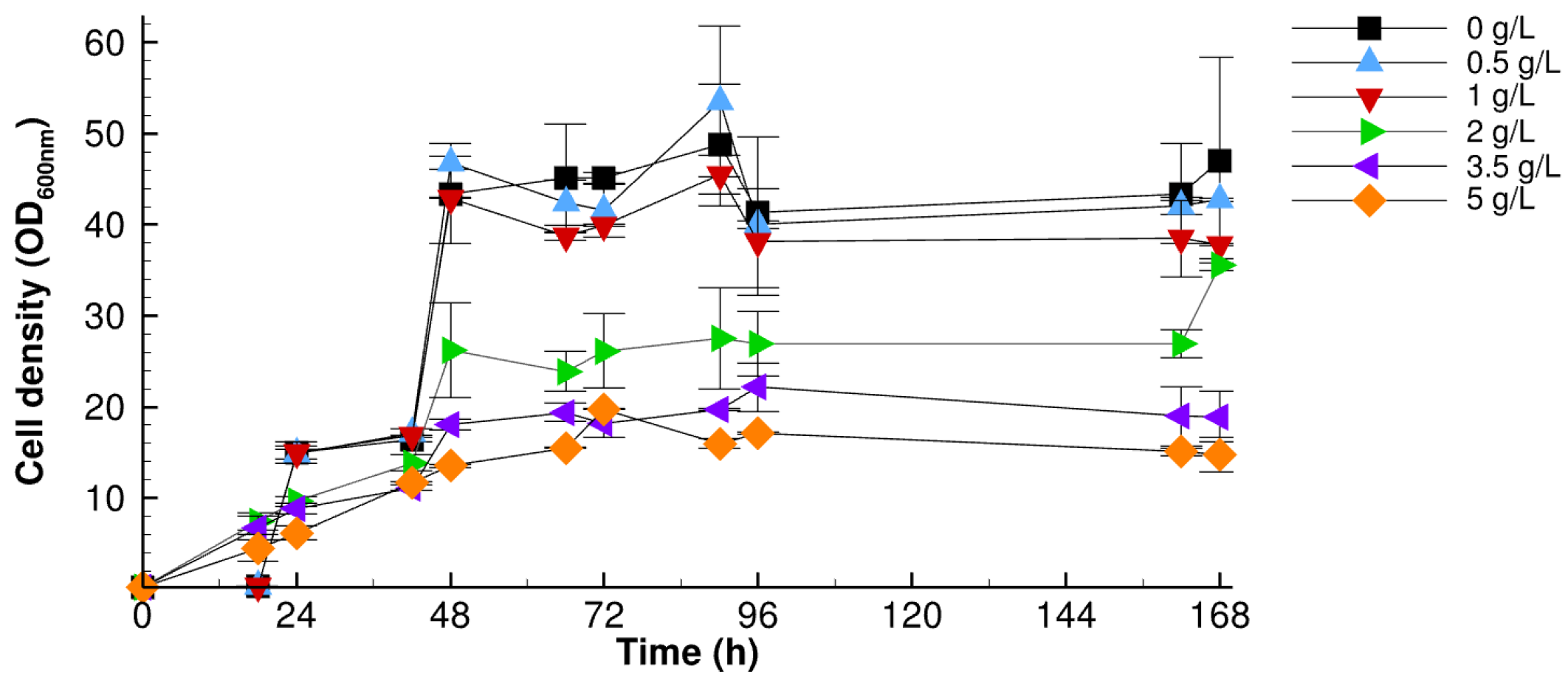
3.2.1. Impact of Sodium Propionate
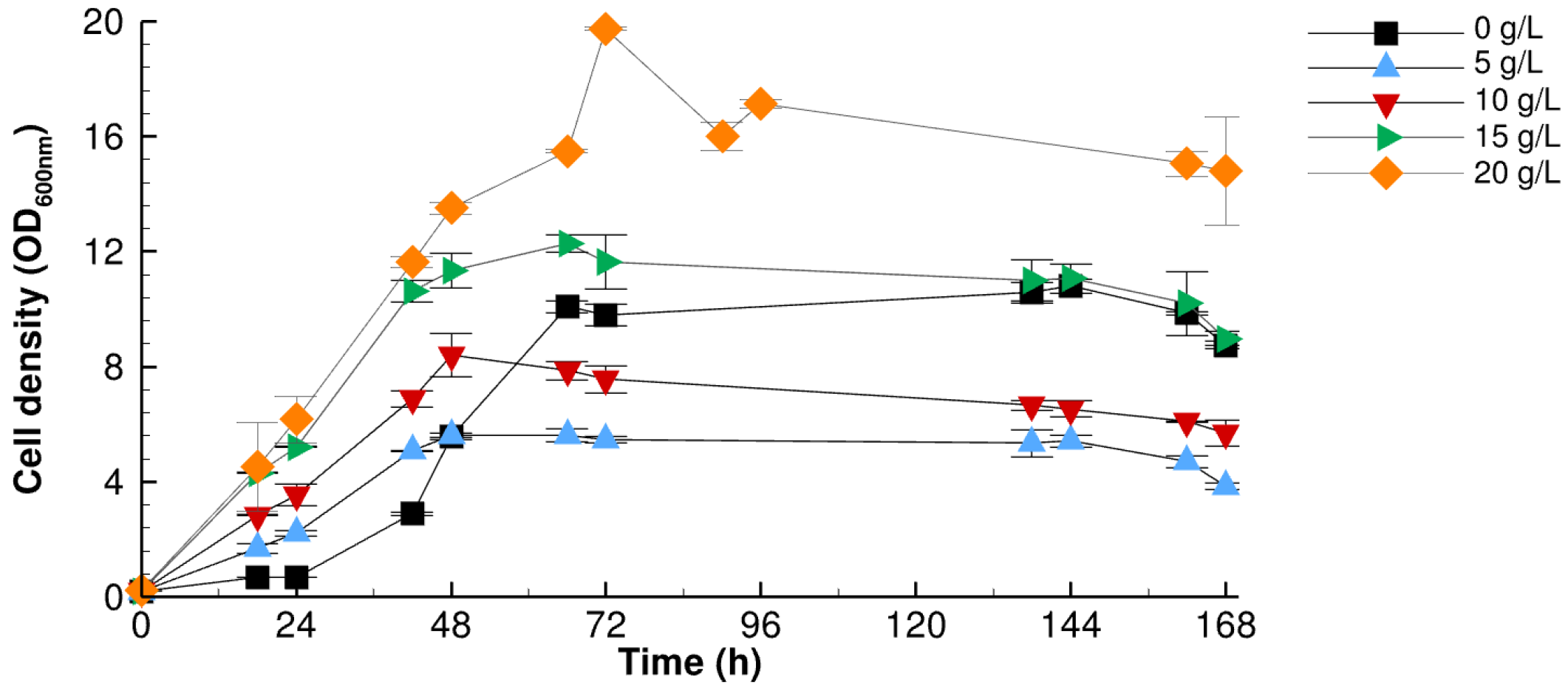
3.2.2. Impact of Sodium Acetate
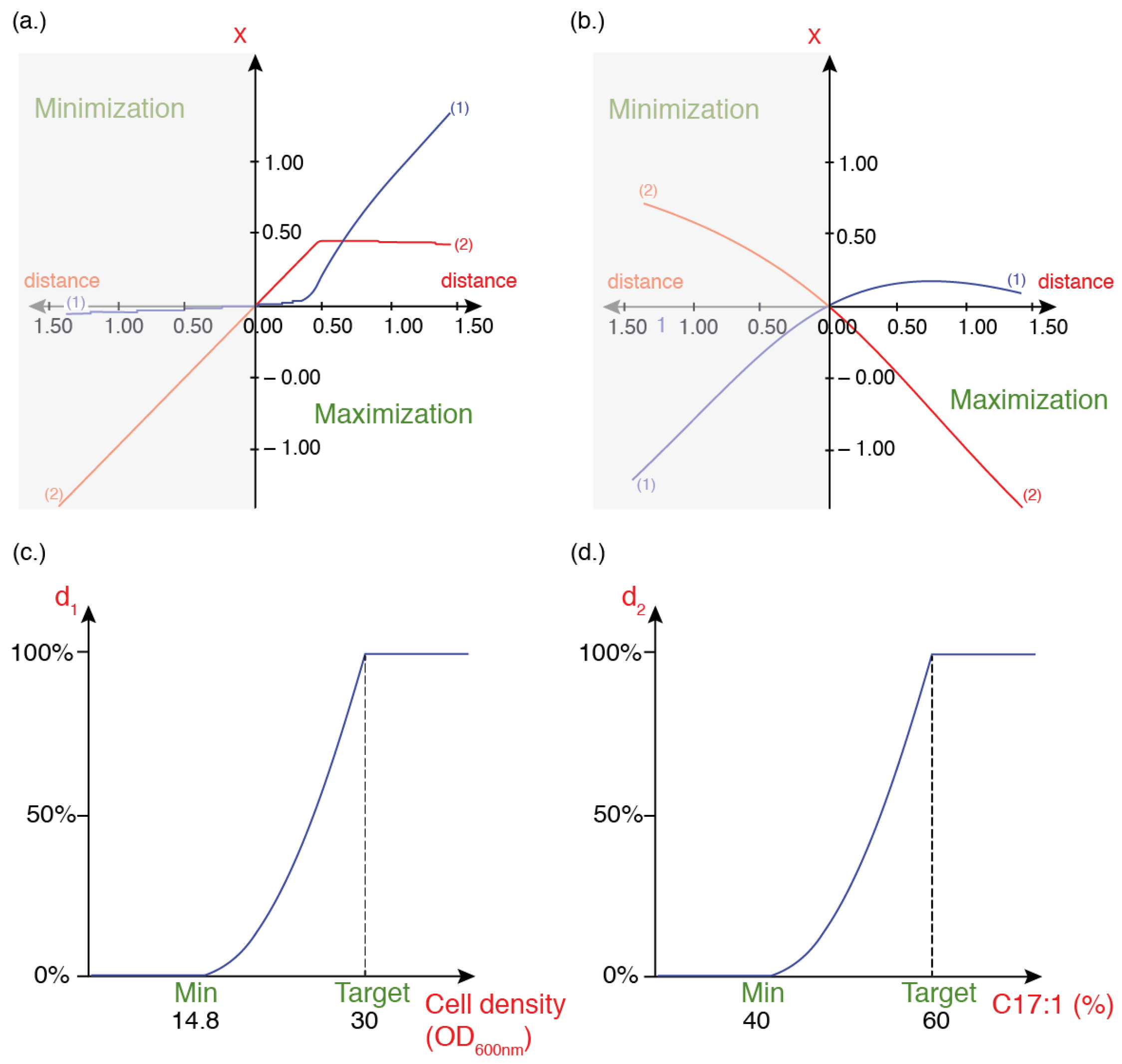
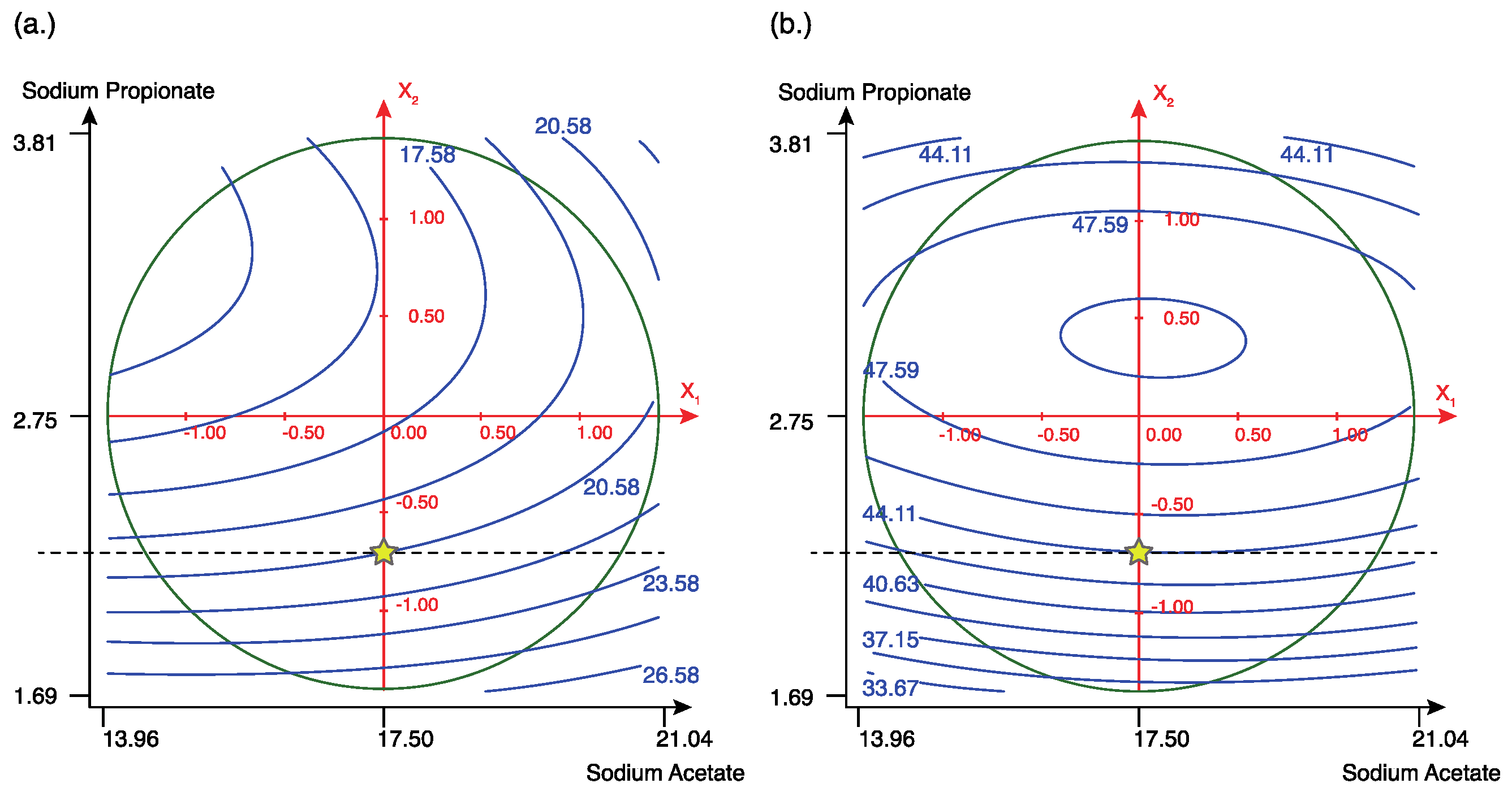
3.3. Optimization of Concentration of FA Precursors
3.3.1. Model’s Validation and Fitting
3.3.2. Optimal Culture Condition Analyses
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El Kantar, S.; Khelfa, A.; Vorobiev, E.; Koubaa, M. Strategies for Increasing Lipid Accumulation and Recovery from Y. Lipolytica: A Review. OCL 2021, 28, 51. [Google Scholar] [CrossRef]
- Ratledge, C. Yeasts, Moulds, Algae and Bacteria as Sources of Lipids. In Technological Advances in Improved and Alternative Sources of Lipids; Kamel, B.S., Kakuda, Y., Eds.; Springer: Boston, MA, USA, 1994; pp. 235–291. ISBN 978-1-4615-2109-9. [Google Scholar]
- Cohen, Z.; Ratledge, C. Single Cell Oils: Microbial and Algal Oils; AOCS Press: Champaign, IL, USA, 2010; ISBN 978-1-893997-73-8. [Google Scholar]
- Ratledge, C. Single Cell Oils for 21st Century. In Single Cell Oils: Microbial and Algal Oils; Cohen, Z., Ratledge, C., Eds.; AOAC Press: Champaign, IL, USA, 2010. [Google Scholar]
- Ratledge, C. Microbial Oils: An Introductory Overview of Current Status and Future Prospects. OCL 2013, 20, D602. [Google Scholar] [CrossRef]
- Fabiszewska, A.U.; Kotyrba, D.; Nowak, D. Assortment of Carbon Sources in Medium for Yarrowia lipolytica Lipase Production: A Statistical Approach. Ann. Microbiol. 2015, 65, 1495–1503. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rywińska, A.; Marcinkiewicz, M.; Cibis, E.; Rymowicz, W. Optimization of Medium Composition for Erythritol Production from Glycerol by Yarrowia lipolytica Using Response Surface Methodology. Prep. Biochem. Biotechnol. 2015, 45, 515–529. [Google Scholar] [CrossRef]
- Ma, Y.; Gao, Z.; Wang, Q.; Liu, Y. Biodiesels from Microbial Oils: Opportunity and Challenges. Bioresour. Technol. 2018, 263, 631–641. [Google Scholar] [CrossRef]
- Ratledge, C. Regulation of Lipid Accumulation in Oleaginous Micro-Organisms. Biochem. Soc. Trans. 2002, 30, 1047–1050. [Google Scholar] [CrossRef]
- Groenewald, M.; Boekhout, T.; Neuvéglise, C.; Gaillardin, C.; van Dijck, P.W.M.; Wyss, M. Yarrowia lipolytica: Safety Assessment of an Oleaginous Yeast with a Great Industrial Potential. Crit. Rev. Microbiol. 2014, 40, 187–206. [Google Scholar] [CrossRef]
- Park, Y.-K.; Ledesma-Amaro, R. What Makes Yarrowia lipolytica Well Suited for Industry? Trends Biotechnol. 2023, 41, 242–254. [Google Scholar] [CrossRef]
- Bao, W.; Li, Z.; Wang, X.; Gao, R.; Zhou, X.; Cheng, S.; Men, Y.; Zheng, L. Approaches to Improve the Lipid Synthesis of Oleaginous Yeast Yarrowia lipolytica: A Review. Renew. Sustain. Energy Rev. 2021, 149, 111386. [Google Scholar] [CrossRef]
- Mitri, S.; Louka, N.; Rossignol, T.; Maroun, R.G.; Koubaa, M. Bioproduction of 2-Phenylethanol by Yarrowia lipolytica on Sugar Beet Molasses as a Low-Cost Substrate. Fermentation 2024, 10, 290. [Google Scholar] [CrossRef]
- Nemer, G.; Louka, N.; Rabiller Blandin, P.; Maroun, R.G.; Vorobiev, E.; Rossignol, T.; Nicaud, J.-M.; Guénin, E.; Koubaa, M. Purification of Natural Pigments Violacein and Deoxyviolacein Produced by Fermentation Using Yarrowia lipolytica. Molecules 2023, 28, 4292. [Google Scholar] [CrossRef] [PubMed]
- Gottardi, D.; Siroli, L.; Vannini, L.; Patrignani, F.; Lanciotti, R. Recovery and Valorization of Agri-Food Wastes and by-Products Using the Non-Conventional Yeast Yarrowia lipolytica. Trends Food Sci. Technol. 2021, 115, 74–86. [Google Scholar] [CrossRef]
- Lazar, Z.; Walczak, E.; Robak, M. Simultaneous Production of Citric Acid and Invertase by Yarrowia lipolytica SUC+ Transformants. Bioresour. Technol. 2011, 102, 6982–6989. [Google Scholar] [CrossRef] [PubMed]
- Celińska, E.; Borkowska, M.; Białas, W. Enhanced Production of Insect Raw-Starch-Digesting Alpha-Amylase Accompanied by High Erythritol Synthesis in Recombinant Yarrowia lipolytica Fed-Batch Cultures at High-Cell-Densities. Process Biochem. 2017, 52, 78–85. [Google Scholar] [CrossRef]
- Rodriguez, G.M.; Hussain, M.S.; Gambill, L.; Gao, D.; Yaguchi, A.; Blenner, M. Engineering Xylose Utilization in Yarrowia lipolytica by Understanding Its Cryptic Xylose Pathway. Biotechnol. Biofuels 2016, 9, 149. [Google Scholar] [CrossRef]
- El Kantar, S.; Koubaa, M. Valorization of Low-Cost Substrates for the Production of Odd Chain Fatty Acids by the Oleaginous Yeast Yarrowia lipolytica. Fermentation 2022, 8, 284. [Google Scholar] [CrossRef]
- Park, Y.-K.; Dulermo, T.; Ledesma-Amaro, R.; Nicaud, J.-M. Optimization of Odd Chain Fatty Acid Production by Yarrowia lipolytica. Biotechnol. Biofuels 2018, 11, 158. [Google Scholar] [CrossRef]
- Žganjar, M.; Ogrizović, M.; Matul, M.; Čadež, N.; Gunde-Cimerman, N.; González-Fernández, C.; Gostinčar, C.; Tomás-Pejó, E.; Petrovič, U. High-Throughput Screening of Non-Conventional Yeasts for Conversion of Organic Waste to Microbial Oils via Carboxylate Platform. Sci. Rep. 2024, 14, 14233. [Google Scholar] [CrossRef]
- Degwert, J.; Jacob, J.; Steckel, F. Use of Cis-9-Heptadecenoic Acid for Treating Psoriasis and Allergies. U.S. Patent WO1994021247A1, 13 January 1998. [Google Scholar]
- Wu, H.; San, K.-Y. Engineering Escherichia Coli for Odd Straight Medium Chain Free Fatty Acid Production. Appl. Microbiol. Biotechnol. 2014, 98, 8145–8154. [Google Scholar] [CrossRef]
- Fontanille, P.; Kumar, V.; Christophe, G.; Nouaille, R.; Larroche, C. Bioconversion of Volatile Fatty Acids into Lipids by the Oleaginous Yeast Yarrowia lipolytica. Bioresour. Technol. 2012, 114, 443–449. [Google Scholar] [CrossRef]
- Park, Y.-K.; Nicaud, J.-M. Screening a Genomic Library for Genes Involved in Propionate Tolerance in Yarrowia lipolytica. Yeast 2020, 37, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Stukey, J.E.; McDonough, V.M.; Martin, C.E. Isolation and Characterization of OLE1, a Gene Affecting Fatty Acid Desaturation from Saccharomyces Cerevisiae. J. Biol. Chem. 1989, 264, 16537–16544. [Google Scholar] [CrossRef] [PubMed]
- Konzock, O.; Matsushita, Y.; Zaghen, S.; Sako, A.; Norbeck, J. Altering the Fatty Acid Profile of Yarrowia lipolytica to Mimic Cocoa Butter by Genetic Engineering of Desaturases. Microb. Cell Factories 2022, 21, 25. [Google Scholar] [CrossRef] [PubMed]
- Tsai, Y.-Y.; Ohashi, T.; Wu, C.-C.; Bataa, D.; Misaki, R.; Limtong, S.; Fujiyama, K. Delta-9 Fatty Acid Desaturase Overexpression Enhanced Lipid Production and Oleic Acid Content in Rhodosporidium Toruloides for Preferable Yeast Lipid Production. J. Biosci. Bioeng. 2019, 127, 430–440. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; CSHL Press: New York, NY, USA, 2001; Volume 1, ISBN 978-0-87969-576-7. [Google Scholar]
- Barth, G.; Gaillardin, C. Physiology and Genetics of the Dimorphic Fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 1997, 19, 219–237. [Google Scholar] [CrossRef]
- Karatay, S.E.; Dönmez, G. Improving the Lipid Accumulation Properties of the Yeast Cells for Biodiesel Production Using Molasses. Bioresour. Technol. 2010, 101, 7988–7990. [Google Scholar] [CrossRef]
- Al Sahyouni, W.; El Kantar, S.; Khelfa, A.; Park, Y.-K.; Nicaud, J.-M.; Louka, N.; Koubaa, M. Optimization of Cis-9-Heptadecenoic Acid Production from the Oleaginous Yeast Yarrowia lipolytica. Fermentation 2022, 8, 245. [Google Scholar] [CrossRef]
- Park, Y.-K.; Bordes, F.; Letisse, F.; Nicaud, J.-M. Engineering Precursor Pools for Increasing Production of Odd-Chain Fatty Acids in Yarrowia lipolytica. Metab. Eng. Commun. 2021, 12, e00158. [Google Scholar] [CrossRef]
- Qiao, K.; Imam Abidi, S.H.; Liu, H.; Zhang, H.; Chakraborty, S.; Watson, N.; Kumaran Ajikumar, P.; Stephanopoulos, G. Engineering Lipid Overproduction in the Oleaginous Yeast Yarrowia lipolytica. Metab. Eng. 2015, 29, 56–65. [Google Scholar] [CrossRef]
- Friedlander, J.; Tsakraklides, V.; Kamineni, A.; Greenhagen, E.H.; Consiglio, A.L.; MacEwen, K.; Crabtree, D.V.; Afshar, J.; Nugent, R.L.; Hamilton, M.A.; et al. Engineering of a High Lipid Producing Yarrowia lipolytica Strain. Biotechnol. Biofuels 2016, 9, 77. [Google Scholar] [CrossRef]
- Wang, F.; Bi, Y.; Diao, J.; Lv, M.; Cui, J.; Chen, L.; Zhang, W. Metabolic Engineering to Enhance Biosynthesis of Both Docosahexaenoic Acid and Odd-Chain Fatty Acids in Schizochytrium Sp. S31. Biotechnol. Biofuels 2019, 12, 141. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Li, L.; Wang, Z.; Shi, S. Microbial Production of Odd-Chain Fatty Acids. Biotechnol. Bioeng. 2023, 120, 917–931. [Google Scholar] [CrossRef] [PubMed]
- Řezanka, T.; Kolouchová, I.; Sigler, K. Precursor Directed Biosynthesis of Odd-Numbered Fatty Acids by Different Yeasts. Folia Microbiol. 2015, 60, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Kolouchová, I.; Schreiberová, O.; Sigler, K.; Masák, J.; Řezanka, T. Biotransformation of Volatile Fatty Acids by Oleaginous and Non-Oleaginous Yeast Species. FEMS Yeast Res. 2015, 15, fov076. [Google Scholar] [CrossRef]
- Zhang, L.-S.; Xu, P.; Chu, M.-Y.; Zong, M.-H.; Yang, J.-G.; Lou, W.-Y. Using 1-Propanol to Significantly Enhance the Production of Valuable Odd-Chain Fatty Acids by Rhodococcus Opacus PD630. World J. Microbiol. Biotechnol. 2019, 35, 164. [Google Scholar] [CrossRef]
- Gao, R.; Li, Z.; Zhou, X.; Cheng, S.; Zheng, L. Oleaginous Yeast Yarrowia lipolytica Culture with Synthetic and Food Waste-Derived Volatile Fatty Acids for Lipid Production. Biotechnol. Biofuels 2017, 10, 247. [Google Scholar] [CrossRef]
- Zhang, L.-S.; Liang, S.; Zong, M.-H.; Yang, J.-G.; Lou, W.-Y. Microbial Synthesis of Functional Odd-Chain Fatty Acids: A Review. World J. Microbiol. Biotechnol. 2020, 36, 35. [Google Scholar] [CrossRef]
- Gao, R.; Li, Z.; Zhou, X.; Bao, W.; Cheng, S.; Zheng, L. Enhanced Lipid Production by Yarrowia lipolytica Cultured with Synthetic and Waste-Derived High-Content Volatile Fatty Acids under Alkaline Conditions. Biotechnol. Biofuels 2020, 13, 3. [Google Scholar] [CrossRef]
- Koubaa, M. Integrated Biorefinery for a Next-Generation Methanization Process Focusing on Volatile Fatty Acid Valorization: A Critical Review. Molecules 2024, 29, 2477. [Google Scholar] [CrossRef]

| Strain | Description | Plasmid | Auxotrophy |
|---|---|---|---|
| JMY7877 | Po1d Δphd1 Δmfe1 Δtgl4 + pTEF-YlDGA2 + pTEF-YlGPD1 + hp4d-YlLDP1 + pTEF-RePCT + pTEF-ScSUC2-LEU2 ex + pTEF-YlHXK1-URA3 ex | JME2103 JME2347 | U+L+ |
| JMY9031 | Y7877 − URA3 − LEU2 | JME547 | U−L− |
| JMY9112 | Y9031 + pTEF-YlDGA2-URA3 ex | JME1132 | U+L− |
| JMY9178 | Y9112 + pTEF-YlOLE1-LEU2 ex | JME5112 | U+L+ |
| Component | Concentrations |
|---|---|
| Sugar beet molasses * | 20 gsucrose/L |
| Crude glycerol * | 30 g/L |
| Yeast extract | 1 g/L |
| NH4Cl | 0.5 g/L |
| Sodium acetate | [0; 5; 10; 15; 20 g/L] |
| Sodium propionate | [0; 0.5; 1; 2; 3.5; 5 g/L] |
| Na2HPO4 | 0.05 M |
| KH2PO4 | 0.05 M |
| Saline solution ** | 1 mL/L |
| Distilled water | - |
| Sodium Propionate (g/L) | 0 | 0.5 | 1 | 2 | 3.5 | 5 |
|---|---|---|---|---|---|---|
| DCW (g/L) | 19.42 ± 2.9 | 18.31 ± 2.4 | 17.0 ± 0.07 | 15.6 ± 0.71 | 11.3 ± 0.14 | 10.5 ± 1.13 |
| FAMEs (%) | ||||||
| C15:0 | 0.46 ± 0.07 | 1.67 ± 0.05 | 2.75 ± 0.15 | 4.07 ± 0.2 | 2.69 ± 0.34 | 2.07 ± 0.19 |
| C16:0 | 25.25 ± 2.22 | 24.36 ± 1.39 | 19.77 ± 0.08 | 7.21 ± 0.44 | 3.2 ± 0.29 | 1.49 ± 0.46 |
| C16:1 | 11.24 ± 0.13 | 10.95 ± 0.91 | 7.87 ± 0.47 | 4.37 ± 0.04 | 3.05 ± 0.17 | 2.22 ± 0.11 |
| C17:0 | 0.5 ± 0.07 | 2.27 ± 0.03 | 4.44 ± 0.52 | 6.01 ± 0.25 | 3.12 ± 0.04 | 1.63 ± 0.18 |
| C17:1 | 3.67 ± 0.25 | 11.83 ± 0.66 | 20.31 ± 0.05 | 46.17 ± 1.47 | 51.78 ± 0.08 | 50.4 ± 0.18 |
| C18:0 | 6.15 ± 1.73 | 4.87 ± 0.94 | 4.05 ± 0.47 | 1.37 ± 0.32 | 0.69 ± 0.01 | 0.22 ± 0.31 |
| C18:1 | 48.54 ± 3.34 | 40.8 ± 0.68 | 36.8 ± 0.49 | 21.59 ± 0.31 | 19.58 ± 0.02 | 18.65 ± 1.36 |
| C18:2 | 0.09 ± 0.06 | 0.42 ± 0.05 | 1.17 ± 0.01 | 3.77 ± 0/04 | 6.47 ± 0.78 | 6.9 ± 0.9 |
| C19:0 | 4.08 ± 0.28 | 3.02 ± 0.39 | 2.79 ± 0.04 | 2.65 ± 0.01 | 3.37 ± 0.11 | 4.76 ± 0.13 |
| C19:1 | 0 | 0 | 0 | 2.73 ± 0.01 | 5.96 ± 0.13 | 8.51 ± 0.49 |
| Saturated FAs | 36.44 ± 4.36 | 36.21 ± 2.8 | 33.8 ± 1.27 | 21.31 ± 6.07 | 13.08 ± 0.69 | 10.19 ± 1.27 |
| Unsaturated FAs | 63.51 ± 3.81 | 64.01 ± 2.3 | 66.16 ± 1.03 | 78.64 ± 6.05 | 86.86 ± 0.69 | 86.69 ± 3.04 |
| OCFAs | 8.71 ± 0.67 | 18.8 ± 1.12 | 30.3 ± 0.76 | 61.63 ± 4.45 | 66.94 ± 0.35 | 67.39 ± 1.17 |
| Sodium Acetate (g/L) | 0 | 5 | 10 | 15 | 20 |
|---|---|---|---|---|---|
| DCW (g/L) | 6.30 ± 0.48 | 3.94 ± 0.17 | 4.92 ± 0.96 | 6.85 ± 0.03 | 10.5 ± 1.13 |
| FAMES (%) | |||||
| C15:0 | 7.59 ± 0.33 | 6.93 ± 0.25 | 3.47 ± 0.10 | 1.81 ± 0.19 | 2.07 ± 0.19 |
| C16:0 | 5.32 ± 0.37 | 3.83 ± 0.38 | 1.89 ± 0.14 | 1.24 ± 0.15 | 1.49 ± 0.46 |
| C16:1 | 2.93 ± 0.06 | 3.52 ± 0.28 | 2.48 ± 0.06 | 1.94 ± 0.10 | 2.22 ± 0.11 |
| C17:0 | 7.93 ± 0.90 | 2.32 ± 0.23 | 1.42 ± 0.27 | 1.29 ± 0.08 | 1.63 ± 0.18 |
| C17:1 | 53.42 ± 0.69 | 50.49 ± 0.88 | 50.40 ± 1.73 | 49.11 ± 0.06 | 50.4 ± 0.18 |
| C18:0 | 1.26 ± 0.01 | 0.71 ± 0.09 | 0.37 ± 0.00 | 0.44 ± 0.09 | 0.22 ± 0.31 |
| C18:1 | 14.83 ± 0.18 | 18.71 ± 0.59 | 20.17 ± 1.22 | 18.09 ± 1.34 | 18.65 ± 1.36 |
| C18:2 | 2.32 ± 0.07 | 2.20 ± 0.40 | 6.15 ± 0.49 | 9.93 ± 0.70 | 6.9 ± 0.9 |
| C19:0 | 2.07 ± 0.27 | 5.83 ± 0.59 | 5.65 ± 0.01 | 4.83 ± 0.13 | 4.76 ± 0.13 |
| C19:1 | 2.32 ± 0.37 | 5.44 ± 0.54 | 7.97 ± 0.61 | 11.66 ± 1.76 | 8.51 ± 0.49 |
| Saturated FAs | 24.15 | 19.61 | 12.80 | 9.59 | 10.19 |
| Unsaturated FAs | 75.81 | 80.35 | 87.16 | 90.72 | 86.69 |
| OCFAs | 73.31 | 71.00 | 68.91 | 68.69 | 67.39 |
| ECFAs | 26.69 | 29 | 31.09 | 31.31 | 32.61 |
| Factors | Measured Responses | Predicted Responses | |||||
|---|---|---|---|---|---|---|---|
| Experiment | Sodium Acetate (g/L) | Sodium Propionate (g/L) | Cell Density (OD600nm) | DCW (g/L) | C17:1 (%) | Cell Density (OD600nm) | C17:1 (%) |
| 1 | 15 (−1) | 2 (−1) | 21.88 | 10.44 | 41.02 | 22.070 | 39.260 |
| 2 | 15 (−1) | 2 (−1) | 20.99 | 9.92 | 40.71 | 22.070 | 39.260 |
| 3 | 20 (+1) | 2 (−1) | 28.12 | 11.14 | 44.74 | 24.040 | 40.220 |
| 4 | 20 (+1) | 2 (−1) | 23.17 | 12.04 | 41.68 | 24.040 | 40.220 |
| 5 | 15 (−1) | 3.5 (+1) | 11.58 | 7.47 | 41.16 | 13.925 | 47.011 |
| 6 | 15 (−1) | 3.5 (+1) | 17.42 | 8.12 | 46.06 | 13.925 | 47.011 |
| 7 | 20 (+1) | 3.5 (+1) | 23.17 | 9.52 | 43.44 | 19.599 | 46.906 |
| 8 | 20 (+1) | 3.5 (+1) | 21.88 | 9.69 | 46.23 | 19.599 | 46.906 |
| 9 | 13.96 (−α) | 2.75 (0) | 15.74 | 9.37 | 47.35 | 15.393 | 46.798 |
| 10 | 13.96 (−α) | 2.75 (0) | 16.43 | 8.97 | 48.18 | 15.393 | 46.798 |
| 11 | 21.04 (+α) | 2.75 (0) | 17.82 | 9.85 | 45.56 | 20.798 | 47.403 |
| 12 | 21.04 (+α) | 2.75 (0) | 18.22 | 10.27 | 47.56 | 20.798 | 47.403 |
| 13 | 17.5 (0) | 1.69 (−α) | 24.75 | 13.78 | 30.19 | 26.172 | 34.494 |
| 14 | 17.5 (0) | 1.69 (−α) | 27.42 | 13.51 | 31.48 | 26.172 | 34.494 |
| 15 | 17.5 (0) | 3.81 (+α) | 15.25 | 9.09 | 48.34 | 17.272 | 44.702 |
| 16 | 17.5 (0) | 3.81 (+α) | 15.05 | 9.21 | 49.28 | 17.272 | 44.702 |
| 17 | 17.5 (0) | 2.75 (0) | 20.59 | 9.47 | 49.19 | 17.269 | 48.811 |
| 18 | 17.5 (0) | 2.75 (0) | 16.34 | 9.74 | 47.96 | 17.269 | 48.811 |
| 19 | 17.5 (0) | 2.75 (0) | 15.94 | 9.52 | 47.46 | 17.269 | 48.811 |
| 20 | 15.97 (−0.612) | 2.48 (−0.36) | 16.53 | 9.84 | 48.38 | 17.845 | 46.450 |
| 21 | 19.03 (+0.612) | 2.48 (−0.36) | 19.5 | 9.70 | 48.25 | 19.784 | 46.828 |
| 22 | 17.5 (0) | 3.28 (+0.707) | 16.34 | 8.99 | 48.14 | 16.157 | 49.060 |
| (a) | |||||
| Source of Variation | Sum of Squares | Degrees of Freedom | Mean Squares | F-Ratio | p-Value |
| Regression | 280.508 | 5 | 56.101 | 9.059 | 0.0307 * |
| Residue | 99.088 | 16 | 6.193 | ||
| Lack of fit | 51.372 | 6 | 8.562 | 1.794 | 19.7 |
| Pure error | 47.716 | 10 | 4.771 | ||
| Total | 379.596 | 21 | |||
| (b) | |||||
| Source of Variation | Sum of Squares | Degrees of Freedom | Mean Squares | F-Ratio | p-Value |
| Regression | 428.727 | 5 | 85.745 | 33.195 | <0.01 * |
| Residue | 152.087 | 16 | 9.505 | ||
| Lack of fit | 126.257 | 6 | 21.042 | 8.147 | 0.219 * |
| Pure error | 25.830 | 10 | 2.583 | ||
| Total | 5880.815 | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabaa Chalabi, N.; El Kantar, S.; Pires De Souza, C.; Khelfa, A.; Nicaud, J.-M.; Debs, E.; Louka, N.; Koubaa, M. Improving the Synthesis of Odd-Chain Fatty Acids in the Oleaginous Yeast Yarrowia lipolytica. Fermentation 2024, 10, 597. https://doi.org/10.3390/fermentation10120597
Tabaa Chalabi N, El Kantar S, Pires De Souza C, Khelfa A, Nicaud J-M, Debs E, Louka N, Koubaa M. Improving the Synthesis of Odd-Chain Fatty Acids in the Oleaginous Yeast Yarrowia lipolytica. Fermentation. 2024; 10(12):597. https://doi.org/10.3390/fermentation10120597
Chicago/Turabian StyleTabaa Chalabi, Nour, Sally El Kantar, Camilla Pires De Souza, Anissa Khelfa, Jean-Marc Nicaud, Espérance Debs, Nicolas Louka, and Mohamed Koubaa. 2024. "Improving the Synthesis of Odd-Chain Fatty Acids in the Oleaginous Yeast Yarrowia lipolytica" Fermentation 10, no. 12: 597. https://doi.org/10.3390/fermentation10120597
APA StyleTabaa Chalabi, N., El Kantar, S., Pires De Souza, C., Khelfa, A., Nicaud, J.-M., Debs, E., Louka, N., & Koubaa, M. (2024). Improving the Synthesis of Odd-Chain Fatty Acids in the Oleaginous Yeast Yarrowia lipolytica. Fermentation, 10(12), 597. https://doi.org/10.3390/fermentation10120597









