Optimized Amino Acid-Enhanced Medium for Efficient L-Asparaginase II Production in E. coli: From Shake Flask to Bioreactor
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganism, Pre-Inoculum Preparation, and Fermentation Conditions
2.1.1. Inoculum Preparation and Cultivation Conditions
2.1.2. Shake Flask Experiments for 25 Full Factorial Design
2.1.3. Shake Flask Experiments for Central Composite Rotatable Design (CCRD)
2.2. Bioreactor Experiments
Soluble L-ASNase Extraction
2.3. Statistical Analysis and Modeling
2.3.1. 25 Full Factorial Design
2.3.2. Central Composite Rotatable Design (CCRD)
2.4. Analyses
2.4.1. L-ASNase Activity Assay
2.4.2. Determination of Cell Dry Weight
2.4.3. Glucose Quantification
2.5. Extraction of Metabolites from Spent Media
2.6. Determination of Amino Acids by LC-MS/MS
3. Results and Discussion
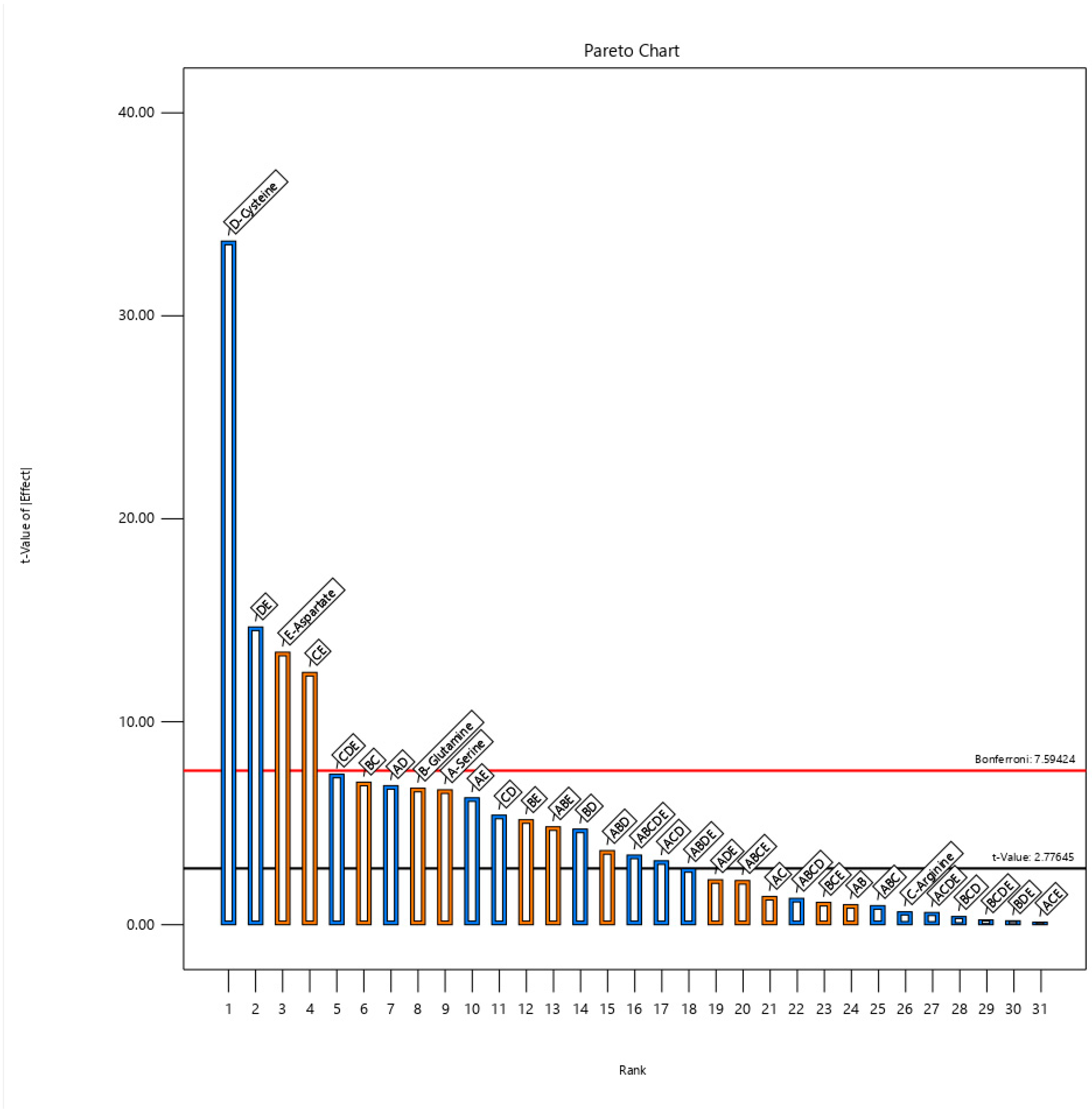
3.1. Screening of Amino Acids Using 25 Full Factorial Design
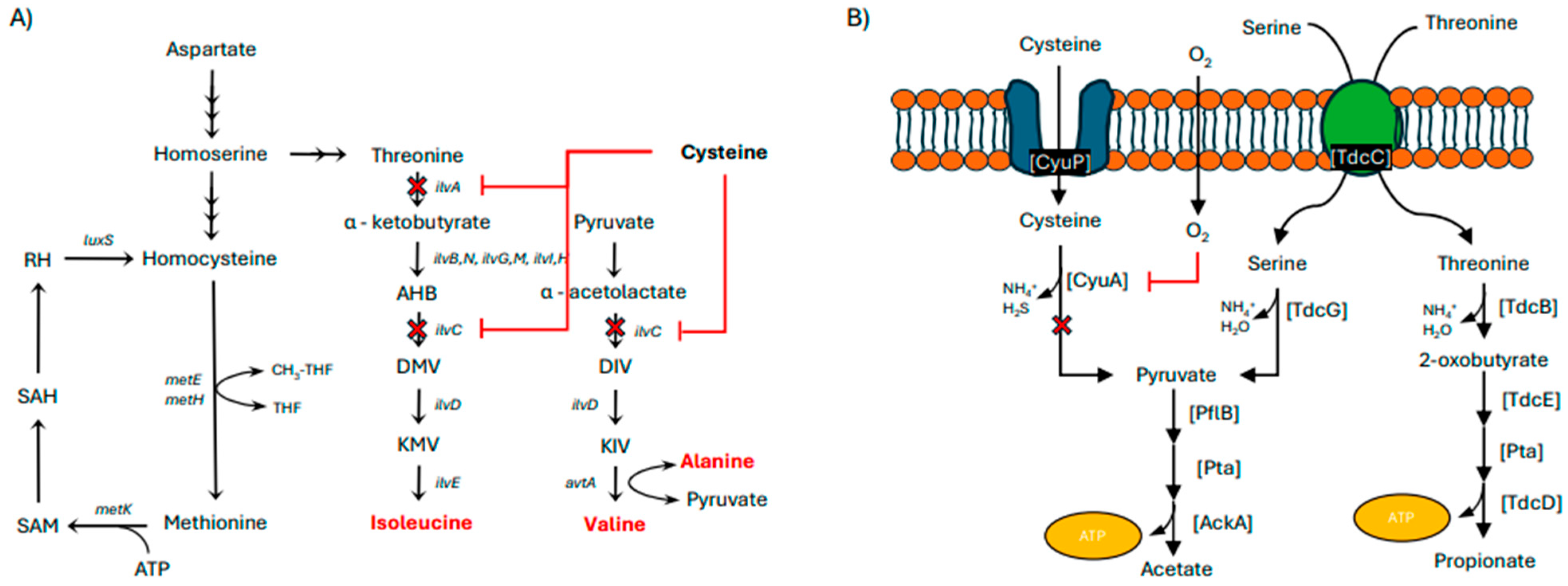
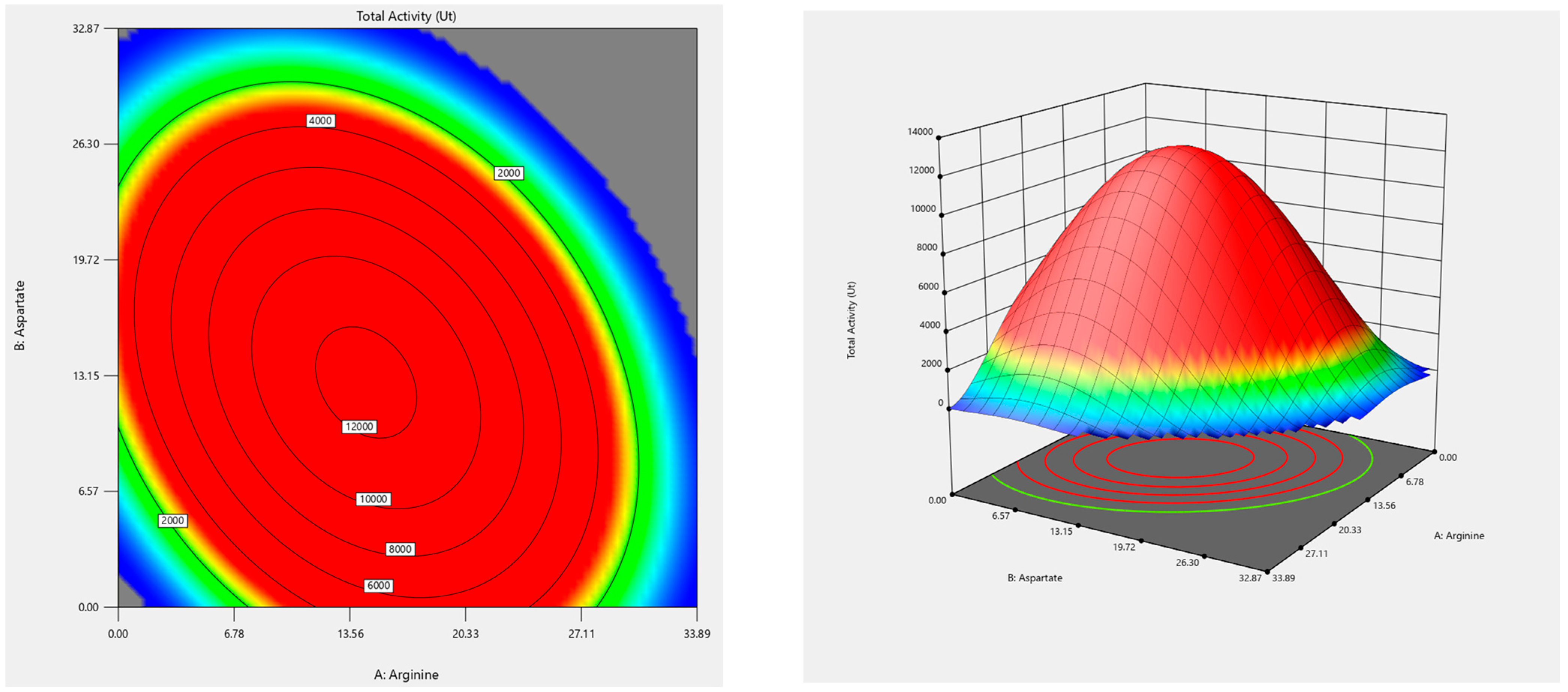
3.2. Optimization of Amino Acids in Defined Medium
3.3. Validation
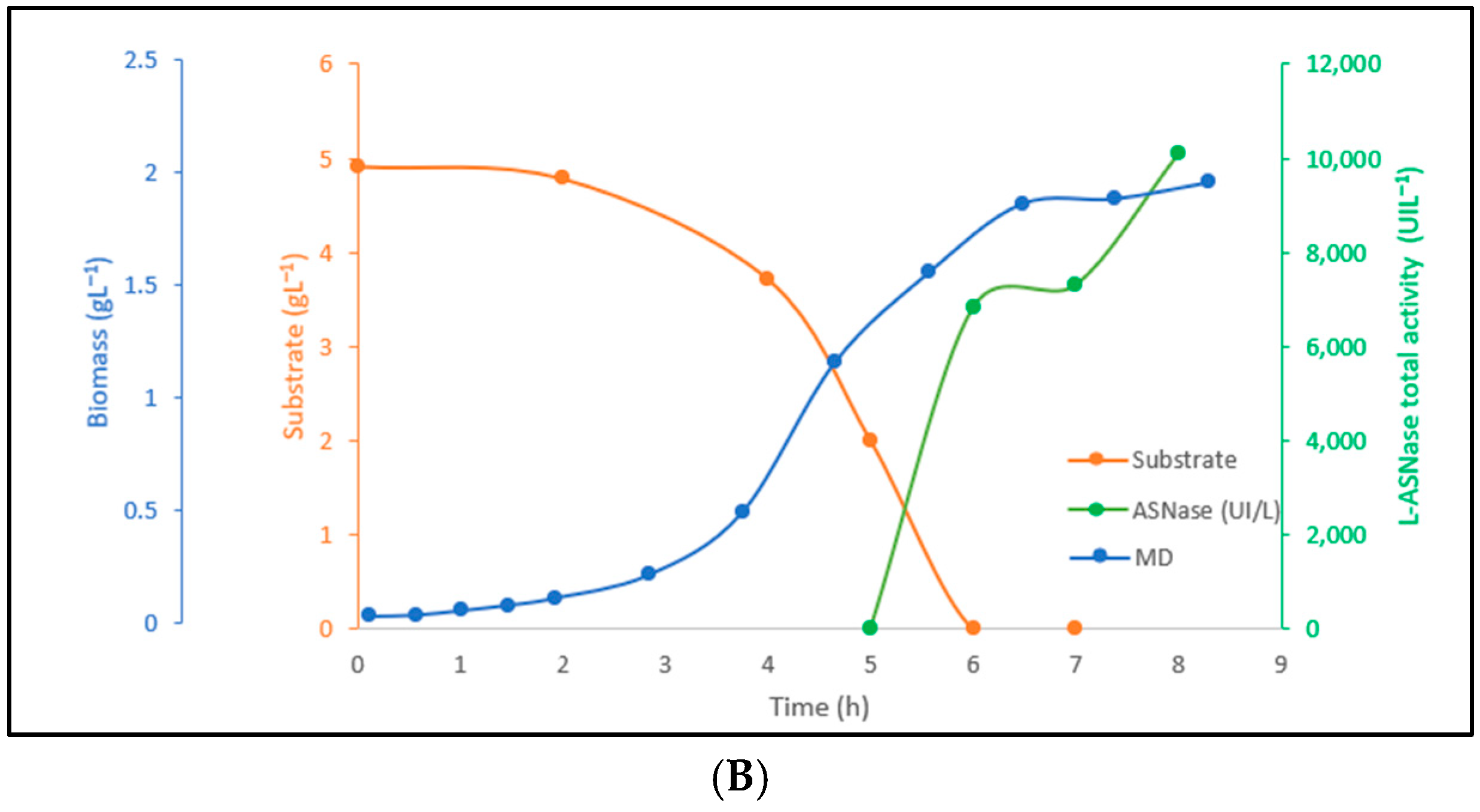
3.4. Evaluation of Induction Conditions and Amino Acids Addition in Bioreactor
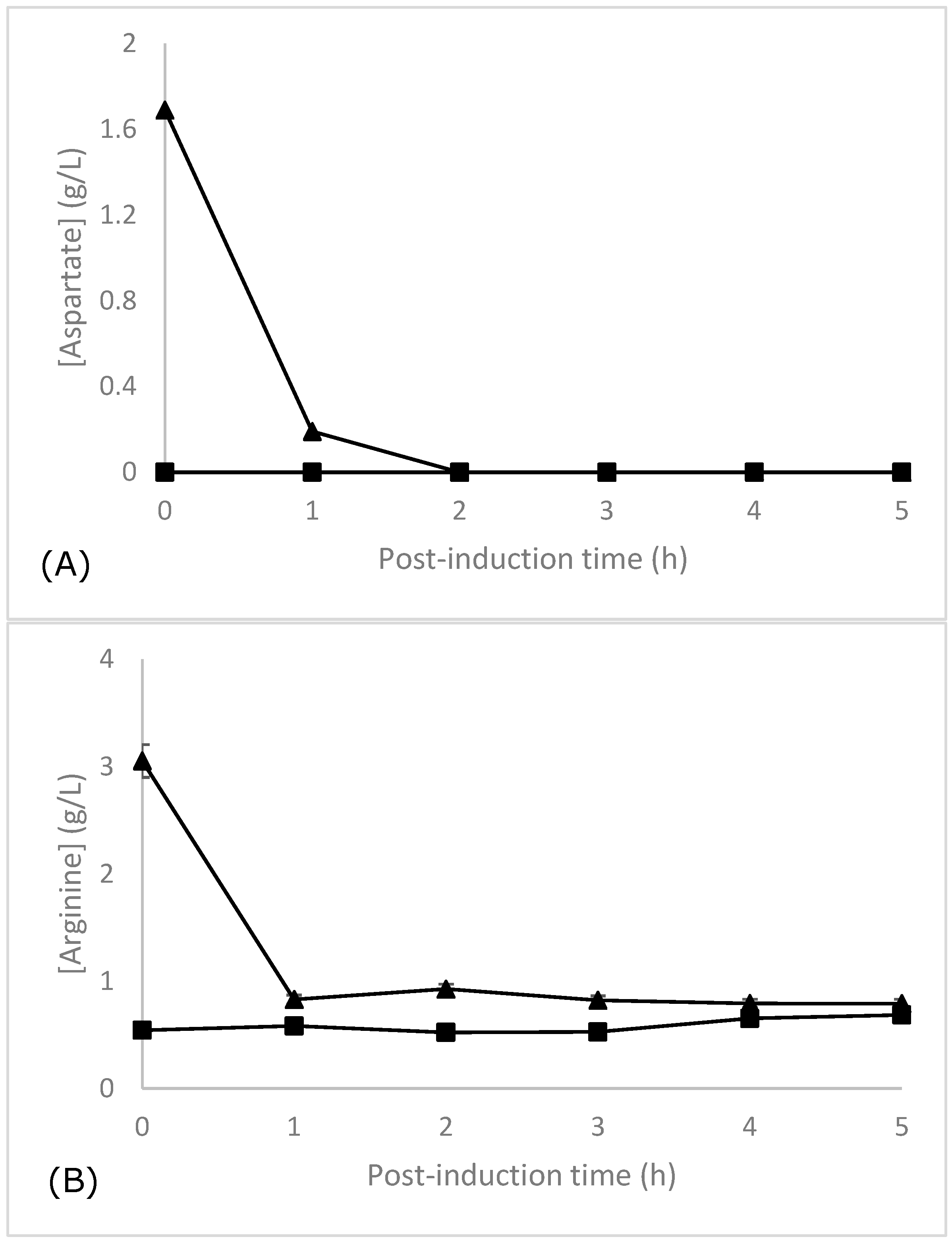
3.5. Amino Acid Analysis by LC-MS/MS
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Xu, W.; Wu, H.; Zhang, W.; Guang, C.; Mu, W. Microbial production, molecular modification, and practical application of l-Asparaginase: A review. Int. J. Biol. Macromol. 2021, 186, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Jangra, S.; Srivastava, S. Microbial Enzymes in Food Industries: Enhancing Quality and Sustainability. In Food Microbial Sustainability: Integration of Food Production and Food Safety; Karnwal, A., Mohammad Said Al-Tawaha, A.R., Eds.; Springer Nature: Singapore, 2023; pp. 193–221. [Google Scholar] [CrossRef]
- Ofori-Boateng, C. (Ed.) Global Market Profile of Bioproducts from Thermochemical Conversion Technologies. In Sustainability of Thermochemical Waste Conversion Technologies; Springer International Publishing: Cham, Switzerland, 2024; pp. 29–54. [Google Scholar] [CrossRef]
- Chand, S.; Mahajan, R.V.; Prasad, J.P.; Sahoo, D.K.; Mihooliya, K.N.; Dhar, M.S.; Sharma, G. A comprehensive review on microbial l-asparaginase: Bioprocessing, characterization, and industrial applications. Biotechnol. Appl. Biochem. 2020, 67, 619–647. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, V.M.; Campani, G.; Santos, M.P.; Silva, G.G.; Pires, M.C.; Gonçalves, V.M.; Giordano, R.d.C.; Sargo, C.R.; Horta, A.C.; Zangirolami, T.C. Cost analysis based on bioreactor cultivation conditions: Production of a soluble recombinant protein using Escherichia coli BL21(DE3). Biotechnol. Rep. 2020, 26, e00441. [Google Scholar] [CrossRef]
- Tripathi, N.K.; Shrivastava, A. Recent Developments in Bioprocessing of Recombinant Proteins: Expression Hosts and Process Development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef]
- Kante, R.K.; Vemula, S.; Somavarapu, S.; Mallu, M.R.; Boje Gowd, B.H.; Ronda, S.R. Optimized upstream and downstream process conditions for the improved production of recombinant human asparaginase (rhASP) from Escherichia coli and its characterization. Biologicals 2018, 56, 45–53. [Google Scholar] [CrossRef]
- Rosano, G.L.; Morales, E.S.; Ceccarelli, E.A. New tools for recombinant protein production in Escherichia coli: A 5-year update. Protein Sci. 2019, 28, 1412–1422. [Google Scholar] [CrossRef]
- Tripathi, N.K. Production and Purification of Recombinant Proteins from Escherichia coli. ChemBioEng Rev. 2016, 3, 116–133. [Google Scholar] [CrossRef]
- Doran, P.M. Bioprocess Engineering Principles; Elsevier: Amsterdam, The Netherlands, 1995. [Google Scholar]
- Gamboa-Suasnavart, R.A.; Marín-Palacio, L.D.; Martínez-Sotelo, J.A.; Espitia, C.; Servín-González, L.; Valdez-Cruz, N.A.; Trujillo-Roldán, M.A. Scale-up from shake flasks to bioreactor, based on power input and Streptomyceslividans morphology, for the production of recombinant APA (45/47 kDa protein) from Mycobacteriumtuberculosis. World J. Microbiol. Biotechnol. 2013, 29, 1421–1429. [Google Scholar] [CrossRef]
- Behravan, A.; Hashemi, A.; Marashi, S.A. A Constraint-based modeling approach to reach an improved chemically defined minimal medium for recombinant antiEpEX-scFv production by Escherichia coli. Biochem. Eng. J. 2022, 179, 108339. [Google Scholar] [CrossRef]
- Kumar, J.; Chauhan, A.S.; Shah, R.L.; Gupta, J.A.; Rathore, A.S. Amino acid supplementation for enhancing recombinant protein production in E. coli. Biotechnol. Bioeng. 2020, 117, 2420–2433. [Google Scholar] [CrossRef]
- Maser, A.; Peebo, K.; Vilu, R.; Nahku, R. Amino acids are key substrates to Escherichia coli BW25113 for achieving high specific growth rate. Res. Microbiol. 2020, 171, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Kaleta, C.; Schäuble, S.; Rinas, U.; Schuster, S. Metabolic costs of amino acid and protein production in Escherichia coli. Biotechnol. J. 2013, 8, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Brumano, L.P.; da Silva, F.V.S.; Costa-Silva, T.A.; Apolinário, A.C.; Santos, J.H.P.M.; Kleingesinds, E.K.; Monteiro, G.; Rangel-Yagui, C.d.O.; Benyahia, B.; Junior, A.P. Development of L-Asparaginase Biobetters: Current Research Status and Review of the Desirable Quality Profiles. Front. Bioeng. Biotechnol. 2019, 6, 212. [Google Scholar] [CrossRef]
- Papaneophytou, C. Design of Experiments As a Tool for Optimization in Recombinant Protein Biotechnology: From Constructs to Crystals. Mol. Biotechnol. 2019, 61, 873–891. [Google Scholar] [CrossRef]
- Uhoraningoga, A.; Kinsella, G.K.; Henehan, G.T.; Ryan, B.J. The goldilocks approach: A review of employing design of experiments in prokaryotic recombinant protein production. Bioengineering 2018, 5, 89. [Google Scholar] [CrossRef]
- Lee, K.M.; Gilmore, D.F. Statistical experimental design for bioprocess modeling and optimization analysis. Appl. Biochem. Biotechnol. 2006, 135, 101–115. [Google Scholar] [CrossRef]
- Shakambari, G.; Ashokkumar, B.; Varalakshmi, P. L-asparaginase—A promising biocatalyst for industrial and clinical applications. Biocatal. Agric. Biotechnol. 2019, 17, 213–224. [Google Scholar] [CrossRef]
- Battistel, A.P.; da Rocha, B.S.; dos Santos, M.T.; Daudt, L.E.; Michalowski, M.B. Allergic reactions to asparaginase: Retrospective cohort study in pediatric patients with acute lymphoid leukemia. Hematol. Transfus. Cell Ther. 2021, 43, 9–14. [Google Scholar] [CrossRef]
- Jia, R.; Wan, X.; Geng, X.; Xue, D.; Xie, Z.; Chen, C. Microbial L-asparaginase for application in acrylamide mitigation from food: Current research status and future perspectives. Microorganisms 2021, 9, 1659. [Google Scholar] [CrossRef]
- Burke, M.J. How to manage asparaginase hypersensitivity in acute lymphoblastic leukemia. Future Oncol. 2014, 10, 2615–2627. [Google Scholar] [CrossRef]
- Parizotto, L.d.A.; Kleingesinds, E.K.; da Rosa, L.M.P.; Effer, B.; Lima, G.M.; Herkenhoff, M.E.; Li, Z.; Rinas, U.; Monteiro, G.; Pessoa, A.; et al. Increased glycosylated L-asparaginase production through selection of Pichia pastoris platform and oxygen-methanol control in fed-batches. Biochem. Eng. J. 2021, 173, 108083. [Google Scholar]
- Effer, B.; Kleingesinds, E.K.; Lima, G.M.; Costa, I.M.; Sánchez-Moguel, I.; Pessoa, A.; Santiago, V.F.; Palmisano, G.; Farías, J.G.; Monteiro, G. Glycosylation of Erwinase results in active protein less recognized by antibodies. Biochem. Eng. J. 2020, 163, 107750. [Google Scholar] [CrossRef]
- Kishore, V.; Nishita, K.P.; Manonmani, H.K. Cloning, expression and characterization of l-asparaginase from Pseudomonas fluorescens for large scale production in E. coli BL21. 3 Biotech 2015, 5, 975–981. [Google Scholar] [CrossRef]
- Costa, I.M.; Moura, D.C.; Lima, G.M.; Pessoa, A.; dos Santos, C.O.; de Oliveira, M.A.; Monteiro, G. Engineered asparaginase from enhances asparagine hydrolase activity and diminishes enzyme immunoreactivity—A new promise to treat acute lymphoblastic leukemia. J. Chem. Technol. Biotechnol. 2022, 97, 228–239. [Google Scholar] [CrossRef]
- Riesenberg, D.; Schulz, V.; Knorre, W.A.; Pohl, H.D.; Korz, D.; Sanders, E.A.; Roß, A.; Deckwer, W.-D. High cell density cultivation of Escherichia coli at controlled specific growth rate. J. Biotechnol. 1991, 20, 17–27. [Google Scholar] [CrossRef]
- Bren, A.; Park, J.O.; Towbin, B.D.; Dekel, E.; Rabinowitz, J.D.; Alon, U. Glucose becomes one of the worst carbon sources for E. coli on poor nitrogen sources due to suboptimal levels of cAMP. Sci. Rep. 2016, 6, 24834. [Google Scholar] [CrossRef]
- Simas, R.G.; Krebs Kleingesinds, E.; Pessoa Junior, A.; Long, P.F. An improved method for simple and accurate colorimetric determination of l-asparaginase enzyme activity using Nessler’s reagent. J. Chem. Technol. Biotechnol. 2021, 96, 1326–1332. [Google Scholar] [CrossRef]
- Magri, A.; Soler, M.F.; Lopes, A.M.; Cilli, E.M.; Barber, P.S.; Pessoa, A.; Pereira, J.F.B. A critical analysis of L-asparaginase activity quantification methods—Colorimetric methods versus high-performance liquid chromatography. Anal. Bioanal. Chem. 2018, 410, 6985–6990. [Google Scholar] [CrossRef]
- Santos, J.C.F. Cultivo de Escherichia coli BL21 (DE3) para Produção de L-Asparaginase II. Master’s Thesis, Faculdade de Ciências Farmacêuticas, Universidade de São Paulo, São Paulo, Brazil, 2017. [Google Scholar]
- Thomas, J.A.; Schlender, K.K.; Larner, J. A rapid filter paper assay for UDPglucose-glycogen glucosyltransferase, including an improved biosynthesis of UDP-14C-glucose. Anal. Biochem. 1968, 25, 486–499. [Google Scholar] [CrossRef]
- Zhou, Y.; Imlay, J.A. Escherichia coli Uses a Dedicated Importer and Desulfidase to Ferment Cysteine. mBio 2022, 13, e02965-21. [Google Scholar] [CrossRef]
- Harris, C.L. Cysteine and growth inhibition of Escherichia coli: Threonine deaminase as the target enzyme. J. Bacteriol. 1981, 145, 1031–1035. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, M.A.; Pedersen, S. Cysteine, even in low concentrations, induces transient amino acid starvation in Escherichia coli. J. Bacteriol. 1991, 173, 5244–5246. [Google Scholar] [CrossRef] [PubMed]
- Tuite, N.L.; Fraser, K.R.; O’Byrne, C.P. Homocysteine Toxicity in Escherichia coli Is Caused by a Perturbation of Branched-Chain Amino Acid Biosynthesis. J. Bacteriol. 2005, 187, 4362–4371. [Google Scholar] [CrossRef]
- Reitzer, L. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 2003, 57, 155–176. [Google Scholar] [CrossRef]
- Schneider, B.L.; Kiupakis, A.K.; Reitzer, L.J. Arginine Catabolism and the Arginine Succinyltransferase Pathway in Escherichia coli. J. Bacteriol. 1998, 180, 4278–4286. [Google Scholar] [CrossRef]
- Strecker, A.; Schubert, C.; Zedler, S.; Steinmetz, P.; Unden, G. DcuA of aerobically grown Escherichia coli serves as a nitrogen shuttle (L-aspartate/fumarate) for nitrogen uptake. Mol. Microbiol. 2018, 109, 801–811. [Google Scholar] [CrossRef]
- Schubert, C. L-Aspartate Is a High-Quality Nitrogen Source of Escherichia coli: Regulation and Physiology. Ph.D. Thesis, Johannes Gutenberg-Universität Mainz, Mainz, Germany, 2021. [Google Scholar]
- Barros, T.d.S. Desenvolvimento de Processos para Produção em Escala da Enzima L-Asparaginase Por Escherichia coli BL21 (DE3) AspB. Master’s Thesis, Universidade de Brasília, Brasília, Brazil, 2020. [Google Scholar]
- Flores-Santos, J.C.; Moguel, I.S.; Monteiro, G.; Pessoa, A.; Vitolo, M. Improvement in extracellular secretion of recombinant l-asparaginase II by Escherichia coli BL21 (DE3) using glycine and n-dodecane. Braz. J. Microbiol. 2021, 52, 1247–1255. [Google Scholar] [CrossRef]
- Guennadi, S.; Danièle, J.P.; Richard, D. Escherichia coli Physiology in Luria-Bertani Broth. J. Bacteriol. 2007, 189, 8746–8749. [Google Scholar] [CrossRef]



| Conditions | Control | Set 1 | Set 2 | Set 3 |
|---|---|---|---|---|
| AA’s addition phase | - | Initial culture | Middle exponential | Final exponential |
| AA’s concentration | - | 1X | 1X | 1X |
| IPTG addition phase | Middle exponential | Middle exponential | Middle exponential | Final exponential |
| Variables | Code | Experimental Values (mM) | |
|---|---|---|---|
| Lower (−1) | Upper (+1) | ||
| Essential amino acids | |||
| Serine | A | 2.5 | 7.5 |
| Glutamine | B | 2.5 | 7.5 |
| Arginine | C | 2.5 | 7.5 |
| Cysteine | D | 2.5 | 7.5 |
| Aspartate | E | 2.5 | 7.5 |
| Code | Factors | Range y Levels | ||||
|---|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | ||
| A | Arginine (mM) | 0.1 | 2.5 | 5 | 7.5 | 9.9 |
| B | Aspartate (mM) | 0.1 | 2.5 | 5 | 7.5 | 9.9 |
| C | Cysteine (mM) | 0.1 | 2.5 | 5 | 7.5 | 9.9 |
| Std | Run | A:Arginine | D:Aspartate | E:Cysteine | Total Activity |
|---|---|---|---|---|---|
| mM | mM | mM | UT | ||
| 1 | 15 | 2.5 | 2.5 | 2.5 | 288 |
| 2 | 14 | 7.5 | 2.5 | 2.5 | 2165 |
| 3 | 18 | 2.5 | 7.5 | 2.5 | 1818 |
| 4 | 19 | 7.5 | 7.5 | 2.5 | 3044 |
| 5 | 11 | 2.5 | 2.5 | 7.5 | 207 |
| 6 | 7 | 7.5 | 2.5 | 7.5 | 27 |
| 7 | 17 | 2.5 | 7.5 | 7.5 | 50 |
| 8 | 13 | 7.5 | 7.5 | 7.5 | 57 |
| 9 | 20 | 0.1 | 5 | 5 | 230 |
| 10 | 9 | 9.9 | 5 | 5 | 786 |
| 11 | 3 | 5 | 0.1 | 5 | 270 |
| 12 | 6 | 5 | 9.9 | 5 | 811 |
| 13 | 5 | 5 | 5 | 0.1 | 3228 |
| 14 | 10 | 5 | 5 | 9.9 | 56 |
| 15 | 1 | 5 | 5 | 5 | 686 |
| 16 | 16 | 5 | 5 | 5 | 917 |
| 17 | 8 | 5 | 5 | 5 | 607 |
| 18 | 2 | 5 | 5 | 5 | 754 |
| 19 | 4 | 5 | 5 | 5 | 840 |
| 20 | 12 | 5 | 5 | 5 | 874 |
| Std | Run | A:Serine | B:Glutamine | C:Arginine | D:Cysteine | E:Aspartate | Total Activity |
|---|---|---|---|---|---|---|---|
| mM | mM | mM | mM | mM | UT | ||
| 1 | 11 | −1.000 | −1.000 | −1.000 | −1.000 | −1.000 | 1318 |
| 2 | 24 | 1.000 | −1.000 | −1.000 | −1.000 | −1.000 | 2272 |
| 3 | 28 | −1.000 | 1.000 | −1.000 | −1.000 | −1.000 | 1526 |
| 4 | 13 | 1.000 | 1.000 | −1.000 | −1.000 | −1.000 | 2063 |
| 5 | 5 | −1.000 | −1.000 | 1.000 | −1.000 | −1.000 | 49 |
| 6 | 1 | 1.000 | −1.000 | 1.000 | −1.000 | −1.000 | 1778 |
| 7 | 31 | −1.000 | 1.000 | 1.000 | −1.000 | −1.000 | 1201 |
| 8 | 16 | 1.000 | 1.000 | 1.000 | −1.000 | −1.000 | 1640 |
| 9 | 15 | −1.000 | −1.000 | −1.000 | 1.000 | −1.000 | 1074 |
| 10 | 3 | 1.000 | −1.000 | −1.000 | 1.000 | −1.000 | 1129 |
| 11 | 8 | −1.000 | 1.000 | −1.000 | 1.000 | −1.000 | 562 |
| 12 | 29 | 1.000 | 1.000 | −1.000 | 1.000 | −1.000 | 913 |
| 13 | 26 | −1.000 | −1.000 | 1.000 | 1.000 | −1.000 | 376 |
| 14 | 17 | 1.000 | −1.000 | 1.000 | 1.000 | −1.000 | 426 |
| 15 | 23 | −1.000 | 1.000 | 1.000 | 1.000 | −1.000 | 426 |
| 16 | 10 | 1.000 | 1.000 | 1.000 | 1.000 | −1.000 | 609 |
| 17 | 27 | −1.000 | −1.000 | −1.000 | −1.000 | 1.000 | 1949 |
| 18 | 25 | 1.000 | −1.000 | −1.000 | −1.000 | 1.000 | 1993 |
| 19 | 32 | −1.000 | 1.000 | −1.000 | −1.000 | 1.000 | 2328 |
| 20 | 36 | 1.000 | 1.000 | −1.000 | −1.000 | 1.000 | 2284 |
| 21 | 6 | −1.000 | −1.000 | 1.000 | −1.000 | 1.000 | 2649 |
| 22 | 20 | 1.000 | −1.000 | 1.000 | −1.000 | 1.000 | 2614 |
| 23 | 33 | −1.000 | 1.000 | 1.000 | −1.000 | 1.000 | 3260 |
| 24 | 9 | 1.000 | 1.000 | 1.000 | −1.000 | 1.000 | 4131 |
| 25 | 19 | −1.000 | −1.000 | −1.000 | 1.000 | 1.000 | 917 |
| 26 | 21 | 1.000 | −1.000 | −1.000 | 1.000 | 1.000 | 421 |
| 27 | 18 | −1.000 | 1.000 | −1.000 | 1.000 | 1.000 | 473 |
| 28 | 30 | 1.000 | 1.000 | −1.000 | 1.000 | 1.000 | 824 |
| 29 | 12 | −1.000 | −1.000 | 1.000 | 1.000 | 1.000 | 522 |
| 30 | 35 | 1.000 | −1.000 | 1.000 | 1.000 | 1.000 | 108 |
| 31 | 4 | −1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 991 |
| 32 | 7 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 845 |
| 33 | 34 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 952 |
| 34 | 2 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 944 |
| 35 | 14 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 745 |
| 36 | 22 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 743 |
| Sum of Squares | df | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|
| Model | 2.948 × 107 | 31 | 9.511 × 105 | 68.53 | 0.0025 b |
| A-Serine | 6.128 × 105 | 1 | 6.128 × 105 | 44.16 | 0.0069 b |
| B-Glutamine | 6.269 × 105 | 1 | 6.269 × 105 | 45.17 | 0.0067 b |
| C-Arginine | 5551.25 | 1 | 5551.25 | 0.4000 | 0.5720 a |
| D-Cysteine | 1.574 × 107 | 1 | 1.574 × 107 | 1133.99 | <0.0001 b |
| E-Aspartic Acid | 2.500 × 106 | 1 | 2.500 × 106 | 180.10 | 0.0009 b |
| AB | 13,442.20 | 1 | 13,442.20 | 0.9685 | 0.3976 a |
| AC | 26,771.85 | 1 | 26,771.85 | 1.93 | 0.2590 a |
| AD | 6.504 × 105 | 1 | 6.504 × 105 | 46.87 | 0.0064 b |
| AE | 5.424 × 105 | 1 | 5.424 × 105 | 39.08 | 0.0083 b |
| BC | 6.840 × 105 | 1 | 6.840 × 105 | 49.29 | 0.0059 b |
| BD | 3.082 × 105 | 1 | 3.082 × 105 | 22.21 | 0.0181 b |
| BE | 3.708 × 105 | 1 | 3.708 × 105 | 26.72 | 0.0141 b |
| CD | 4.041 × 105 | 1 | 4.041 × 105 | 29.12 | 0.0125 b |
| CE | 2.143 × 106 | 1 | 2.143 × 106 | 154.41 | 0.0011 b |
| DE | 2.985 × 106 | 1 | 2.985 × 106 | 215.11 | 0.0007 b |
| ABC | 12,123.11 | 1 | 12,123.11 | 0.8735 | 0.4189 a |
| ABD | 1.849 × 105 | 1 | 1.849 × 105 | 13.32 | 0.0355 b |
| ABE | 3.226 × 105 | 1 | 3.226 × 105 | 23.25 | 0.0170 b |
| ACD | 1.376 × 105 | 1 | 1.376 × 105 | 9.92 | 0.0513 a |
| ACE | 209.98 | 1 | 209.98 | 0.0151 | 0.9099 a |
| ADE | 68,040.94 | 1 | 68,040.94 | 4.90 | 0.1137 a |
| BCD | 2205.93 | 1 | 2205.93 | 0.1589 | 0.7168 a |
| BCE | 16,700.86 | 1 | 16,700.86 | 1.20 | 0.3528 a |
| BDE | 500.17 | 1 | 500.17 | 0.0360 | 0.8616 a |
| CDE | 7.627 × 105 | 1 | 7.627 × 105 | 54.95 | 0.0051 b |
| ABCD | 23,275.35 | 1 | 23,275.35 | 1.68 | 0.2860 a |
| ABCE | 65,651.01 | 1 | 65,651.01 | 4.73 | 0.1179 a |
| ABDE | 1.059 × 105 | 1 | 1.059 × 105 | 7.63 | 0.0700 a |
| ACDE | 5019.30 | 1 | 5019.30 | 0.3617 | 0.5900 a |
| BCDE | 759.35 | 1 | 759.35 | 0.0547 | 0.8301 a |
| ABCDE | 1.630 × 105 | 1 | 1.630 × 105 | 11.74 | 0.0416 b |
| Curvature | 0.0000 | 0 | |||
| Curvature | 9.554 × 105 | 1 | 9.554 × 105 | 68.84 | 0.0037 |
| Error | 41,636.36 | 3 | 13,878.79 | ||
| Cor Total | 3.048 × 107 | 35 |
| Sum of Squares | df | Mean Square | F-Value | p-Value | |
|---|---|---|---|---|---|
| Model | 4282.16 | 9 | 475.8 | 28.1 | <0.0001 b |
| A-Arginine | 218.92 | 1 | 218.92 | 12.93 | 0.0049 b |
| B-Aspartate | 178.73 | 1 | 178.73 | 10.56 | 0.0087 a |
| C-Cysteine | 3192.44 | 1 | 3192.44 | 188.55 | <0.0001 b |
| AB | 6.83 | 1 | 6.83 | 0.4034 | 0.5396 a |
| AC | 323.26 | 1 | 323.26 | 19.09 | 0.0014 b |
| BC | 193.02 | 1 | 193.02 | 11.4 | 0.0070 b |
| A2 | 74.73 | 1 | 74.73 | 4.41 | 0.0620 a |
| B2 | 57.05 | 1 | 57.05 | 3.37 | 0.0963 a |
| C2 | 21.81 | 1 | 21.81 | 1.29 | 0.2828 a |
| Residual | 169.31 | 10 | 16.93 | ||
| Lack of Fit | 146 | 5 | 29.2 | 6.26 | 0.0327 b |
| Pure Error | 23.31 | 5 | 4.66 | ||
| Cor Total | 4451.47 | 19 |
| Culture Medium | Yxs | µx (h−1) | Exponential Phase | tg (h) | Xmax (g L−1) | Ypx | qp (h−1) |
|---|---|---|---|---|---|---|---|
| CDM | 0.822 | 0.502 | 0.5 h ≤ t ≤ 6 h | 1.381 | 1.149 | 1683.77 | 275.46 |
| CDM-S | 1.16 | 0.74 | 0.5 h ≤ t ≤ 5 h | 0.937 | 1.96 | 13,891.27 | 3600.62 |
| LB | - | 1.23 | 0.5 h ≤ t ≤ 3 h | 0.564 | 1.09 | - | - |
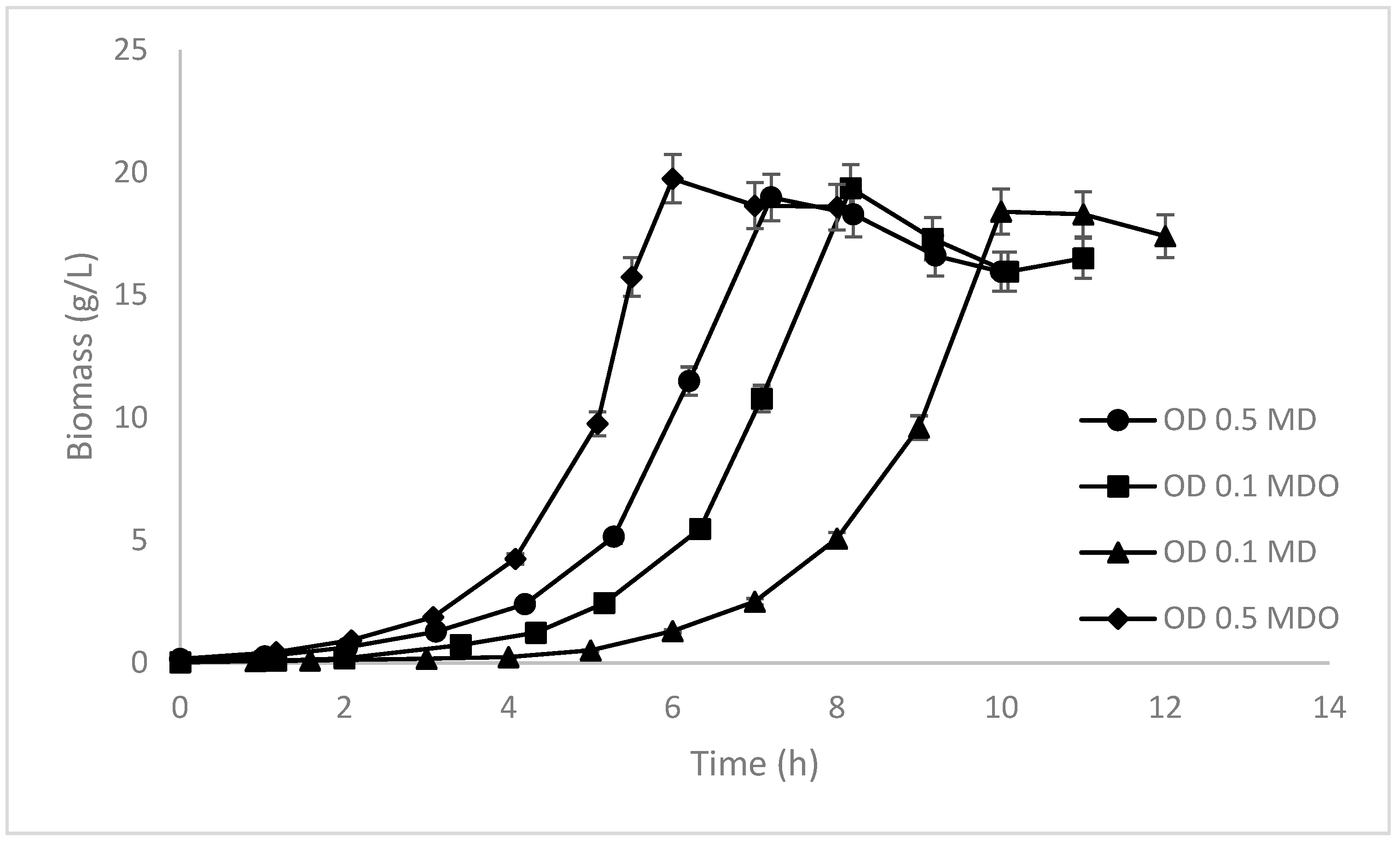
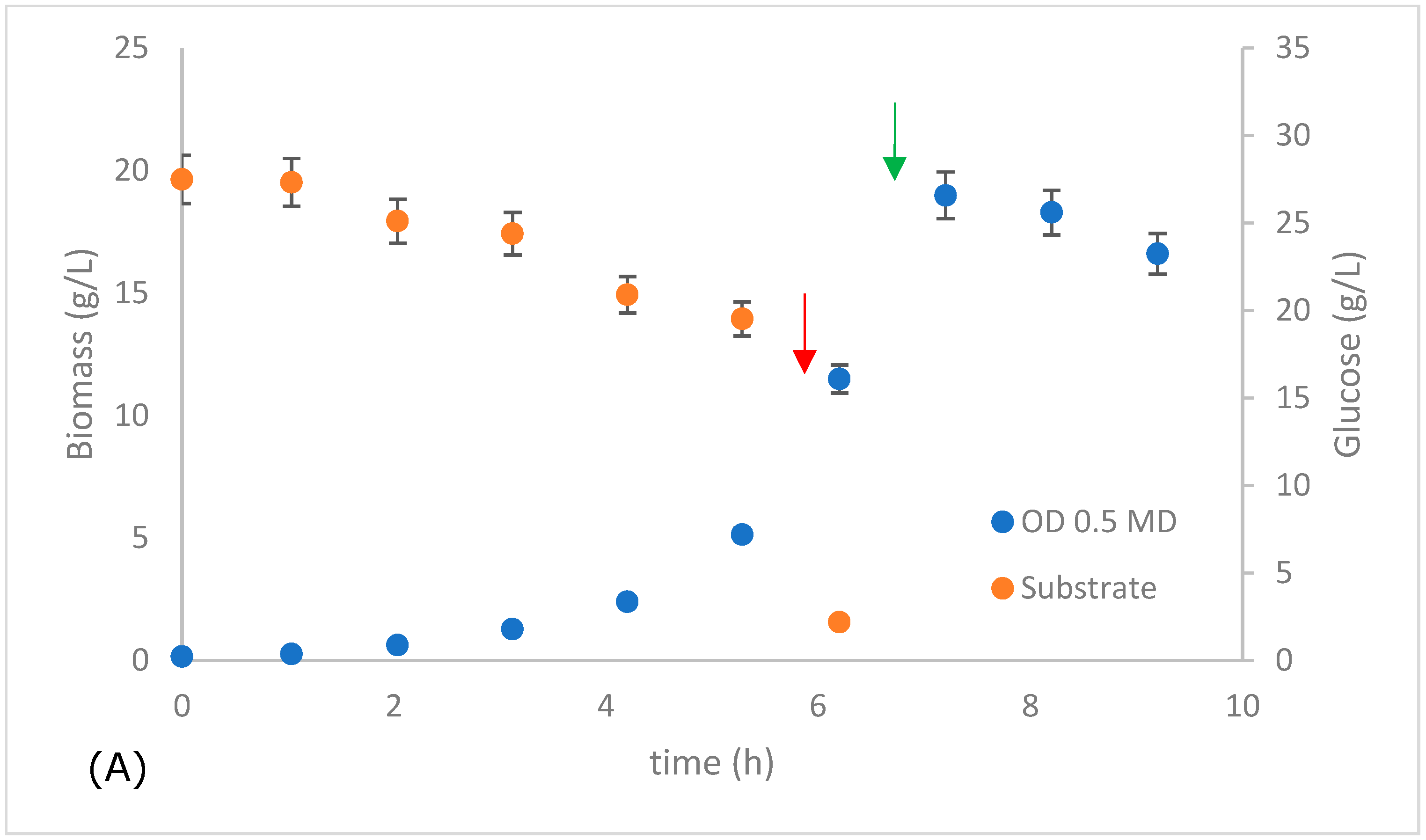
| Kinetic Parameters | MD ODinitial 0.1 | MD ODinitial 0.5 | Laboratory Data (OD 0.1) |
|---|---|---|---|
| mx (h−1) | 0.6157 | 0.707 | 0.7 |
| td (h) | 1.126 | 0.98 | 0.99 |
| YX/s (gg−1) | 0.4 | 0.448 | 0.52 |
| Xmáx (g L−1) | 18.397 | 18.986 | 14 |
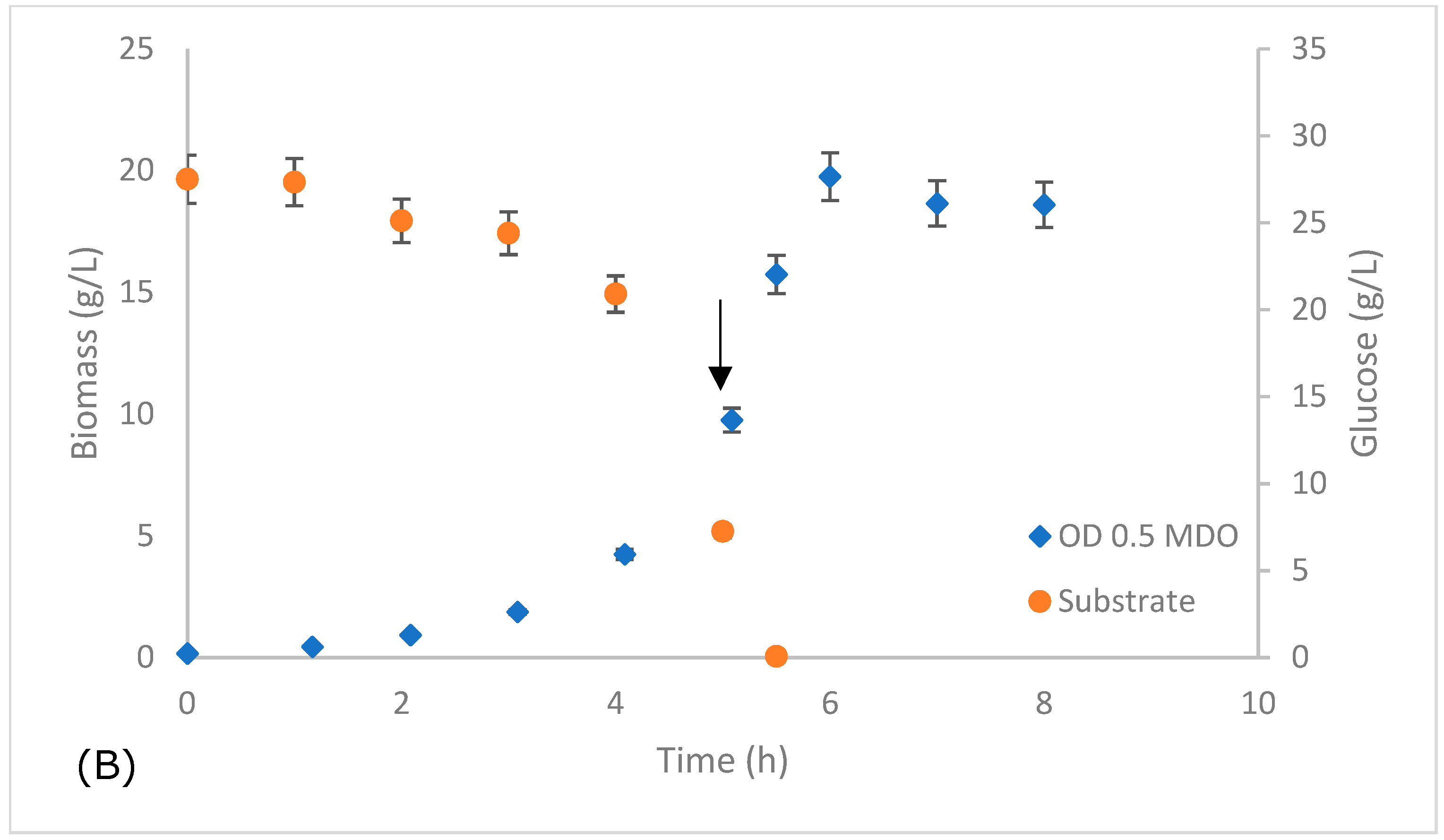
| Kinetic Parameters | MDO ODinitial 0.1 | MDO ODinitial 0.5 |
|---|---|---|
| mx (h−1) | 0.755 | 0.832 |
| tg (h) | 0.918 | 0.835 |
| YX/s (gg−1) | 0.703 | 0.712 |
| Xmáx (g L−1) | 19.36 | 19.75 |
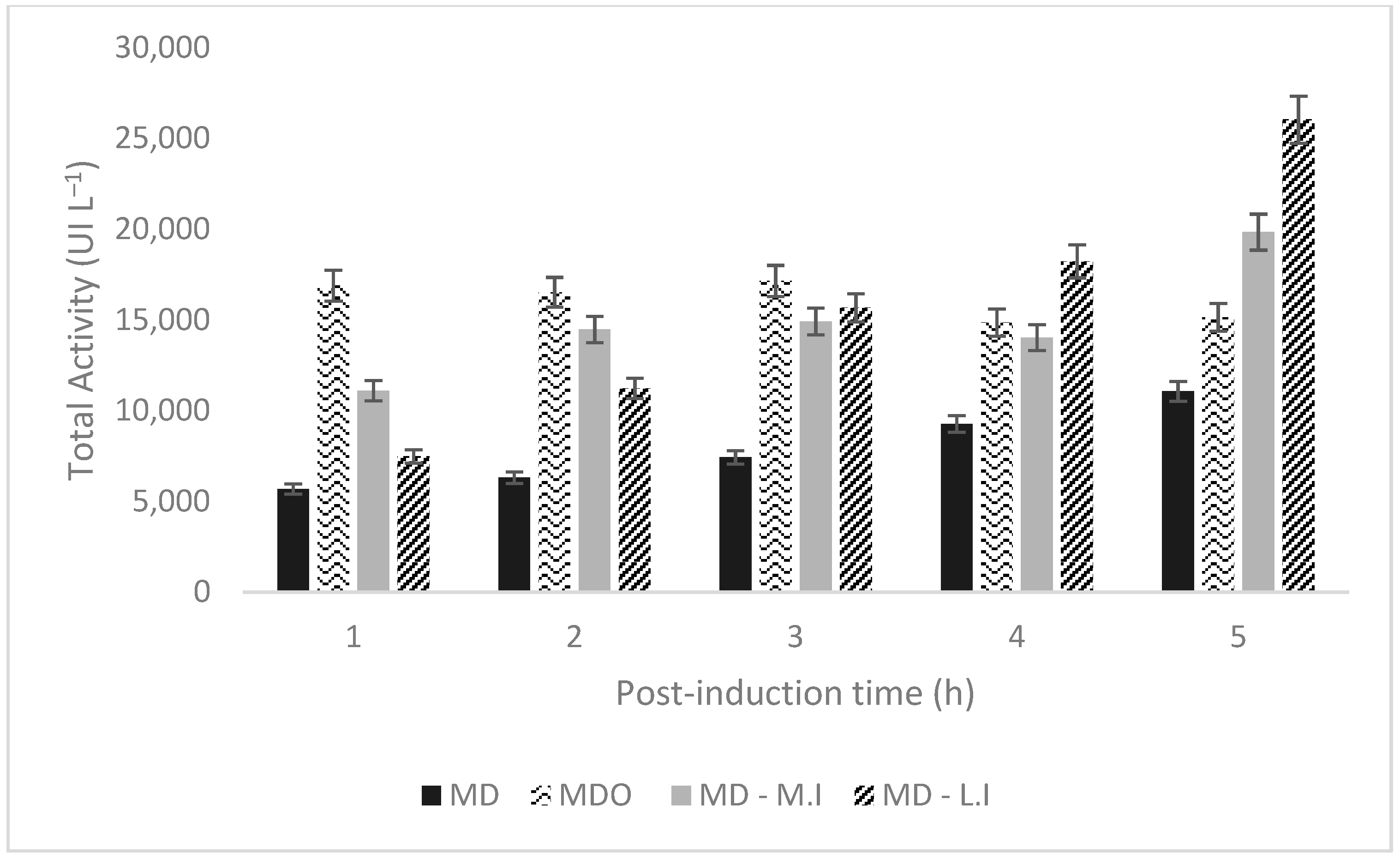
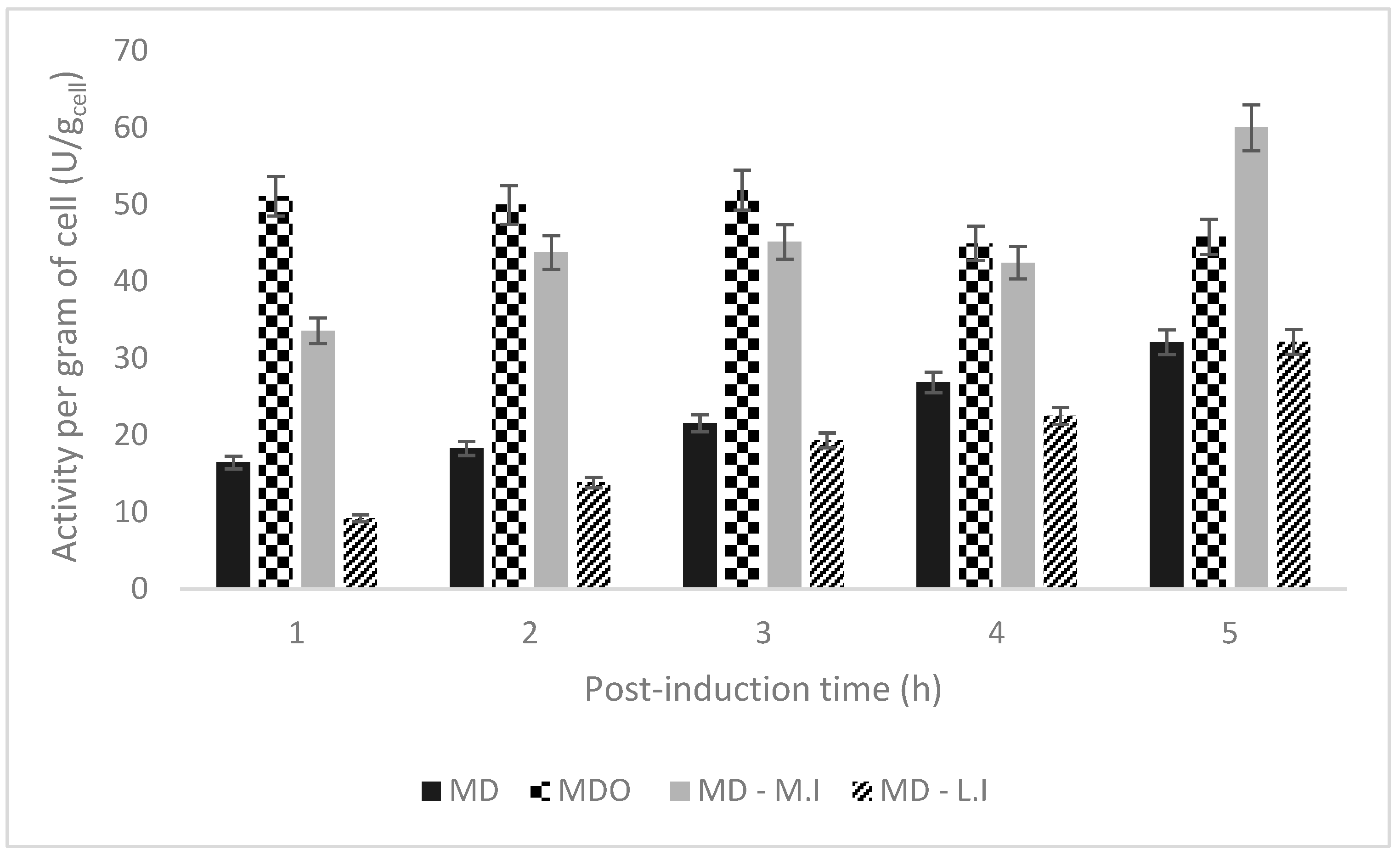
| Culture Strategie | Xmax (g L−1) | Total Activity (U L−1) | Activity Per Dry Mass (U/gcell) | Time of Maximum Activity (h) | U/gglucose | Qp (U/L.h) |
|---|---|---|---|---|---|---|
| MD | 11.5 | 11,063.1 | 32.07 | 11 (5 h post-induction) | 402.3 | 1005.74 |
| MDO | 11.5 | 15,147.15 | 45.81 | 6.5 (1 h post-induction) | 550.8 | 2330.33 |
| MD–M.I | 11.5 | 19,836.43 | 60 | 11 (5 h post-induction) | 721.32 | 1803.31 |
| MD–L.I | 18 | 26,032.96 | 32.14 | 12 (5 h post-induction) | 946.65 | 2169.41 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lefin, N.; Miranda, J.; Munhoz Costa, I.; Pedroso Reynaldo, A.; Monteiro, G.; Zamorano, M.; Pessoa, A., Jr.; Farias, J.G. Optimized Amino Acid-Enhanced Medium for Efficient L-Asparaginase II Production in E. coli: From Shake Flask to Bioreactor. Fermentation 2025, 11, 239. https://doi.org/10.3390/fermentation11050239
Lefin N, Miranda J, Munhoz Costa I, Pedroso Reynaldo A, Monteiro G, Zamorano M, Pessoa A Jr., Farias JG. Optimized Amino Acid-Enhanced Medium for Efficient L-Asparaginase II Production in E. coli: From Shake Flask to Bioreactor. Fermentation. 2025; 11(5):239. https://doi.org/10.3390/fermentation11050239
Chicago/Turabian StyleLefin, Nicolás, Javiera Miranda, Iris Munhoz Costa, Alejandro Pedroso Reynaldo, Gisele Monteiro, Mauricio Zamorano, Adalberto Pessoa, Jr., and Jorge G. Farias. 2025. "Optimized Amino Acid-Enhanced Medium for Efficient L-Asparaginase II Production in E. coli: From Shake Flask to Bioreactor" Fermentation 11, no. 5: 239. https://doi.org/10.3390/fermentation11050239
APA StyleLefin, N., Miranda, J., Munhoz Costa, I., Pedroso Reynaldo, A., Monteiro, G., Zamorano, M., Pessoa, A., Jr., & Farias, J. G. (2025). Optimized Amino Acid-Enhanced Medium for Efficient L-Asparaginase II Production in E. coli: From Shake Flask to Bioreactor. Fermentation, 11(5), 239. https://doi.org/10.3390/fermentation11050239









