Effect of Temperature and pH on Microbial Communities Fermenting a Dairy Coproduct Mixture
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
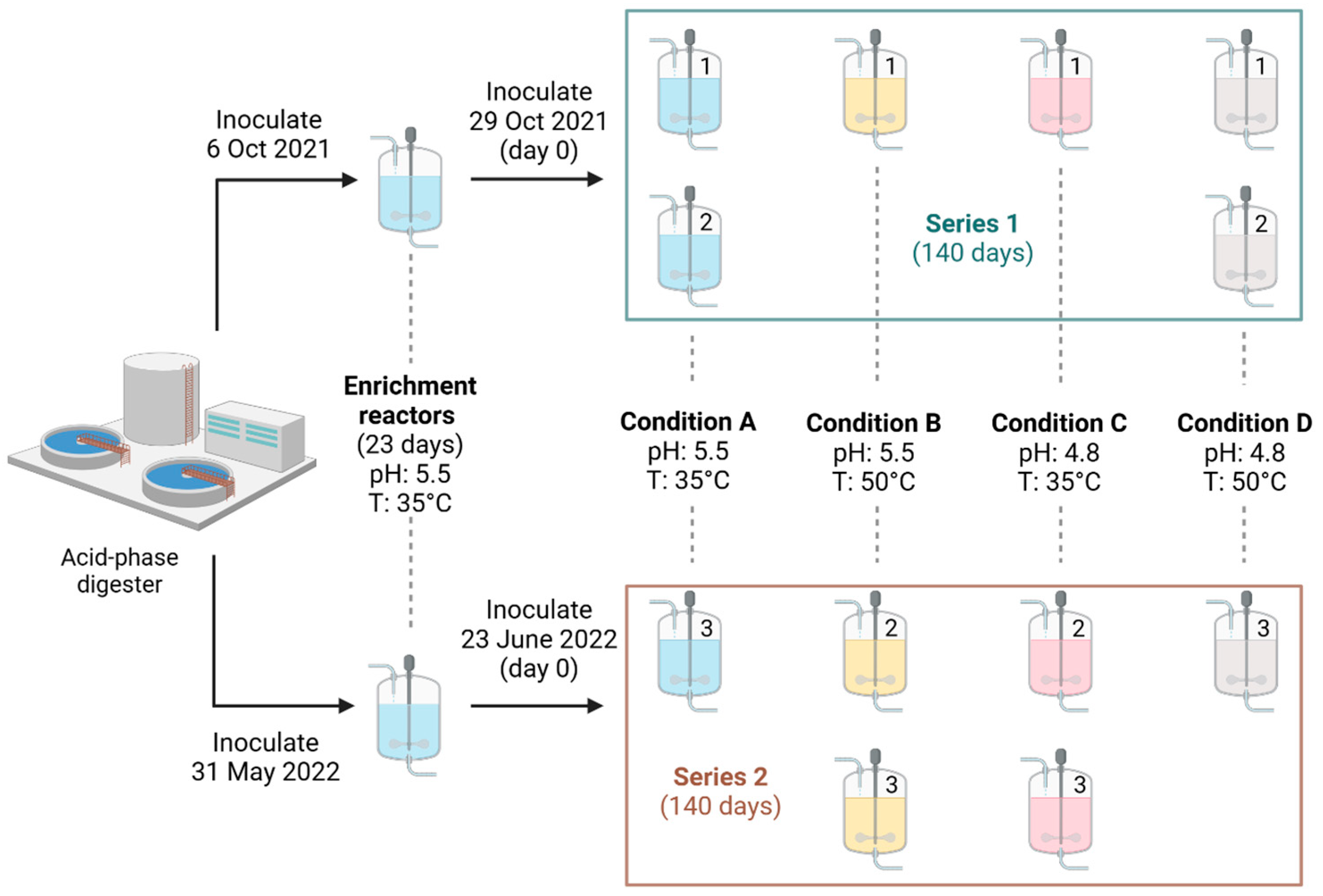
2.2. Experimental Design and Bioreactor System
2.3. Sample Collection and Storage
2.4. Analytical Tests
2.5. DNA Sequencing and Metagenome Assembly
2.6. Microbial Community Analysis
2.7. Machine Learning Functional Classifications
2.8. Gene and Metabolic Pathway Predictions
2.9. Phylogenetic Tree Generation
3. Results
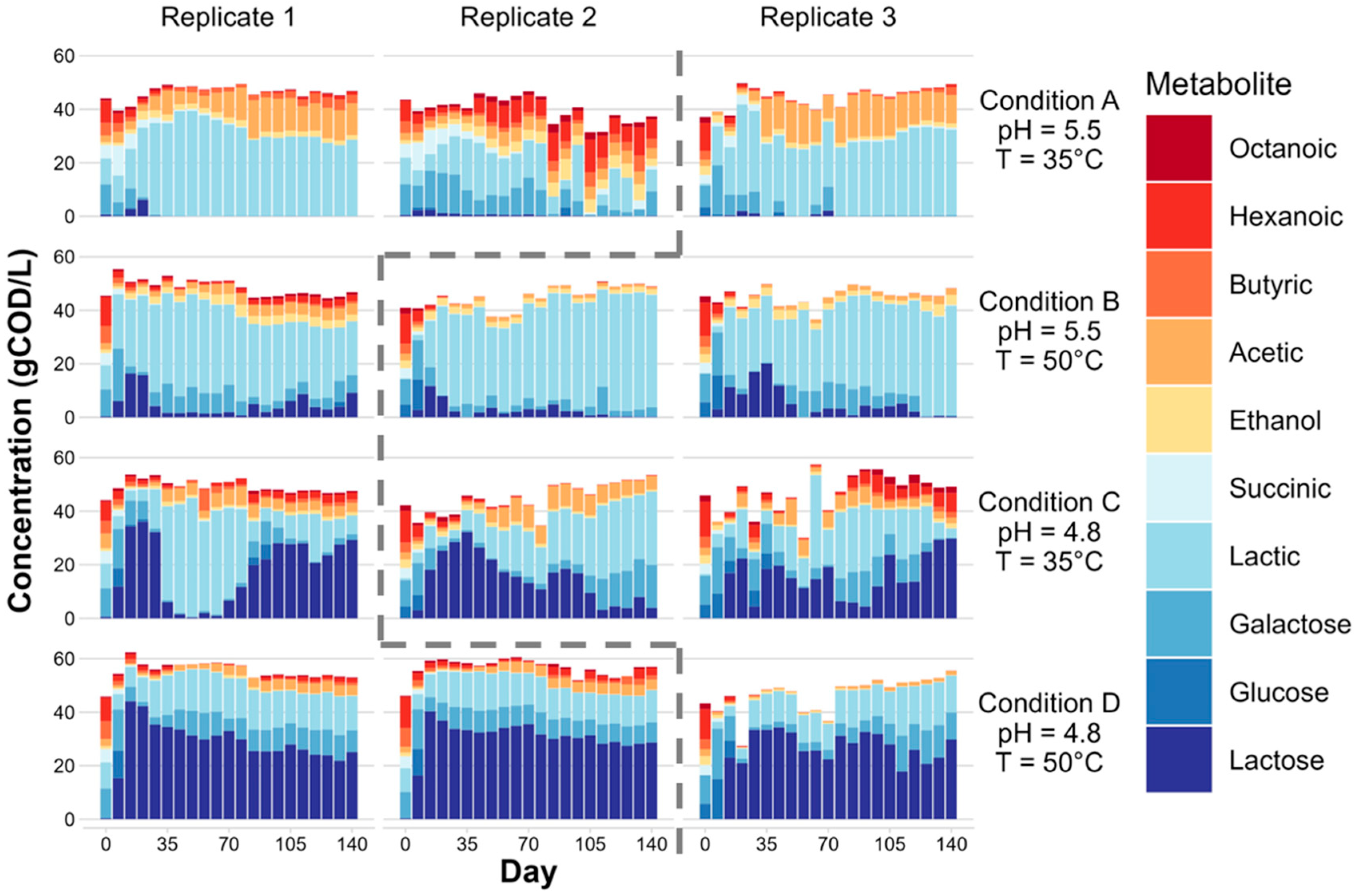
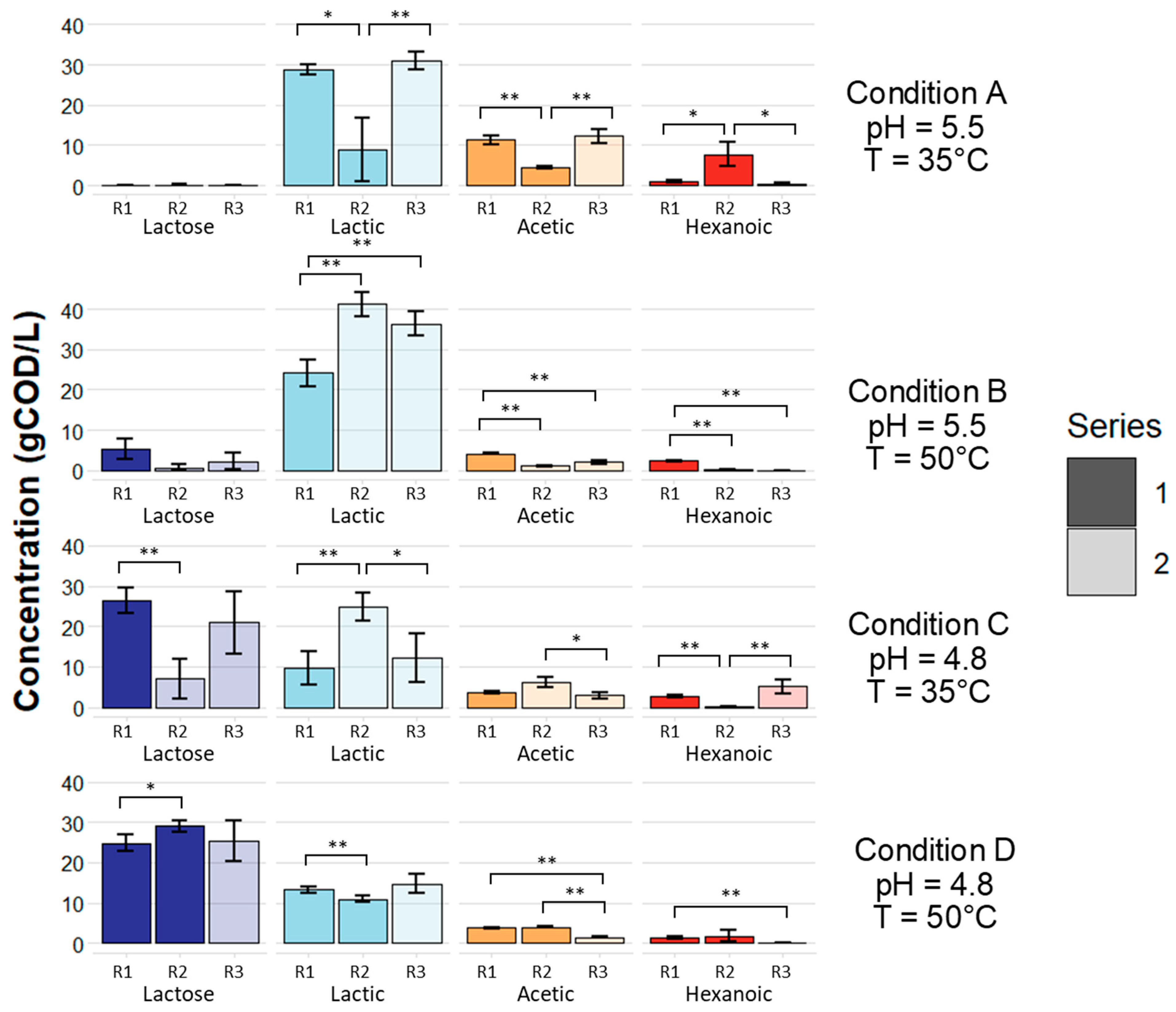
3.1. Microbial Fermentation of Dairy Coproduct Mixtures
3.2. Metagenomic Analysis of Bioreactor Microbial Communities
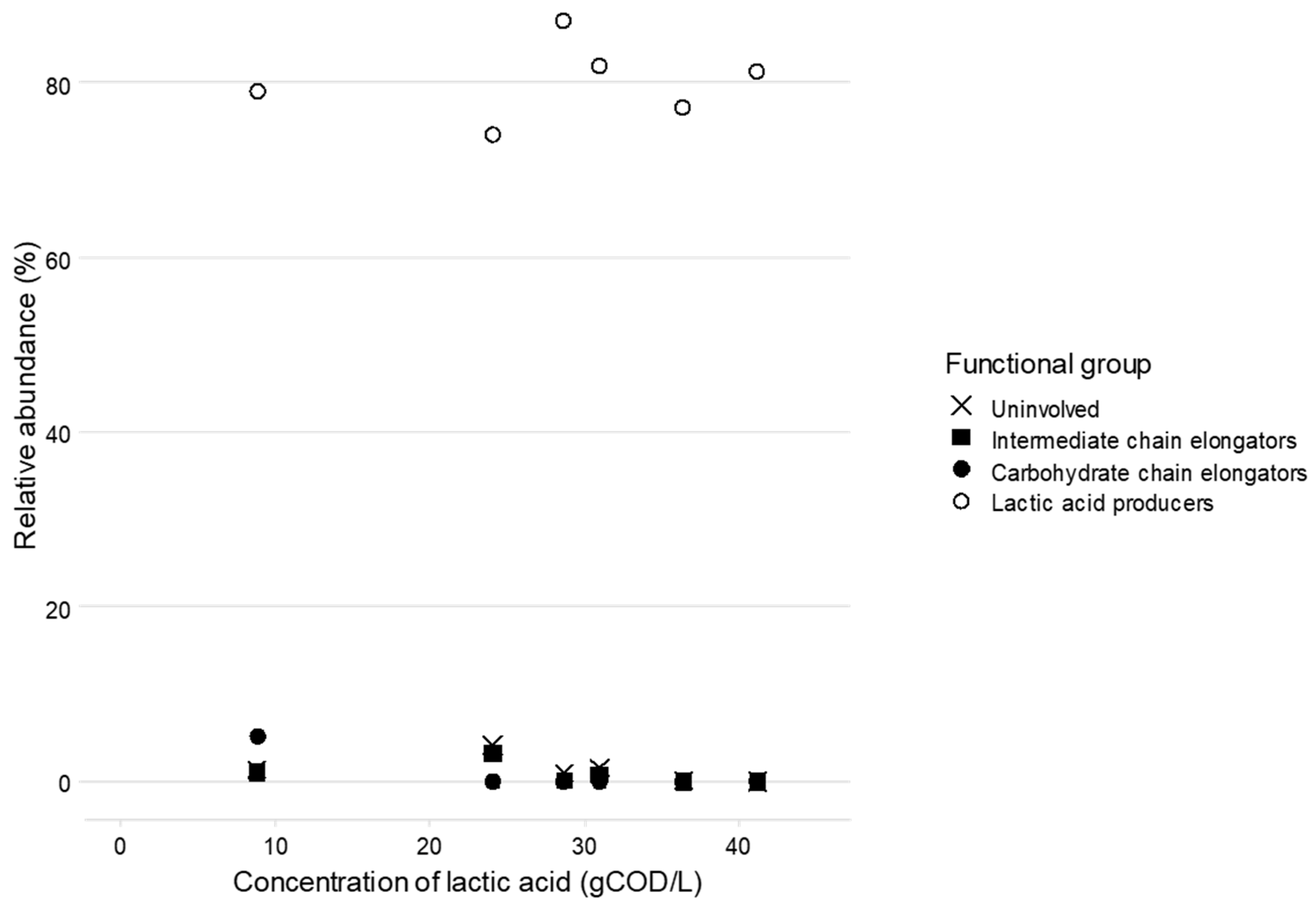
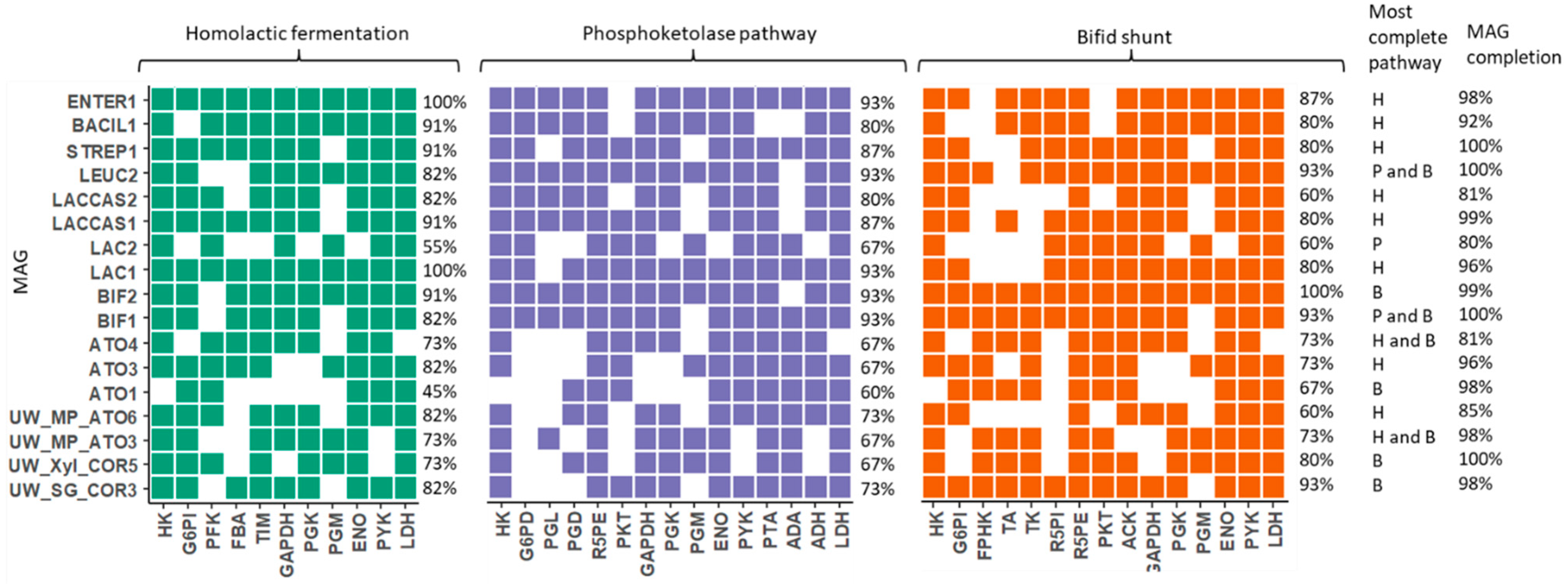
3.3. Lactic Acid Pathway Analysis
4. Discussion
4.1. Communities Enriched for Lactic Acid Producing Organisms
4.2. The Presence of Chain Elongators Were Associated with Lower Lactic Acid Accumulation
4.3. Communities Exhibited a High Degree of Functional Redundancy
4.4. Current Questions of Pathway Utilization
4.5. Comparison to UFMP-Fed Bioreactor
4.6. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Venkata Mohan, S.; Modestra, J.A.; Amulya, K.; Butti, S.K.; Velvizhi, G. A Circular Bioeconomy with Biobased Products from CO2 Sequestration. Trends Biotechnol. 2016, 34, 506–519. [Google Scholar] [CrossRef]
- Wadler, C.S.; Wolters, J.F.; Fortney, N.W.; Throckmorton, K.O.; Zhang, Y.; Miller, C.R.; Schneider, R.M.; Wendt-Pienkowski, E.; Currie, C.R.; Donohue, T.J.; et al. Utilization of Lignocellulosic Biofuel Conversion Residue by Diverse Microorganisms. Biotechnol. Biofuels Bioprod. 2022, 15, 70. [Google Scholar] [CrossRef]
- Perez, J.M.; Sener, C.; Misra, S.; Umana, G.E.; Coplien, J.; Haak, D.; Li, Y.; Maravelias, C.T.; Karlen, S.D.; Ralph, J.; et al. Integrating Lignin Depolymerization with Microbial Funneling Processes Using Agronomically Relevant Feedstocks. Green Chem. 2022, 24, 2795–2811. [Google Scholar] [CrossRef]
- Uçkun Kiran, E.; Trzcinski, A.P.; Ng, W.J.; Liu, Y. Bioconversion of Food Waste to Energy: A Review. Fuel 2014, 134, 389–399. [Google Scholar] [CrossRef]
- Scarborough, M.J.; Lawson, C.E.; DeCola, A.C.; Gois, I.M. Microbiomes for Sustainable Biomanufacturing. Curr. Opin. Microbiol. 2022, 65, 8–14. [Google Scholar] [CrossRef]
- Han, W.; He, P.; Shao, L.; Lü, F. Road to Full Bioconversion of Biowaste to Biochemicals Centering on Chain Elongation: A Mini Review. J. Environ. Sci. 2019, 86, 50–64. [Google Scholar] [CrossRef]
- Duber, A.; Jaroszynski, L.; Zagrodnik, R.; Chwialkowska, J.; Juzwa, W.; Ciesielski, S.; Oleskowicz-Popiel, P. Exploiting the Real Wastewater Potential for Resource Recovery—n-Caproate Production from Acid Whey. Green Chem. 2018, 20, 3790–3803. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, W.; Tan, L.; Wang, Y.; He, G. Optimization of Metabolic Pathways for Bioconversion of Lignocellulose to Ethanol through Genetic Engineering. Biotechnol. Adv. 2009, 27, 593–598. [Google Scholar] [CrossRef]
- Cheng, K.-K.; Wang, G.-Y.; Zeng, J.; Zhang, J.-A. Improved Succinate Production by Metabolic Engineering. Biomed Res. Int. 2013, 2013, 538790. [Google Scholar] [CrossRef]
- Spirito, C.M.; Daly, S.E.; Werner, J.J.; Angenent, L.T. Redundancy in Anaerobic Digestion Microbiomes during Disturbances by the Antibiotic Monensin. Appl. Environ. Microbiol. 2018, 84, e02692-17. [Google Scholar] [CrossRef]
- Louca, S.; Polz, M.F.; Mazel, F.; Albright, M.B.N.; Huber, J.A.; O’Connor, M.I.; Ackermann, M.; Hahn, A.S.; Srivastava, D.S.; Crowe, S.A.; et al. Function and Functional Redundancy in Microbial Systems. Nat. Ecol. Evol. 2018, 2, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Girvan, M.S.; Campbell, C.D.; Killham, K.; Prosser, J.I.; Glover, L.A. Bacterial Diversity Promotes Community Stability and Functional Resilience after Perturbation. Environ. Microbiol. 2005, 7, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Stams, A.J.M.; Plugge, C.M. Electron Transfer in Syntrophic Communities of Anaerobic Bacteria and Archaea. Nat. Rev. Microbiol. 2009, 7, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Sieuwerts, S.; De Bok, F.A.M.; Hugenholtz, J.; Van Hylckama Vlieg, J.E.T. Unraveling Microbial Interactions in Food Fermentations: From Classical to Genomics Approaches. Appl. Environ. Microbiol. 2008, 74, 4997–5007. [Google Scholar] [CrossRef]
- Mee, M.T.; Collins, J.J.; Church, G.M.; Wang, H.H. Syntrophic Exchange in Synthetic Microbial Communities. Proc. Natl. Acad. Sci. USA 2014, 111, E2149–E2156. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, R.; van Loosdrecht, M.C. Mixed Culture Biotechnology for Bioenergy Production. Curr. Opin. Biotechnol. 2007, 18, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.Q.; Mu, Y.; Fang, H.H.P. Thermodynamic Analysis of Product Formation in Mesophilic Acidogenesis of Lactose. Biotechnol. Bioeng. 2004, 87, 813–822. [Google Scholar] [CrossRef]
- Liu, B.; Kleinsteuber, S.; Centler, F.; Harms, H.; Sträuber, H. Competition between Butyrate Fermenters and Chain-Elongating Bacteria Limits the Efficiency of Medium-Chain Carboxylate Production. Front. Microbiol. 2020, 11, 336. [Google Scholar] [CrossRef] [PubMed]
- Candry, P.; Ganigué, R. Chain Elongators, Friends, and Foes. Curr. Opin. Biotechnol. 2021, 67, 99–110. [Google Scholar] [CrossRef]
- Prazeres, A.R.; Carvalho, F.; Rivas, J. Cheese Whey Management: A Review. J. Environ. Manag. 2012, 110, 48–68. [Google Scholar] [CrossRef]
- Menchik, P.; Zuber, T.; Zuber, A.; Moraru, C.I. Short Communication: Composition of Coproduct Streams from Dairy Processing: Acid Whey and Milk Permeate. J. Dairy Sci. 2019, 102, 3978–3984. [Google Scholar] [CrossRef]
- Asunis, F.; De Gioannis, G.; Dessì, P.; Isipato, M.; Lens, P.N.L.; Muntoni, A.; Polettini, A.; Pomi, R.; Rossi, A.; Spiga, D. The Dairy Biorefinery: Integrating Treatment Processes for Cheese Whey Valorisation. J. Environ. Manag. 2020, 276, 111240. [Google Scholar] [CrossRef]
- Tarapata, J.; Zieliński, M.; Zulewska, J. Valorization of Dairy By-Products: Efficiency of Energy Production from Biogas Obtained in Anaerobic Digestion of Ultrafiltration Permeates. Energies 2022, 15, 6829. [Google Scholar] [CrossRef]
- Walters, K.A.; Mohan, G.; Myers, K.S.; Ingle, A.T.; Donohue, T.J.; Noguera, D.R. A Metagenome-Level Analysis of a Microbial Community Fermenting Ultra-Filtered Milk Permeate. Front. Bioeng. Biotechnol. 2023, 11, 1173656. [Google Scholar] [CrossRef]
- Biddy, M.J.; Scarlata, C.; Kinchin, C. Chemicals from Biomass: A Market Assessment of Bioproducts with Near-Term Potential. 2016. Available online: www.nrel.gov/publications (accessed on 11 November 2021).
- Dwidar, M.; Park, J.-Y.; Mitchell, R.J.; Sang, B.-I. The Future of Butyric Acid in Industry. Sci. World J. 2012, 2012, 471417. [Google Scholar] [CrossRef]
- Spirito, C.M.; Richter, H.; Rabaey, K.; Stams, A.J.M.; Angenent, L.T. Chain Elongation in Anaerobic Reactor Microbiomes to Recover Resources from Waste. Curr. Opin. Biotechnol. 2014, 27, 115–122. [Google Scholar] [CrossRef]
- Komesu, A.; de Oliveira, J.A.R.; da Silva Martins, L.H.; Maciel, M.R.W.; Filho, R.M. Lactic Acid Production to Purification: A Review. BioResources 2017, 12, 4364–4383. [Google Scholar] [CrossRef]
- Xu, J.; Hao, J.; Guzman, J.J.L.; Spirito, C.M.; Harroff, L.A.; Angenent, L.T. Temperature-Phased Conversion of Acid Whey Waste Into Medium-Chain Carboxylic Acids via Lactic Acid: No External e-Donor. Joule 2018, 2, 280–295. [Google Scholar] [CrossRef]
- Kim, M.S.; Na, J.G.; Lee, M.K.; Ryu, H.; Chang, Y.K.; Triolo, J.M.; Yun, Y.M.; Kim, D.H. More Value from Food Waste: Lactic Acid and Biogas Recovery. Water Res. 2016, 96, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Hollister, E.B.; Forrest, A.K.; Wilkinson, H.H.; Ebbole, D.J.; Tringe, S.G.; Malfatti, S.A.; Holtzapple, M.T.; Gentry, T.J. Mesophilic and Thermophilic Conditions Select for Unique but Highly Parallel Microbial Communities to Perform Carboxylate Platform Biomass Conversion. PLoS ONE 2012, 7, e39689. [Google Scholar] [CrossRef]
- Kim, D.H.; Lim, W.T.; Lee, M.K.; Kim, M.S. Effect of Temperature on Continuous Fermentative Lactic Acid (LA) Production and Bacterial Community, and Development of LA-Producing UASB Reactor. Bioresour. Technol. 2012, 119, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Tada, K.; Kanno, T.; Horiuchi, J.I. Selective Production of Lactic Acid in Continuous Anaerobic Acidogenesis by Extremely Low PH Operation. J. Biosci. Bioeng. 2012, 114, 537–539. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Wang, X.C.; Hu, Y.; Zhang, Y.; Li, Y. Effect of PH on Lactic Acid Production from Acidogenic Fermentation of Food Waste with Different Types of Inocula. Bioresour. Technol. 2017, 224, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Li, H.; Zheng, C. Shifting Product Spectrum by PH Adjustment during Long-Term Continuous Anaerobic Fermentation of Food Waste. Bioresour. Technol. 2018, 270, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Candry, P.; Radic, L.; Favere, J.; Carvajal-arroyo, J.M.; Rabaey, K.; Ganigué, R. Mildly Acidic PH Selects for Chain Elongation to Caproic Acid over Alternative Pathways during Lactic Acid Fermentation. Water Res. 2020, 186, 116396. [Google Scholar] [CrossRef] [PubMed]
- Fortney, N.W.; Hanson, N.J.; Rosa, P.R.F.; Donohue, T.J.; Noguera, D.R. Diverse Profile of Fermentation Byproducts from Thin Stillage. Front. Bioeng. Biotechnol. 2021, 9, 695306. [Google Scholar] [CrossRef] [PubMed]
- Ganigué, R.; Sánchez-Paredes, P.; Bañeras, L.; Colprim, J. Low Fermentation PH Is a Trigger to Alcohol Production, but a Killer to Chain Elongation. Front. Microbiol. 2016, 7, 702. [Google Scholar] [CrossRef] [PubMed]
- Jankowska, E.; Chwiałkowska, J.; Stodolny, M.; Oleskowicz-Popiel, P. Effect of PH and Retention Time on Volatile Fatty Acids Production during Mixed Culture Fermentation. Bioresour. Technol. 2015, 190, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Walters, K.A.; Myers, K.S.; Donohue, T.J.; Noguera, D.R. Metagenome-Assembled Genomes from Microbiomes Fermenting Dairy Coproducts. Microbiol. Resour. Announc. 2024, 13, e0017324. [Google Scholar] [CrossRef]
- Olm, M.R.; Brown, C.T.; Brooks, B.; Banfield, J.F. DRep: A Tool for Fast and Accurate Genomic Comparisons That Enables Improved Genome Recovery from Metagenomes through De-Replication. ISME J. 2017, 11, 2864–2868. [Google Scholar] [CrossRef]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Myers, K.S.; Ingle, A.T.; Walters, K.A.; Fortney, N.W.; Scarborough, M.J.; Donohue, T.J.; Noguera, D.R. Comparison of Metagenomes from Fermentation of Various Agroindustrial Residues Suggests a Common Model of Community Organization. Front. Bioeng. Biotechnol. 2023, 11, 1197175. [Google Scholar] [CrossRef]
- Ingle, A.T.; Fortney, N.W.; Walters, K.A.; Donohue, T.J.; Noguera, D.R. Mixed Acid Fermentation of Carbohydrate-Rich Dairy Manure Hydrolysate. Front. Bioeng. Biotechnol. 2021, 9, 724304. [Google Scholar] [CrossRef] [PubMed]
- Scarborough, M.J.; Lawson, C.E.; Hamilton, J.J.; Donohue, T.J.; Noguera, D.R. Metatranscriptomic and Thermodynamic Insights into Medium-Chain Fatty Acid Production Using an Anaerobic Microbiome. mSystems 2018, 3, e00221-18. [Google Scholar] [CrossRef] [PubMed]
- Fortney, N.W.; Myers, K.S.; Ingle, A.T.; Walters, K.A.; Scarborough, M.J.; Donohue, T.J.; Noguera, D.R. Metagenomes and Metagenome-Assembled Genomes from Microbiomes Metabolizing Thin Stillage from an Ethanol Biorefinery. Microbiol. Resour. Announc. 2022, 11, e0029022. [Google Scholar] [CrossRef]
- Ingle, A.T.; Fortney, N.W.; Myers, K.S.; Walters, K.A.; Scarborough, M.J.; Donohue, T.J.; Noguera, D.R. Metagenome-Assembled Genomes from a Microbiome Grown in Dairy Manure Hydrolysate. Microbiol. Resour. Announc. 2022, 11, e0029222. [Google Scholar] [CrossRef]
- Scarborough, M.J.; Myers, K.S.; Donohue, T.J.; Noguera, D.R. Medium-Chain Fatty Acid Synthesis by “Candidatus Weimeria Bifida” Gen. Nov., Sp. Nov., and “Candidatus Pseudoramibacter Fermentans” Sp. Nov. Appl. Environ. Microbiol. 2020, 86, e02242-19. [Google Scholar] [CrossRef]
- Scarborough, M.J.; Myers, K.S.; Fortney, N.W.; Ingle, A.T.; Donohue, T.J.; Noguera, D.R. Metagenome-Assembled Genomes from a Microbiome Converting Xylose to Medium-Chain Carboxylic Acids. Microbiol. Resour. Announc. 2022, 11, e0115121. [Google Scholar] [CrossRef] [PubMed]
- Walters, K.A.; Myers, K.S.; Wang, H.; Fortney, N.W.; Ingle, A.T.; Scarborough, M.J.; Donohue, T.J.; Noguera, D.R. Metagenomes and Metagenome-Assembled Genomes from Microbial Communities Fermenting Ultrafiltered Milk Permeate. Microbiol. Resour. Announc. 2022, 11, e0029322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Si, S.; Hsieh, C.-J. GPU-Acceleration for Large-Scale Tree Boosting. arXiv 2017, arXiv:1706.08359. [Google Scholar]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and Standardized Annotation of Bacterial Genomes via Alignment-Free Sequence Identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef] [PubMed]
- Gerts, E.M.; Yu, Y.K.; Agarwala, R.; Schäffer, A.A.; Altschul, S.F. Composition-Based Statistics and Translated Nucleotide Searches: Improving the TBLASTN Module of BLAST. BMC Biol. 2006, 4, 41. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Bai, Z.; Xu, T.; He, B. Glucokinase Contributes to Glucose Phosphorylation in D-Lactic Acid Production by Sporolactobacillus Inulinus Y2-8. J. Ind. Microbiol. Biotechnol. 2012, 39, 1685–1692. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Nanda, A.; Khadka, B. Novel Molecular, Structural and Evolutionary Characteristics of the Phosphoketolases from Bifidobacteria and Coriobacteriales. PLoS ONE 2017, 12, e0172176. [Google Scholar] [CrossRef]
- Weghoff, M.C.; Bertsch, J.; Müller, V. A Novel Mode of Lactate Metabolism in Strictly Anaerobic Bacteria. Environ. Microbiol. 2015, 17, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Buckel, W.; Thauer, R.K. Energy Conservation via Electron Bifurcating Ferredoxin Reduction and Proton/Na+ Translocating Ferredoxin Oxidation. Biochim. Biophys. Acta—Bioenerg. 2013, 1827, 94–113. [Google Scholar] [CrossRef] [PubMed]
- Costas, A.M.G.; Poudel, S.; Miller, A.-F.; Schut, G.J.; Ledbetter, R.N.; Fixen, K.R.; Seefeldt, L.C.; Adams, M.W.W.; Harwood, C.S.; Boyd, E.S.; et al. Defining Electron Bifurcation in the Electron-Transferring Flavoprotein Family. J. Bacteriol. 2017, 199, e00440-17. [Google Scholar] [CrossRef]
- Ledbetter, R.N.; Garcia Costas, A.M.; Lubner, C.E.; Mulder, D.W.; Tokmina-Lukaszewska, M.; Artz, J.H.; Patterson, A.; Magnuson, T.S.; Jay, Z.J.; Duan, H.D.; et al. The Electron Bifurcating FixABCX Protein Complex from Azotobacter Vinelandii: Generation of Low-Potential Reducing Equivalents for Nitrogenase Catalysis. Biochemistry 2017, 56, 4177–4190. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Hinderberger, J.; Seedorf, H.; Zhang, J.; Buckel, W.; Thauer, R.K. Coupled Ferredoxin and Crotonyl Coenzyme A (CoA) Reduction with NADH Catalyzed by the Butyryl-CoA Dehydrogenase/Etf Complex from Clostridium Kluyveri. J. Bacteriol. 2008, 190, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D.; Latendresse, M.; Caspi, R. The Pathway Tools Pathway Prediction Algorithm. Stand. Genom. Sci. 2011, 5, 424–429. [Google Scholar] [CrossRef]
- Karp, P.D.; Midford, P.E.; Billington, R.; Kothari, A.; Krummenacker, M.; Latendresse, M.; Ong, W.K.; Subhraveti, P.; Caspi, R.; Fulcher, C.; et al. Pathway Tools Version 23.0 Update: Software for Pathway/Genome Informatics and Systems Biology. Brief. Bioinform. 2021, 22, 109–126. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Bianchini, G.; Sánchez-Baracaldo, P. TreeViewer—Cross-Platform Software to Draw Phylogenetic Trees, Version 2.1.0; CERN: Geneva, Switzerland, 2023. [CrossRef]
- Davis, T.L.; Dirks, B.; Carnero, E.A.; Corbin, K.D.; Krakoff, J.; Parrington, S.; Lee, D.; Smith, S.R.; Rittmann, B.E.; Krajmalnik-Brown, R.; et al. Chemical Oxygen Demand Can Be Converted to Gross Energy for Food Items Using a Linear Regression Model. J. Nutr. 2021, 151, 445–453. [Google Scholar] [CrossRef]
- Zhu, X.; Tao, Y.; Liang, C.; Li, X.; Wei, N.; Zhang, W.; Zhou, Y.; Yang, Y.; Bo, T. The Synthesis of N-Caproate from Lactate: A New Efficient Process for Medium-Chain Carboxylates Production. Sci. Rep. 2015, 5, 14360. [Google Scholar] [CrossRef]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum Information about a Single Amplified Genome (MISAG) and a Metagenome-Assembled Genome (MIMAG) of Bacteria and Archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef]
- Pokusaeva, K.; Fitzgerald, G.F.; Van Sinderen, D. Carbohydrate Metabolism in Bifidobacteria. Genes Nutr. 2011, 6, 285–306. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ruan, L.; Sun, M.; Gänzle, M. A Genomic View of Lactobacilli and Pediococci Demonstrates That Phylogeny Matches Ecology and Physiology. Appl. Environ. Microbiol. 2015, 81, 7233–7243. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A Taxonomic Note on the Genus Lactobacillus: Description of 23 Novel Genera, Emended Description of the Genus Lactobacillus Beijerinck 1901, and Union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.E. Mesophilic and Thermophilic Cultures Used in Traditional Cheesemaking. In Cheese and Microbes; Wiley: Hoboken, NJ, USA, 2014; pp. 73–94. [Google Scholar] [CrossRef]
- Gänzle, M.G. Lactic Metabolism Revisited: Metabolism of Lactic Acid Bacteria in Food Fermentations and Food Spoilage. Curr. Opin. Food Sci. 2015, 2, 106–117. [Google Scholar] [CrossRef]
- Su, F.; Xu, P. Genomic Analysis of Thermophilic Bacillus Coagulans Strains: Efficient Producers for Platform Bio-Chemicals. Sci. Rep. 2014, 4, 3926. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sun, Y.; Liu, Z.; Dong, F.; Li, Y.; Wang, Y. Genome-Scale Modeling for Bacillus coagulans to Understand the Metabolic Characteristics. Biotechnol. Bioeng. 2020, 117, 3545–3558. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gu, Y.; Zhou, W.; Zhao, D.; Qiao, Z.; Zheng, J.; Gao, J.; Chen, X.; Ren, C.; Xu, Y. Adaptability of a Caproate-Producing Bacterium Contributes to Its Dominance in an Anaerobic Fermentation System. Appl. Environ. Microbiol. 2021, 87, e0120321. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Gu, Y.; Zhao, D.; Qiao, Z.; Zheng, J.; Gao, J.; Ren, C.; Xu, Y. Caproicibacterium lactatifermentans Sp. Nov., Isolated from Pit Clay Used for the Production of Chinese Strong Aroma-Type Liquor. Int. J. Syst. Evol. Microbiol. 2022, 72, 005206. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zhu, X.; Lin, F.; Shen, C.; Li, Y.; Ao, L.; Fan, W.; Ren, C.; Xu, Y. Caproicibacterium amylolyticum Gen. Nov., Sp. Nov., a Novel Member of the Family Oscillospiraceae Isolated from Pit Clay Used for Making Chinese Strong Aroma-Type Liquor. Int. J. Syst. Evol. Microbiol. 2021, 71, 004789. [Google Scholar] [CrossRef]
- Brady, C.; Hunter, G.; Kirk, S.; Arnold, D.; Denman, S. Rahnella victoriana Sp. Nov., Rahnella bruchi Sp. Nov., Rahnella woolbedingensis Sp. Nov., Classification of Rahnella Genomospecies 2 and 3 as Rahnella variigena Sp. Nov. and Rahnella inusitata Sp. Nov., Respectiv. Syst. Appl. Microbiol. 2014, 37, 545–552. [Google Scholar] [CrossRef]
- Cotta, M.; Forster, R. The Family Lachnospiraceae, Including the Genera Butyrivibrio, Lachnospira and Roseburia. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 965–981. [Google Scholar] [CrossRef]
- Dwidar, M.; Kim, S.; Jeon, B.S.; Um, Y.; Mitchell, R.J.; Sang, B.I. Co-Culturing a Novel Bacillus Strain with Clostridium tyrobutyricum ATCC 25755 to Produce Butyric Acid from Sucrose. Biotechnol. Biofuels 2013, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.J.; de Fatima Borges, M.; de Freitas Rosa, M.; Castro-Gómez, R.J.H.; Spinosa, W.A. Acetic Acid Bacteria in the Food Industry: Systematics, Characteristics and Applications. Food Technol. Biotechnol. 2018, 56, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Chen, W.J.; Adeolu, M.; Chai, Y. Molecular Signatures for the Class Coriobacteriia and Its Different Clades; Proposal for Division of the Class Coriobacteriia into the Emended Order Coriobacteriales, Containing the Emended Family Coriobacteriaceae and Atopobiaceae fam. nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 3379–3397. [Google Scholar] [CrossRef] [PubMed]
- Scholz, C.F.P.; Kilian, M. The Natural History of Cutaneous Propionibacteria, and Reclassification of Selected Species within the Genus Propionibacterium to the Proposed Novel Genera Acidipropionibacterium Gen. Nov., Cutibacterium Gen. Nov. and Pseudopropionibacterium gen. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 4422–4432. [Google Scholar] [CrossRef] [PubMed]
- DeMoss, R.D.; Bard, R.C.; Gunsalus, I.C. The Mechanism of the Heterolactic Fermentation: A New Route of Ethanol Formation. J. Bacteriol. 1951, 62, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, C.; Assavasirijinda, N.; Yu, B.; Wang, L.; Ma, Y. Non-Sterilized Fermentation of High Optically Pure d-Lactic Acid by a Genetically Modified Thermophilic Bacillus Coagulans Strain. Microb. Cell Fact. 2017, 16, 213. [Google Scholar] [CrossRef]
- Konings, W.N. The Cell Membrane and the Struggle for Life of Lactic Acid Bacteria. Antonie Leeuwenhoek 2002, 82, 3–27. [Google Scholar] [CrossRef] [PubMed]
- Vanwonterghem, I.; Jensen, P.D.; Dennis, P.G.; Hugenholtz, P.; Rabaey, K.; Tyson, G.W. Deterministic Processes Guide Long-Term Synchronised Population Dynamics in Replicate Anaerobic Digesters. ISME J. 2014, 8, 2015–2028. [Google Scholar] [CrossRef] [PubMed]
- Dini-Andreote, F.; Stegen, J.C.; Van Elsas, J.D.; Salles, J.F. Disentangling Mechanisms That Mediate the Balance between Stochastic and Deterministic Processes in Microbial Succession. Proc. Natl. Acad. Sci. USA 2015, 112, E1326–E1332. [Google Scholar] [CrossRef]
- Candry, P.; Chadwick, G.L.; Cara, J.M.; Candry, P.; Chadwick, G.L.; Caravajal-arroyo, J.M.; Lacoere, T.; Winkler, M.H. Trophic Interactions Shape the Spatial Organization of Medium- Chain Carboxylic Acid Producing Granular Biofilm Communities. ISME J. 2023, 17, 2014–2022. [Google Scholar] [CrossRef]
- Chwialkowska, J.; Duber, A.; Zagrodnik, R.; Walkiewicz, F.; Łężyk, M.; Oleskowicz-Popiel, P. Caproic Acid Production from Acid Whey via Open Culture Fermentation—Evaluation of the Role of Electron Donors and Downstream Processing. Bioresour. Technol. 2019, 279, 74–83. [Google Scholar] [CrossRef]
- Carvajal-Arroyo, J.M.; Candry, P.; Andersen, S.J.; Props, R.; Seviour, T.; Ganigué, R.; Rabaey, K. Granular Fermentation Enables High Rate Caproic Acid Production from Solid-Free Thin Stillage. Green Chem. 2019, 21, 1330–1339. [Google Scholar] [CrossRef]
- Crognale, S.; Braguglia, C.M.; Gallipoli, A.; Gianico, A.; Rossetti, S.; Montecchio, D. Direct Conversion of Food Waste Extract into Caproate: Metagenomics Assessment of Chain Elongation Process. Microorganisms 2021, 9, 327. [Google Scholar] [CrossRef]
- Liu, B.; Sträuber, H.; Saraiva, J.; Harms, H.; Silva, S.G.; Kasmanas, J.C.; Kleinsteuber, S.; Nunes da Rocha, U. Machine Learning-Assisted Identification of Bioindicators Predicts Medium-Chain Carboxylate Production Performance of an Anaerobic Mixed Culture. Microbiome 2022, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Belenguer, A.; Duncan, S.H.; Calder, A.G.; Holtrop, G.; Louis, P.; Lobley, G.E.; Flint, H.J. Two Routes of Metabolic Cross-Feeding between Bifidobacterium adolescentis and Butyrate-Producing Anaerobes from the Human Gut. Appl. Environ. Microbiol. 2006, 72, 3593–3599. [Google Scholar] [CrossRef] [PubMed]
- Ulčar, B.; Regueira, A.; Podojsteršek, M.; Boon, N.; Ganigué, R. Why Do Lactic Acid Bacteria Thrive in Chain Elongation Microbiomes? Front. Bioeng. Biotechnol. 2023, 11, 1291007. [Google Scholar] [CrossRef] [PubMed]
- Hayek, S.; Ibrahim, S. Current Limitations and Challenges with Lactic Acid Bacteria: A Review. Food Nutr. Sci. 2013, 4, 73–87. [Google Scholar] [CrossRef]
- Kieliszek, M.; Pobiega, K.; Piwowarek, K.; Kot, A.M. Characteristics of the Proteolytic Enzymes Produced by Lactic Acid Bacteria. Molecules 2021, 26, 1858. [Google Scholar] [CrossRef] [PubMed]
- Teusink, B.; Molenaar, D. Systems Biology of Lactic Acid Bacteria: For Food and Thought. Curr. Opin. Syst. Biol. 2017, 6, 7–13. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | Value a |
|---|---|
| Total chemical oxygen demand (g COD/L) | 63 ± 2 (n = 4) |
| Soluble chemical oxygen demand (g COD/L) | 64 ± 2 (n = 4) |
| Lactose (g COD/L) | 52 ± 3 (n = 4) |
| Lactic acid (g COD/L) | 3 ± 0 (n = 4) |
| Total soluble protein (g COD/L) b | 4 ± 0 (n = 5) |
| Total nitrogen (mg/L) | 680 ± 10 (n = 5) |
| pH | 4.93 ± 0.02 (n = 3) |
| MAG ID | Phylum a | Classification a | Reference Genome a | ANI a | Quality b | Completeness c (%) | Contamination c (%) | MAG Size c (bp) | Contigs c | N50 c (bp) | %GC c | NCBI Genome Accession Number | Reactor of Origin d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ATO1 | Actinomycetota | Tractidigestivibacter scatoligenes | GCF_001494635.1 | 0.9565 | MQ | 98.39 | 0 | 2,241,335 | 40 | 140,862 | 62.75 | GCA_036454705.1 | A2 |
| ATO2 | Actinomycetota | UBA7741 | N/A | N/A | MQ | 98.39 | 0.27 | 2,092,240 | 63 | 45,270 | 69.72 | GCA_036454645.1 | S2 |
| ATO3 | Actinomycetota | UBA7748 sp900314535 | GCA_900314535.1 | 0.9723 | MQ | 95.97 | 0.81 | 2,022,229 | 69 | 34,988 | 60.23 | GCA_036454665.1 | B1 |
| ATO4 | Actinomycetota | Olegusella sp002407925 | GCA_002407925.1 | 0.9865 | MQ | 80.65 | 0 | 1,498,345 | 8 | 252,912 | 55.53 | GCA_036454505.1 | A1 |
| COR1 | Actinomycetota | CAIFEU01 sp903789505 | GCA_903789505.1 | 0.9735 | MQ | 95.16 | 1.41 | 2,881,083 | 38 | 127,573 | 69.49 | GCA_036453905.1 | S1 |
| EGG1 | Actinomycetota | CAIFEB01 sp903789375 | GCA_903789375.1 | 0.9674 | MQ | 97.9 | 0 | 2,264,117 | 63 | 44,458 | 64.89 | GCA_036453845.1 | S2 |
| BIF1 | Actinomycetota | Bifidobacterium mongoliense | GCF_000741285.1 | 0.9757 | MQ | 99.73 | 1.14 | 2,108,758 | 7 | 1,311,668 | 63.18 | GCA_036454425.1 | B3 |
| BIF2 | Actinomycetota | Bifidobacterium crudilactis | GCF_000738005.1 | 0.9749 | HQ | 99.08 | 2.12 | 2,461,645 | 7 | 508,638 | 57.61 | GCA_036454125.1 | B3 |
| BIF3 | Actinomycetota | Bifidobacterium subtile | GCF_000741775.1 | 0.9894 | MQ | 99.44 | 2.73 | 2,748,131 | 16 | 283,464 | 61.21 | GCA_036454005.1 | B1 |
| BIF4 | Actinomycetota | Bifidobacterium thermophilum | GCA_000771265.1 | 0.9551 | MQ | 100 | 0.15 | 2,049,272 | 27 | 111,917 | 60.52 | GCA_036453945.1 | A2 |
| BIF5 | Actinomycetota | Bifidobacterium sp022649135 | GCA_022649135.1 | 0.9934 | MQ | 98.31 | 1.53 | 2,450,011 | 41 | 95,366 | 61.5 | GCA_036453965.1 | A1 |
| PROP1 | Actinomycetota | Acidipropionibacterium acidipropionici | GCF_001441165.1 | 0.9883 | MQ | 100 | 0 | 3,480,401 | 29 | 187,533 | 68.94 | GCA_036452805.1 | A3 |
| LAC1 | Bacillota | Lactobacillus delbrueckii | GCF_001433875.1 | 0.9759 | MQ | 95.78 | 0 | 1,796,983 | 54 | 45,732 | 50.08 | GCA_036453405.1 | A1 |
| LAC2 | Bacillota | Lactobacillus absiana | GCA_017309015.1 | 0.9782 | MQ | 79.99 | 0.97 | 1,391,865 | 75 | 19,352 | 52.73 | GCA_036455685.1 | A1 |
| LAC3 | Bacillota | Lactobacillus amylovorus | GCF_002706375.1 | 0.9756 | MQ | 79.28 | 0.09 | 1,343,105 | 73 | 21,953 | 38.61 | GCA_036453345.1 | A1 |
| LACCAS1 | Bacillota | Lacticaseibacillus paracasei | GCF_000829035.1 | 0.9836 | HQ | 99.46 | 0 | 2,787,648 | 39 | 102,065 | 46.5 | GCA_036453305.1 | A3 |
| LACCAS2 | Bacillota | Lacticaseibacillus rhamnosus | GCF_900636965.1 | 0.9749 | MQ | 80.98 | 0 | 2,071,044 | 23 | 132,814 | 46.82 | GCA_036453285.1 | B1 |
| LEUC1 | Bacillota | Leuconostoc gelidum | GCF_000166715.1 | 0.9875 | MQ | 99.44 | 0.19 | 1,870,646 | 8 | 442,939 | 36.63 | GCA_036453045.1 | B2 |
| LEUC2 | Bacillota | Leuconostoc mesenteroides | GCF_000014445.1 | 0.9903 | MQ | 100 | 0 | 1,800,527 | 24 | 92,862 | 37.7 | GCA_036452885.1 | A2 |
| LENLAC1 | Bacillota | Lentilactobacillus sunkii | GCF_001435575.1 | 0.966 | MQ | 98.89 | 0 | 2,748,557 | 25 | 142,530 | 42.13 | GCA_036453105.1 | B1 |
| STREP1 | Bacillota | Lactococcus lactis | GCF_900099625.1 | 0.9846 | MQ | 100 | 0.28 | 2,510,060 | 20 | 233,864 | 34.87 | GCA_036452675.1 | A2 |
| STREP2 | Bacillota | Lactococcus cremoris | GCF_001591705.1 | 0.9826 | MQ | 85.85 | 0.38 | 1,744,274 | 56 | 36,778 | 35.7 | GCA_036452495.1 | B1 |
| BACIL1 | Bacillota | Weizmannia coagulans | GCF_000290615.1 | 0.9832 | MQ | 91.61 | 0 | 2,447,334 | 70 | 48,544 | 47.87 | GCA_036454485.1 | B2 |
| ERY1 | Bacillota | Bulleidia | N/A | N/A | MQ | 96.19 | 0.45 | 2,113,669 | 59 | 50,393 | 47.89 | GCA_036453465.1 | A2 |
| ERY2 | Bacillota | Bulleidia sp900319505 | GCA_900319505.1 | 0.9523 | MQ | 92.52 | 0.32 | 1,691,123 | 66 | 30,478 | 47.86 | GCA_036453425.1 | S1 |
| LCO1 | Bacillota A | Bilifractor sp900319355 | GCA_900319355.1 | 0.9934 | MQ | 97.13 | 0.32 | 2,309,795 | 40 | 80,513 | 52.52 | GCA_036453235.1 | S2 |
| LCO2 | Bacillota A | CAG-791 sp900315055 | GCA_900315055.1 | 0.9889 | MQ | 94.91 | 0.4 | 2,906,087 | 106 | 37,633 | 55.88 | GCA_036453225.1 | A2 |
| LCO3 | Bacillota A | Bilifractor | N/A | N/A | MQ | 91.56 | 1.27 | 2,395,184 | 17 | 174,318 | 50.59 | GCA_036453165.1 | S1 |
| LCO4 | Bacillota A | UBA1066 | N/A | N/A | MQ | 89.81 | 0 | 2,176,431 | 67 | 54,455 | 55.06 | GCA_036453195.1 | A2 |
| ACUT1 | Bacillota A | Caproicibacterium sp022483045 | GCA_022483045.1 | 0.9999 | MQ | 82.55 | 0 | 1,598,564 | 22 | 86,104 | 51.34 | GCA_036454745.1 | A3 |
| ACUT2 | Bacillota A | Caproicibacterium sp902809935 | GCA_902809935.1 | 0.9948 | MQ | 82.7 | 0.34 | 1,580,463 | 82 | 22,595 | 42.65 | GCA_036454725.1 | S2 |
| ENTER1 | Pseudomonadota | Rahnella inusitata | GCF_003263515.1 | 0.9896 | MQ | 98.03 | 0.08 | 5,063,486 | 10 | 2,624,760 | 53.06 | GCA_036453825.1 | S2 |
| ACET1 | Pseudomonadota | Acetobacter sp012517935 | GCA_012517935.1 | 0.958 | MQ | 100 | 0.25 | 2,793,541 | 28 | 121,035 | 53.7 | GCA_036452275.1 | A2 |
| PSEU1 | Pseudomonadota | Pseudomonas E helleri | GCF_001043025.1 | 0.9728 | MQ | 75.79 | 0 | 5,159,292 | 131 | 53,203 | 58.58 | GCA_036452715.1 | S2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walters, K.A.; Myers, K.S.; Ingle, A.T.; Donohue, T.J.; Noguera, D.R. Effect of Temperature and pH on Microbial Communities Fermenting a Dairy Coproduct Mixture. Fermentation 2024, 10, 422. https://doi.org/10.3390/fermentation10080422
Walters KA, Myers KS, Ingle AT, Donohue TJ, Noguera DR. Effect of Temperature and pH on Microbial Communities Fermenting a Dairy Coproduct Mixture. Fermentation. 2024; 10(8):422. https://doi.org/10.3390/fermentation10080422
Chicago/Turabian StyleWalters, Kevin A., Kevin S. Myers, Abel T. Ingle, Timothy J. Donohue, and Daniel R. Noguera. 2024. "Effect of Temperature and pH on Microbial Communities Fermenting a Dairy Coproduct Mixture" Fermentation 10, no. 8: 422. https://doi.org/10.3390/fermentation10080422
APA StyleWalters, K. A., Myers, K. S., Ingle, A. T., Donohue, T. J., & Noguera, D. R. (2024). Effect of Temperature and pH on Microbial Communities Fermenting a Dairy Coproduct Mixture. Fermentation, 10(8), 422. https://doi.org/10.3390/fermentation10080422






