Ordered Changes in Methane Production Performance and Metabolic Pathway Transition of Methanogenic Archaea under Gradually Increasing Sodium Propionate Stress Intensity
Abstract
1. Introduction
2. Materials and Methods
2.1. Medium and Inoculum
2.2. Experimental System and Protocol
2.3. Analytical Methods
2.3.1. Physicochemical Properties
2.3.2. Microbial Community Structure and Functional Predictions
2.4. Model Application
2.5. Data Analysis
3. Results and Discussion
3.1. Batch Reactor’s Daily and Cumulative Methane Yields
3.2. GM and FO Modeling
3.3. pH, NH4+-N, and VFA Contents
3.4. Microbiological Communities
3.5. Microbial Functions for Methane Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dahiya, S.; Kumar, A.N.; Shanthi Sravan, J.; Chatterjee, S.; Sarkar, O.; Mohan, S.V. Food Waste Biorefinery: Sustainable Strategy for Circular Bioeconomy. Bioresour. Technol. 2018, 248, 2–12. [Google Scholar] [CrossRef]
- Niknejad, P.; Azizi, S.M.M.; Hillier, K.; Gupta, R.; Dhar, B.R. Biodegradability and Transformation of Biodegradable Disposables in High-Solids Anaerobic Digestion Followed by Hydrothermal Liquefaction. Resour. Conserv. Recycl. 2023, 193, 106979. [Google Scholar] [CrossRef]
- Wang, W.; Lee, D.-J.; Lei, Z. Integrating Anaerobic Digestion with Microbial Electrolysis Cell for Performance Enhancement: A Review. Bioresour. Technol. 2022, 344, 126321. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, L.; Yang, P.; Peng, Y.; Peng, X. Exploring Instability Mechanisms of Overloaded Anaerobic Digestion Based on the Combination of Degradation Dynamics of Acetate and Propionate and Metagenomics Assay. Biochem. Eng. J. 2023, 198, 109032. [Google Scholar] [CrossRef]
- Yuan, H.; Zhu, N. Progress in Inhibition Mechanisms and Process Control of Intermediates and By-Products in Sewage Sludge Anaerobic Digestion. Renew. Sustain. Energy Rev. 2016, 58, 429–438. [Google Scholar] [CrossRef]
- Wong, B.-T.; Show, K.-Y.; Su, A.; Wong, R.; Lee, D.-J. Effect of Volatile Fatty Acid Composition on Upflow Anaerobic Sludge Blanket (UASB) Performance. Energy Fuels 2008, 22, 108–112. [Google Scholar] [CrossRef]
- Han, Y.; Green, H.; Tao, W. Reversibility of Propionic Acid Inhibition to Anaerobic Digestion: Inhibition Kinetics and Microbial Mechanism. Chemosphere 2020, 255, 126840. [Google Scholar] [CrossRef]
- Speece, R.E. Anaerobic Biotechnology and Odor/Corrosion Control for Municipalities and Industries; Archae Press: Nashville, TN, USA, 2008. [Google Scholar]
- Zheng, Z.; Liu, J.; Yuan, X.; Wang, X.; Zhu, W.; Yang, F.; Cui, Z. Effect of Dairy Manure to Switchgrass Co-Digestion Ratio on Methane Production and the Bacterial Community in Batch Anaerobic Digestion. Appl. Energy 2015, 151, 249–257. [Google Scholar] [CrossRef]
- Lay, J.; Li, Y.; Noike, T. Analysis of Environmental Factors Affecting Methane Production from High-Solids Organic Waste. Water Sci. Technol. 1997, 36, 493–500. [Google Scholar] [CrossRef]
- Zhang, W.; Wei, Q.; Wu, S.; Qi, D.; Li, W.; Zuo, Z.; Dong, R. Batch Anaerobic Co-Digestion of Pig Manure with Dewatered Sewage Sludge under Mesophilic Conditions. Appl. Energy 2014, 128, 175–183. [Google Scholar] [CrossRef]
- Sitthi, S.; Hatamoto, M.; Watari, T.; Yamaguchi, T. Accelerating Anaerobic Propionate Degradation and Studying Microbial Community Using Modified Polyvinyl Alcohol Beads during Anaerobic Digestion. Bioresour. Technol. Rep. 2022, 17, 100907. [Google Scholar] [CrossRef]
- Zhou, M.; Yan, B.; Wong, J.W.C.; Zhang, Y. Enhanced Volatile Fatty Acids Production from Anaerobic Fermentation of Food Waste: A Mini-Review Focusing on Acidogenic Metabolic Pathways. Bioresour. Technol. 2018, 248, 68–78. [Google Scholar] [CrossRef]
- Abbassi-Guendouz, A.; Trably, E.; Hamelin, J.; Dumas, C.; Steyer, J.P.; Delgenès, J.-P.; Escudié, R. Microbial Community Signature of High-Solid Content Methanogenic Ecosystems. Bioresour. Technol. 2013, 133, 256–262. [Google Scholar] [CrossRef]
- Wu, D.; Li, L.; Zhen, F.; Liu, H.; Xiao, F.; Sun, Y.; Peng, X.; Li, Y.; Wang, X. Thermodynamics of Volatile Fatty Acid Degradation during Anaerobic Digestion under Organic Overload Stress: The Potential to Better Identify Process Stability. Water Res. 2022, 214, 118187. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Schnürer, A.; Westerholm, M. Enrichment and Description of Novel Bacteria Performing Syntrophic Propionate Oxidation at High Ammonia Level. Environ. Microbiol. 2021, 23, 1620–1637. [Google Scholar] [CrossRef] [PubMed]
- Holtzapple, M.T.; Wu, H.; Weimer, P.J.; Dalke, R.; Granda, C.B.; Mai, J.; Urgun-Demirtas, M. Microbial Communities for Valorizing Biomass Using the Carboxylate Platform to Produce Volatile Fatty Acids: A Review. Bioresour. Technol. 2022, 344, 126253. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, C.; Nges, I.A.; Liu, J. The Effects of Pre-Aeration and Inoculation on Solid-State Anaerobic Digestion of Rice Straw. Bioresour. Technol. 2017, 224, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Yang, S.-T. Extractive Fermentation for Enhanced Propionic Acid Production from Lactose by Propionibacterium Acidipropionici. Biotechnol. Prog. 1998, 14, 457–465. [Google Scholar] [CrossRef]
- Stams, A.J.M.; De Bok, F.A.M.; Plugge, C.M.; Van Eekert, M.H.A.; Dolfing, J.; Schraa, G. Exocellular Electron Transfer in Anaerobic Microbial Communities. Environ. Microbiol. 2006, 8, 371–382. [Google Scholar] [CrossRef]
- Yue, Y.; Wang, J.; Wu, X.; Zhang, J.; Chen, Z.; Kang, X.; Lv, Z. The Fate of Anaerobic Syntrophy in Anaerobic Digestion Facing Propionate and Acetate Accumulation. Waste Manag. 2021, 124, 128–135. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Wu, J. Enhancement of Methane Production in Anaerobic Digestion Process: A Review. Appl. Energy 2019, 240, 120–137. [Google Scholar] [CrossRef]
- Widyasti, E.; Shikata, A.; Hashim, R.; Sulaiman, O.; Sudesh, K.; Wahjono, E.; Kosugi, A. Biodegradation of Fibrillated Oil Palm Trunk Fiber by a Novel Thermophilic, Anaerobic, Xylanolytic Bacterium Caldicoprobacter Sp. CL-2 Isolated from Compost. Enzyme Microb. Technol. 2018, 111, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, J.; Li, Y.; Zhu, T.; Yu, Q.; Wang, T.; Liang, S.; Zhang, Y. Why Do DIETers like Drinking: Metagenomic Analysis for Methane and Energy Metabolism during Anaerobic Digestion with Ethanol. Water Res. 2020, 171, 115425. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Rui, J.; Pei, Z.; Sun, X.; Zhang, S.; Yan, Z.; Wang, Y.; Liu, X.; Zheng, T.; Li, X. Straw- and Slurry-Associated Prokaryotic Communities Differ during Co-Fermentation of Straw and Swine Manure. Appl. Microbiol. Biotechnol. 2014, 98, 4771–4780. [Google Scholar] [CrossRef] [PubMed]
- Imachi, H.; Sakai, S.; Kubota, T.; Miyazaki, M.; Saito, Y.; Takai, K. Sedimentibacter Acidaminivorans Sp. Nov., an Anaerobic, Amino-Acid-Utilizing Bacterium Isolated from Marine Subsurface Sediment. Int. J. Syst. Evol. Microbiol. 2016, 66, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- De Bok, F.A.M.; Harmsen, H.J.M.; Plugge, C.M.; De Vries, M.C.; Akkermans, A.D.L.; De Vos, W.M.; Stams, A.J.M. The First True Obligately Syntrophic Propionate-Oxidizing Bacterium, Pelotomaculum Schinkii Sp. Nov., Co-Cultured with Methanospirillum Hungatei, and Emended Description of the Genus Pelotomaculum. Int. J. Syst. Evol. Microbiol. 2005, 55, 1697–1703. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhu, G. Deciphering the Effects of Antibiotics on Nitrogen Removal and Bacterial Communities of Autotrophic Denitrification Systems in a Three-Dimensional Biofilm Electrode Reactor. Environ. Pollut. 2022, 315, 120476. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tang, Y.; Xiong, P.; Zhang, M.; Deng, Q.; Liang, D.; Zhao, Z.; Feng, Y.; Zhang, Y. High-Efficiency Methanogenesis via Kitchen Wastes Served as Ethanol Source to Establish Direct Interspecies Electron Transfer during Anaerobic Co-Digestion with Waste Activated Sludge. Water Res. 2020, 176, 115763. [Google Scholar] [CrossRef] [PubMed]
- Gieg, L.M.; Fowler, S.J.; Berdugo-Clavijo, C. Syntrophic Biodegradation of Hydrocarbon Contaminants. Curr. Opin. Biotechnol. 2014, 27, 21–29. [Google Scholar] [CrossRef]
- Basak, B.; Patil, S.M.; Kumar, R.; Ahn, Y.; Ha, G.-S.; Park, Y.-K.; Ali Khan, M.; Jin Chung, W.; Woong Chang, S.; Jeon, B.-H. Syntrophic Bacteria- and Methanosarcina-Rich Acclimatized Microbiota with Better Carbohydrate Metabolism Enhances Biomethanation of Fractionated Lignocellulosic Biocomponents. Bioresour. Technol. 2022, 360, 127602. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, L.; Huang, J.; Qu, Y.; Pan, Y.; Liu, L.; Zhu, H. Recovering Short-Chain Fatty Acids from Waste Sludge via Biocarriers and Microfiltration Enhanced Anaerobic Fermentation. Resour. Conserv. Recycl. 2022, 182, 106342. [Google Scholar] [CrossRef]
- Xie, Z.; Huang, S.; Wan, Y.; Deng, F.; Cao, Q.; Liu, X.; Li, D. Power to Biogas Upgrading: Effects of Different H2/CO2 Ratios on Products and Microbial Communities in Anaerobic Fermentation System. Sci. Total Environ. 2023, 865, 161305. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Zhi, L.; Jiang, H.; Chen, R.; Wang, X.; Li, Y.-Y. Characterization of Microbial Evolution in High-Solids Methanogenic Co-Digestion of Canned Coffee Processing Wastewater and Waste Activated Sludge by an Anaerobic Membrane Bioreactor. J. Clean. Prod. 2019, 232, 1442–1451. [Google Scholar] [CrossRef]
- Tuncay, S.; Akcakaya, M.; Icgen, B. Ozonation of Sewage Sludge Prior to Anaerobic Digestion Led to Methanosaeta Dominated Biomethanation. Fuel 2022, 313, 122690. [Google Scholar] [CrossRef]
- Yan, M.; Hu, Z.; Duan, Z.; Sun, Y.; Dong, T.; Sun, X.; Zhen, F.; Li, Y. Microbiome Re-Assembly Boosts Anaerobic Digestion under Volatile Fatty Acid Inhibition: Focusing on Reactive Oxygen Species Metabolism. Water Res. 2023, 246, 120711. [Google Scholar] [CrossRef]
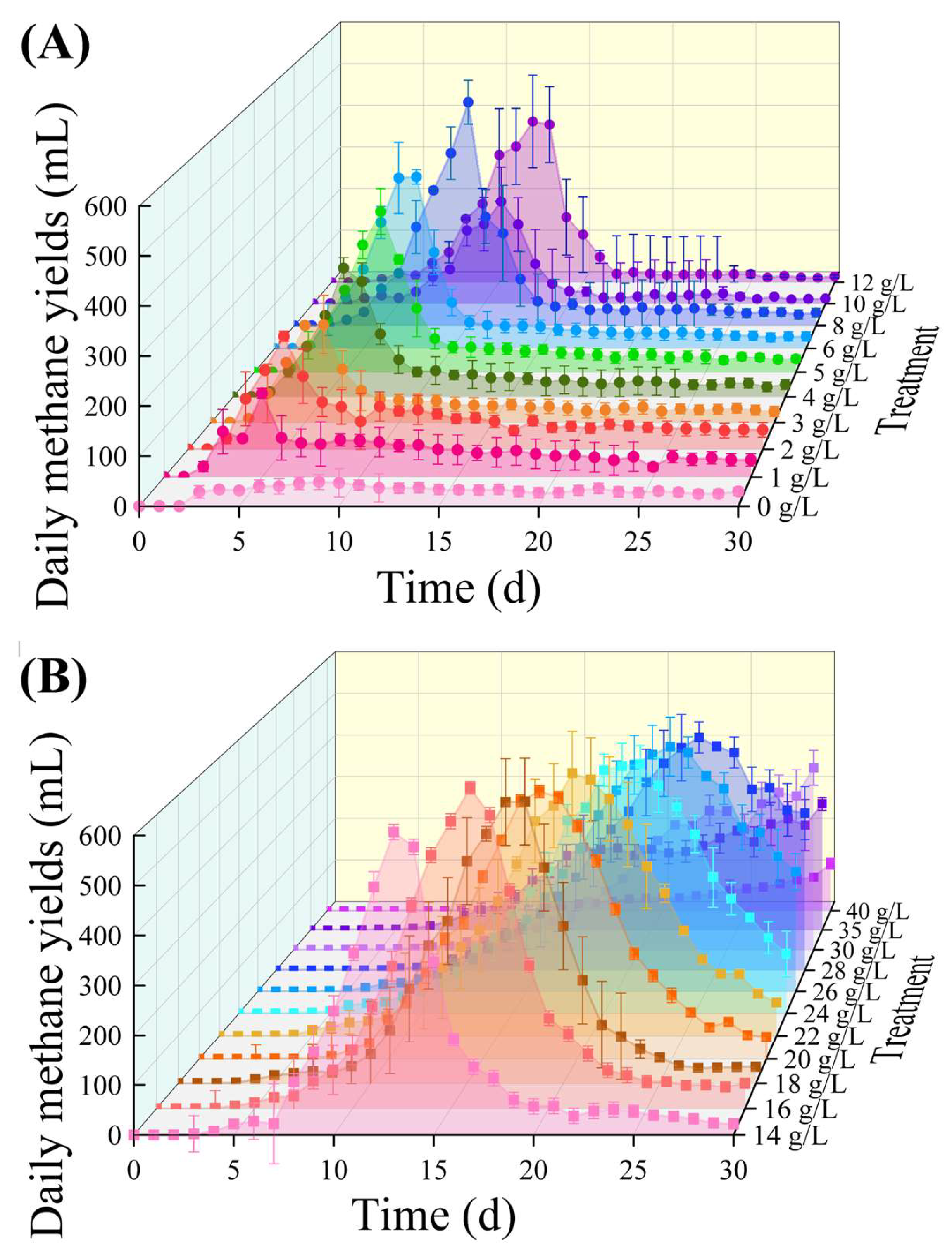


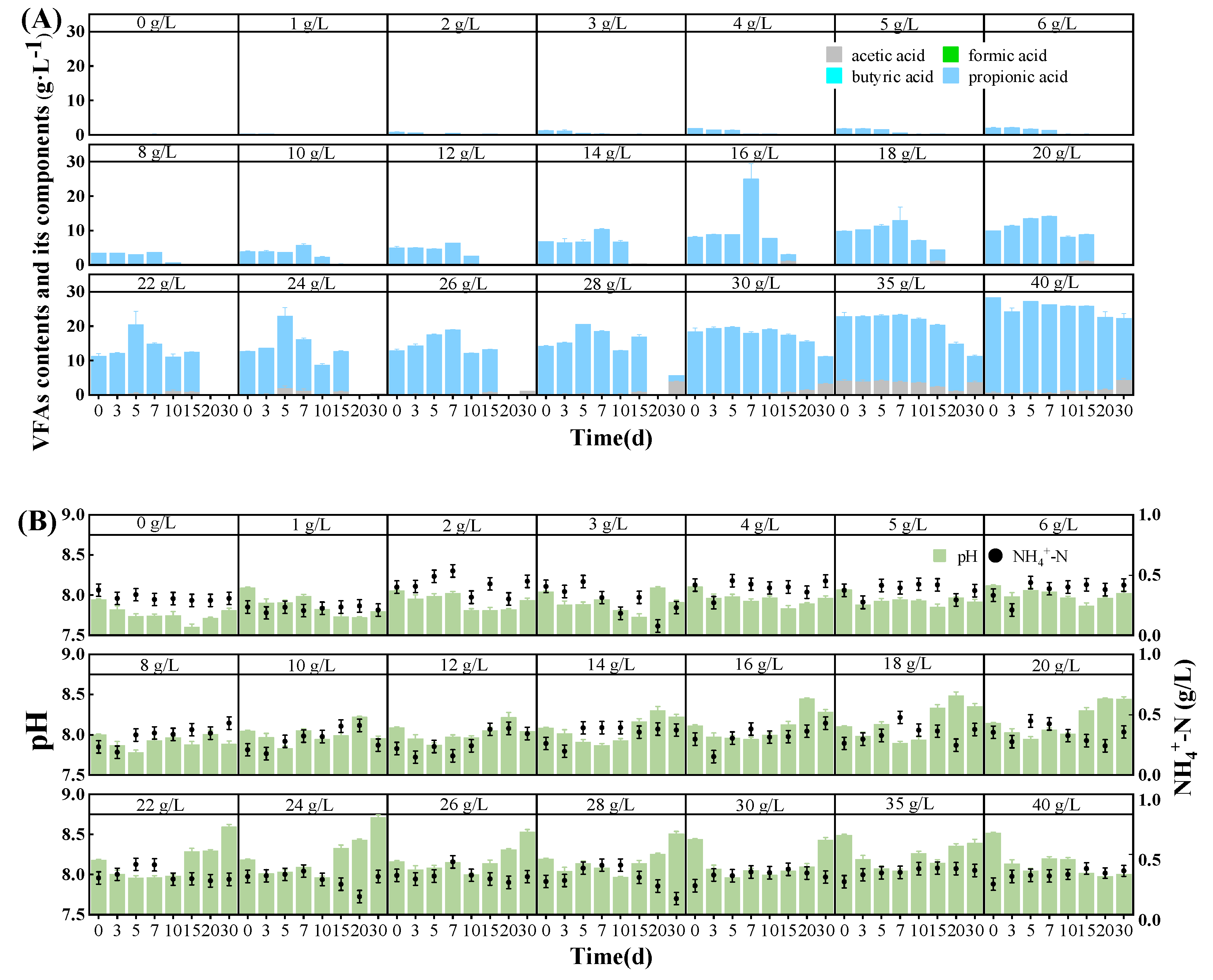
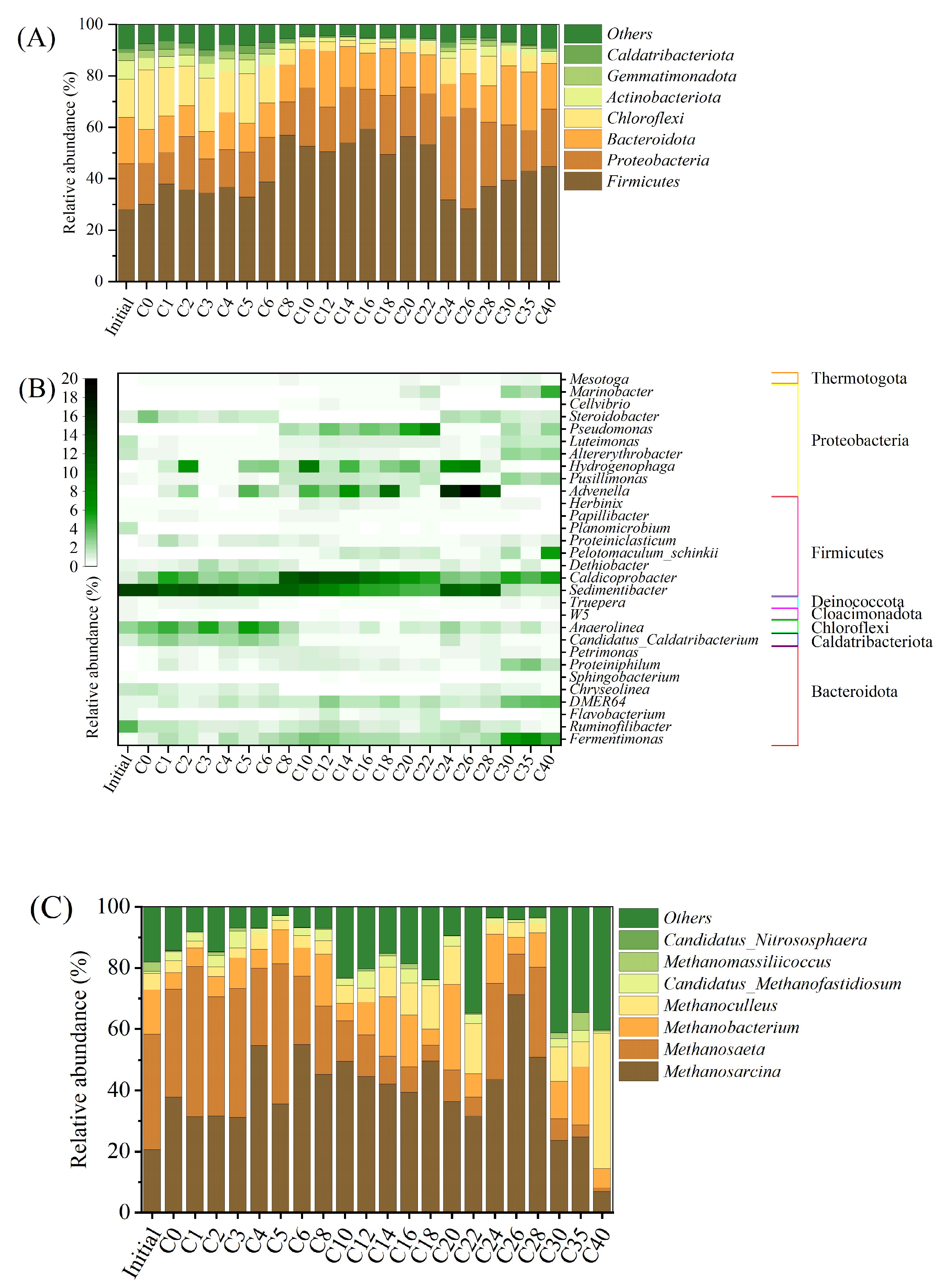

| FO | GM | ||||||
|---|---|---|---|---|---|---|---|
| Sodium Propionate Contents (g/L) | Hydrolytic Acidification Rate Kh | Fitting Index R2 | Predicted Value P (mL) | Experimental Value M (mL) | Maximum Methane Yield Rate Rmax (mL/d) | Delay Period λ (d) | Fitting Index R2 |
| 0 | 0.0720 | 0.94 | 1037.713 | 935.80 | 42.18051 | 3.32 | 0.995 |
| 1 | 0.1032 | 0.91 | 1701.91 | 1663.64 | 80.86 | 1.42 | 0.992 |
| 2 | 0.1242 | 0.84 | 2041.61 | 2069.91 | 106.11 | 0.89 | 0.984 |
| 3 | 0.1505 | 0.84 | 1434.07 | 1538.07 | 97.95 | 1.12 | 0.972 |
| 4 | 0.1540 | 0.93 | 1711.89 | 1851.66 | 143.60 | 2.03 | 0.977 |
| 5 | 0.1719 | 0.90 | 2090.42 | 2264.60 | 192.85 | 2.55 | 0.980 |
| 6 | 0.1862 | 0.92 | 2314.77 | 2501.21 | 257.10 | 3.18 | 0.985 |
| 8 | 0.2210 | 0.77 | 2713.41 | 2891.18 | 349.41 | 5.50 | 0.993 |
| 10 | 0.2082 | 0.80 | 1339.28 | 1412.67 | 181.80 | 6.67 | 0.994 |
| 12 | 0.2244 | 0.88 | 2394.50 | 2452.58 | 359.13 | 7.58 | 0.998 |
| 14 | 0.2245 | 0.66 | 3844.98 | 3959.53 | 516.59 | 8.93 | 0.998 |
| 16 | 0.1602 | 0.85 | 5215.39 | 5234.62 | 599.72 | 10.84 | 0.999 |
| 18 | 0.1464 | 0.85 | 4855.22 | 4786.56 | 561.36 | 11.94 | 0.998 |
| 20 | 0.1096 | 0.83 | 5884.02 | 5629.49 | 545.09 | 12.37 | 0.999 |
| 22 | 0.0809 | 0.82 | 6723.58 | 6084.24 | 507.15 | 12.76 | 0.998 |
| 24 | 0.0564 | 0.78 | 7678.98 | 6314.10 | 488.21 | 14.00 | 0.998 |
| 26 | 0.0375 | 0.76 | 9361.12 | 6601.25 | 488.83 | 15.19 | 0.999 |
| 28 | 0.0290 | 0.73 | 9921.92 | 6233.15 | 479.91 | 16.39 | 0.999 |
| 30 | 0.0151 | 0.73 | 9908.19 | 4141.75 | 348.08 | 18.24 | 0.999 |
| 35 | 0.0262 | 0.84 | 6747.83 | 3891.21 | 228.87 | 13.24 | 0.999 |
| 40 | 0.0035 | 0.75 | 6473.39 | 777.07 | 127.24 | 25.58 | 0.999 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Li, Y.; Zheng, Z.; Li, L.; Hao, J.; Liu, S.; Wang, Y.; Qi, C. Ordered Changes in Methane Production Performance and Metabolic Pathway Transition of Methanogenic Archaea under Gradually Increasing Sodium Propionate Stress Intensity. Fermentation 2024, 10, 201. https://doi.org/10.3390/fermentation10040201
Liu M, Li Y, Zheng Z, Li L, Hao J, Liu S, Wang Y, Qi C. Ordered Changes in Methane Production Performance and Metabolic Pathway Transition of Methanogenic Archaea under Gradually Increasing Sodium Propionate Stress Intensity. Fermentation. 2024; 10(4):201. https://doi.org/10.3390/fermentation10040201
Chicago/Turabian StyleLiu, Mengxi, Yuanyuan Li, Zehui Zheng, Lin Li, Jianjun Hao, Shuang Liu, Yaya Wang, and Chuanren Qi. 2024. "Ordered Changes in Methane Production Performance and Metabolic Pathway Transition of Methanogenic Archaea under Gradually Increasing Sodium Propionate Stress Intensity" Fermentation 10, no. 4: 201. https://doi.org/10.3390/fermentation10040201
APA StyleLiu, M., Li, Y., Zheng, Z., Li, L., Hao, J., Liu, S., Wang, Y., & Qi, C. (2024). Ordered Changes in Methane Production Performance and Metabolic Pathway Transition of Methanogenic Archaea under Gradually Increasing Sodium Propionate Stress Intensity. Fermentation, 10(4), 201. https://doi.org/10.3390/fermentation10040201





