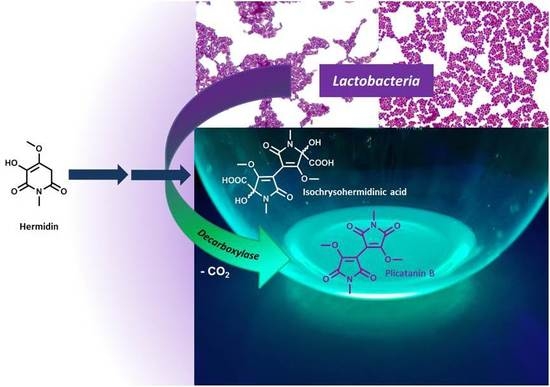
Conversion of Plant Secondary Metabolites upon Fermentation of Mercurialis perennis L. Extracts with two Lactobacteria Strains †
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Culture Media
2.2. Isolation, Gram-Staining and DNA Genotypization of the Lactobacteria
2.3. Plant Material, Extraction (General Procedure)
2.4. Fermentation of Aqueous M. perennis Extracts with Isolated Lactobacteria Strains
2.5. GC-MS Analyses
2.6. NMR Spectroscopy
2.7. Enrichment of Isochrysohermidinic Acid (14) from an Aqueous M. perennis Extract
2.8. RP-HPLC-(DAD)/ESI-MSn Analyses
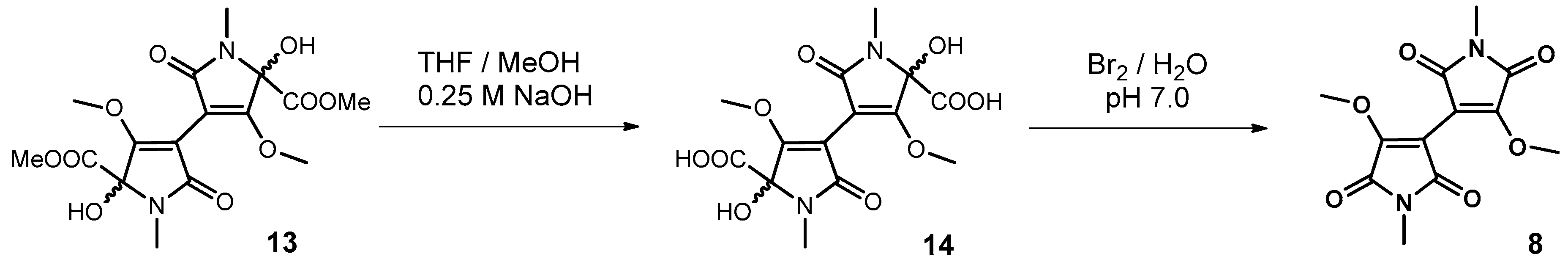
2.9. Isolation of Plicatanin B (8) and N-Methylmaleimide (5) from a Fermentation Broth of M. perennis
2.10. Synthesis of Reference Compounds
2.10.1. Synthesis of Plicatanin B (bis-(3-methoxy-1-N-methyl-maleimide); 8)
2.10.2. Synthesis of 3-Bromo-1N-methylmaleimide
2.10.3. Synthesis of 3-Methoxy-1N-methylmaleimide (5)
2.10.4. Synthesis of Methylarbutin (4-Methoxyphenol-β−D-glucopyranoside; 15)
3. Results and Discussion
3.1. Identification of the Lactobacteria in Fermented Aqueous M. perennis Extracts
3.2. Fermentation of M. perennis with the Lactobacteria Isolates and GC-MS Analysis of the Metabolites
3.3. Approach to the Reaction Mechanism Converting Chrysohermidin (4) into Plicatanin B (8)
4. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Daeschel, M.A.; Andersson, R.E.; Fleming, H.P. Microbial ecology of fermenting plant materials. FEMS Microbiol. Rev. 1987, 46, 357–367. [Google Scholar] [CrossRef]
- Kok, C.R.; Hutkins, R. Yogurt and other fermented foods as sources of health-promoting bacteria. Nutr. Rev. 2018, 76 (Suppl. 1), 4–15. [Google Scholar] [CrossRef]
- Muck, R.E.; Nadeau, E.M.G.; McAllister, T.A.; Contreras-Govea, F.E.; Santos, M.C.; Kung, L., Jr. Silage review: Recent advances and future uses of silage additives. J. Dairy Sci. 2018, 101, 3980–4000. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, M.; Mujumdar, A.S.; Gao, Z. Recent research process of fermented plant extract: A review. Trends Food Sci. Technol. 2017, 65, 40–48. [Google Scholar] [CrossRef]
- Touret, T.; Oliveira, M.; Semedo-Lemsaddek, T. Putative probiotic lactic acid bacteria isolated from sauerkraut fermentations. PLoS ONE 2018, 13, e0203501. [Google Scholar] [CrossRef]
- Hussain, A.; Bose, S.; Wang, J.-H.; Yadav, M.K.; Mahajan, G.B.; Kim, H. Fermentation, a feasible strategy for enhancing bioactivity of herbal medicine. Food Res. Int. 2016, 81, 1–16. [Google Scholar] [CrossRef]
- Lorenz, P.; Stintzing, F.C. Die Fermentation als innovatives Verfahren zur Gewinnung von Arzneipflanzenauszügen. In Anthroposophische Pharmazie, 1st ed.; Meyer, U., Pedersen, P.A., Eds.; Salumed Verlag: Berlin, Germany, 2016; pp. 663–681. [Google Scholar]
- Van Duynhoven, J.; Vaughan, E.E.; van Dorsten, F.; Gomez-Roldan, V.; de Vos, R.; Vervoort, J.; van der Hooft, J.J.; Roger, L.; Draijer, R.; Jacobs, D.M. Interactions of black tea polyphenols with human gut microbiota: Implications for gut and cardiovascular health. Am. J. Clin. Nutr. 2013, 98, 1631S–1641S. [Google Scholar] [CrossRef]
- Luca, S.V.; Macovei, I.; Bujor, A.; Miron, A.; Skalicka-Woźniak, K.; Aprotosoaie, A.C.; Trifan, A. Bioactivity of dietary polyphenols: The role of metabolites. Crit. Rev. Food Sci. Nutr. 2019, 7, 1–34. [Google Scholar] [CrossRef]
- Rizzello, C.G.; Coda, R.; Macías, D.S.; Pinto, D.; Marzani, B.; Filannino, P.; Giuliani, G.; Paradiso, V.M.; Di Cagno, R.; Gobbetti, M. Lactic acid fermentation as a tool to enhance the functional features of Echinacea spp. Microb. Cell. Fact. 2013, 12, 40. [Google Scholar] [CrossRef]
- Lei, V.; Amoa-Awua, W.K.A.; Brimer, L. Degradation of cyanogenic glycosides by Lactobacillus plantarum strains from spontaneous cassava fermentation and other microorganisms. Int. J. Food Microbiol. 1999, 53, 169–184. [Google Scholar] [CrossRef]
- Lee, N.-K.; Paik, H.-D. Bioconversion using lactic acid bacteria: Ginsenosides, GABA, and phenolic compounds. J. Microbiol. Biotechnol. 2017, 27, 869–877. [Google Scholar] [CrossRef]
- Lee, S.O.; Kim, S.J.; Kim, J.S.; Ji, H.; Lee, E.O.; Lee, H.J. Comparison of the main components and bioactivity of Rhus verniciflua Stokes extracts by different detoxification processing methods. BMC Complement. Altern. Med. 2018, 18, 242. [Google Scholar] [CrossRef]
- Millet, A.; Stintzing, F.; Merfort, I. Validation of a GC-FID method for rapid quantification of nicotine in fermented extracts prepared from Nicotiana tabaccum fresh leaves and studies of nicotine mebabolites. J. Pharm. Biomed. Anal. 2009, 49, 1166–1171. [Google Scholar] [CrossRef] [PubMed]
- Millet, A.; Stintzing, F.; Merfort, I. Flavonol quantification and stability of phenolics in fermented extracts from fresh Betula pendula leaves. J. Pharm. Biomed. Anal. 2010, 53, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenberger, M.; Stintzing, F.; Meyer, U.; Lindequist, U. Biochemical, microbiological and phytochemical studies on aqueous-fermented extracts from Atropa belladonna L. Part 1 – biochemistry and microbiology; Part 2 – Phytochemistry. Pharmazie 2012, 67, 331–344. [Google Scholar]
- Duckstein, S.M.; Stintzing, F.C. LC-MS(n) characterization of steroidal saponins in Helleborus niger L. roots and their conversion products during fermentation. Steroids 2015, 93, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Knittel, D.N.; Stintzing, F.C.; Kammerer, D.R. Metabolic fate of cardiac glycosides and flavonoids upon fermentation of aqueous sea squill (Drima maritima L.) extracts. J. Pharm. Biomed. Anal. 2015, 110, 100–109. [Google Scholar] [CrossRef]
- Lorenz, P.; Beckmann, C.; Felenda, J.; Meyer, U.; Stintzing, F.C. Das Waldbingelkraut (Mercurialis perennis L.) - Pharmakognosie einer alten Arzneipflanze. Z. Phytother. 2013, 34, 40–46. [Google Scholar] [CrossRef]
- Rugman, F.; Meecham, J.; Edmondson, J. Mercurialis perennis (dog’s mercury) poisoning: A case of mistaken identity. Br. Med. J. (Clin. Res. Ed.) 1983, 287, 1924. [Google Scholar] [CrossRef][Green Version]
- Watson, P.J. Suspected dog’s mercury (Mercurialis perennis) poisoning in cattle. Vet. Rec. 1998, 142, 116–117. [Google Scholar] [CrossRef]
- Welchman, D.B.; Gibbens, J.C.; Giles, N.; Piercy, D.W.; Skinner, P.H. Suspected annual mercury (Mercurialis annua) poisoning of lambs grazing fallow arable land. Vet. Rec. 1995, 137, 592–593. [Google Scholar] [PubMed]
- Lorenz, P.; Duckstein, S.; Conrad, J.; Knödler, M.; Meyer, U.; Stintzing, F.C. An approach to the chemotaxonomic differentiation of two European dog’s mercury species: Mercurialis annua L. and M. perennis L. Chem. Biodivers. 2012, 9, 282–294. [Google Scholar] [CrossRef] [PubMed]
- Swan, G.A. Hermidin, a chromogen from Mercurialis perennis L. Experientia 1984, 40, 687–688. [Google Scholar] [CrossRef]
- Swan, G.A. Isolation, structure and synthesis of hermidin, a chromogen from Mercurialis perennis L. J. Chem. Soc. Perkin Trans. I 1985, 1757–1766. [Google Scholar] [CrossRef]
- Lorenz, P.; Hradecky, M.; Berger, M.; Bertrams, J.; Meyer, U.; Stintzing, F.C. Lipophilic constituents from aerial and root parts of Mercurialis perennis L. Phytochem. Anal. 2010, 21, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, P.; Conrad, J.; Bertrams, J.; Berger, M.; Duckstein, S.; Meyer, U.; Stintzing, F.C. Investigations into the phenolic constituents of dog’s mercury (Mercurialis perennis L.) by LC-MS/MS and GC/MS analyses. Phytochem. Anal. 2012, 23, 60–71. [Google Scholar] [CrossRef]
- Lorenz, P.; Knödler, M.; Bertrams, J.; Berger, M.; Meyer, U.; Stintzing, F.C. n-Alkylresorcinol occurrence in Mercurialis perennis L. (Euphorbiaceae). Z. Naturforsch. 2010, 65c, 174–179. [Google Scholar] [CrossRef]
- Lorenz, P.; Heinrich, M.; Conrad, J.; Heller, A.; Stintzing, F.C.; Kammerer, D.R. Comprehensive characterization of n-alkylresorcinols and other lipid constituents of Mercurialis tomentosa L. from Alicante, Spain. Chem. Biodiv. 2017, 14. [Google Scholar] [CrossRef]
- Lorenz, P.; Conrad, J.; Stintzing, F.C. Metabolic fate of depsides and alkaloid constituents in aqueous extracts from Mercurialis perennis L. during fermentation. Chem. Biodivers. 2013, 10, 1706–1723. [Google Scholar] [CrossRef]
- Lorenz, P.; Conrad, J.; Duckstein, S.; Kammerer, D.R.; Stintzing, F.C. Chemistry of hermidin: Insights from extraction experiments with the main alkaloid of Mercurialis perennis L. tracked by GC/MS and LC/MSn. Helv. Chim. Acta 2014, 97, 1606–1623. [Google Scholar] [CrossRef]
- GHP, German Homoeopathic Pharmacopoeia, Homöopathisches Arzneibuch, Spezielle Herstellungsvorschriften, Vorschrift 34 c; Deutscher Apotheker-Verlag: Stuttgart, Germany; Govi Pharmazeutischer Verlag: Eschborn, Germany, 2018.
- Stevens, H.; Brinkhoff, T.; Simon, M. Composition of free-living, aggregate-associated and sediment surface-associated bacterial communities in the German Wadden Sea. Aquat. Microb. Ecol. 2005, 38, 15–30. [Google Scholar] [CrossRef][Green Version]
- National Center for Biotechnology Information (NCBI). Available online: http://www.ncbi.nlm.nih.gov/blast/ (accessed on 13 April 2019).
- Agrawal, V.; Desai, S. Centrifugally accelerated thin layer chromatography for isolation of marker compounds and bioactives. J. Pharmacogn. Phytochem. 2015, 3, 145–149. [Google Scholar]
- Tabussum, A.; Riaz, N.; Saleem, M.; Ashraf, M.; Ahmad, M.; Alam, U.; Jabeen, B.; Malik, A.; Jabbar, A. α-Glucosidase inhibitory constituents from Chrozophora plicata. Phytochem. Lett. 2013, 6, 614–619. [Google Scholar] [CrossRef]
- Tedaldi, L.M.; Smith, M.B.; Nathani, R.I.; Baker, J.R. Bromomaleimides; new reagents for selective and reversible modification of cysteine. Chem. Commun. 2009, 43, 6583–6585. [Google Scholar] [CrossRef]
- Gill, G.B.; James, G.D.; Oates, K.V.; Pattenden, G. The synthesis of 5-ylidenepyrrol-2(5H)-ones from maleimides and from pyrrol-2-(5H)-ones. J. Chem. Soc. Perkin Trans. I 1993, 2567–2579. [Google Scholar] [CrossRef]
- Gabrielle, S.-P.; Dafik, L.; Klegraf, E.; Hanessian, S. Stereocontrolled synthesis of phenolic α-D-glucopyranosides. Synthesis 2016, 48, 3575–3588, Suppl. Materials S6. [Google Scholar]
- Lutterbach, R.; Stöckigt, J. Dynamics of the biosynthesis of methylursubin in plant cells employing in vivo 13C NMR without labeling. Phytochemistry 1995, 40, 801–806. [Google Scholar] [CrossRef]
- Simpson, W.J.; Taguchi, H. The genus Pediococcus, with notes on the genera Tetratogenococcus and Aerococcus. In The Genera of Lactic Acid Bacteria; Wood, B.J.B., Holzapfel, W.H., Eds.; Chapman & Hall: London, UK, 1995; pp. 125–172. [Google Scholar]
- Salvetti, E.; Torriani, S.; Felis, G.E. The genus Lactobacillus: A taxonomic update. Probiotics Antimicrob. Proteins 2012, 4, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Hammes, W.P.; Vogel, R.F. The genus Lactobacillus. In The genera of lactic acid bacteria. The lactic acid bacteria, Vol 2; Wood, B.J.B., Holzapfel, W.H., Eds.; Springer: Boston, MA, USA, 1995; pp. 19–54. [Google Scholar]
- Felis, G.E.; Salvetti, E.; Torriani, S. Systematics of Lactic Acid Bacteria: Current Status. In Biotechnology of Lactic Acid Bacteria: Novel Applications; Mozzi, F., Raya, R.R., Vignolo, G.M., Eds.; John Wiley & Sons, Ltd.: Chichester, West Sussex, UK, 2016; pp. 25–31. [Google Scholar]
- Xie, Y.; Husband, J.T.; Torrent-Sucarrat, M.; Yang, H.; Liu, W.; O’Reilly, K.O. Rational design of substituted maleimide dyes with tunable fluorescence and solvafluorochromism. Chem. Commun. 2018, 54, 3339–3342. [Google Scholar] [CrossRef] [PubMed]
- Pink, J.M.; Stewart, R. Mechanism of oxidative decarboxylation of α-hydroxy acids by bromine water. Part I. Oxidation in neutral and alkaline medium. Can. J. Chem. 1971, 49, 649–653. [Google Scholar] [CrossRef]
- Sticher, O.; Soldati, F.; Lehmann, D. High-performance liquid chromatographic separation and quantitative determination of arbutin, methylarbutin, hydroquinone and hydroquinone-monomethylether in Arctostaphylos, Bergenia, Calluna and Vaccinium species. Planta Med. 1979, 35, 253–261. [Google Scholar] [CrossRef]
- Dusková, J.; Dusek, J.; Jahodár, L. Bergenia crassifolia (L.) Fritsch in vitro. Ceska Slov. Farm. 2001, 50, 83–85. [Google Scholar] [PubMed]
- Flores-Bocanegra, L.; Pérez-Vásquez, A.; Torres-Piedra, M.; Bye, R.; Linares, E.; Mata, R. α-Glucosidase inhibitors from Vauquelinia corymbosa. Molecules 2015, 20, 15330–15342. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Oh, J.; Jeong, Y.-S. Lactobacillus plantarum-mediated conversion of flavonoid glycosides into flavonols, quercetin, and kaempferol in Cudrania tricuspidata leaves. Food Sci. Biotechnol. 2015, 24, 1817–1821. [Google Scholar] [CrossRef]
- Yokoyama, M.T.; Carlson, J.R. Production of skatole and para-cresol by a rumen Lactobacillus sp. Appl. Environ. Microbiol. 1981, 41, 71–76. [Google Scholar] [PubMed]
- Barbieri, F.; Montanari, C.; Gardini, F.; Tabanelli, G. Biogenic amine production by lactic acid bacteria: A review. Foods 2019, 8, 17. [Google Scholar] [CrossRef]
- Van Beek, S.; Priest, F.G. Decarboxylation of substituted cinnamic acids by lactic acid bacteria isolated during malt whisky fermentation. Appl. Environ. Microbiol. 2000, 66, 5322–5328. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, H.; Landete, J.M.; Curiel, J.A.; de Las Rivas, B.; Mancheño, J.M.; Muñoz, R. Characterization of the p-coumaric acid decarboxylase from Lactobacillus plantarum CECT 748(T). J. Agric. Food Chem. 2008, 56, 3068–3072. [Google Scholar] [CrossRef]
- Rodríguez, H.; Curiel, J.A.; Landete, J.M.; de las Rivas, B.; López de Felipe, F.; Gómez-Cordovés, C.; Mancheño, J.M.; Muñoz, R. Food phenolics and lactic acid bacteria. Int. J. Food Microbiol. 2009, 132, 79–90. [Google Scholar] [CrossRef]









| Isolate (Strain) | GenBank Accession Number 1 | Sequence Length (bp) | Closest Relative in NCBI 2 | Similarity |
|---|---|---|---|---|
| LP1 | MK841313.1 | 1083 | Lactobacillus plantarum strain 2.7.17, MK611349.1 | 100% |
| PP1 | MK841045.1 | 1441 | Pediococcus pentosaceus strain CMGB-L16, MF348228.1 | 100% |
| Compound | Retention Time (min) | Characteristic Mass Signals, m/z (% BPI) |
|---|---|---|
| mequinol (4-methoxyphenol; 9) | 11.1 | 124 (M+, 100), 109 (98), 95 (2), 81 (46), 65 (5), 53 (15) 1 |
| 3-methoxy-1N-methyl-maleimide (5) | 11.9 | 141 (M+, 23), 123 (8), 113 (12), 112 (25), 84 (23), 69 (100), 56 (9), 53 (7) |
| (–)-cis-myrtanol (10) | 12.8 | 136 ([M-H2O]+, 11), 123 (68), 121 (42), 107 (15), 95 (28), 93 (100), 81 (65), 79 (38), 69 (73), 67 (80), 55 (45) 1, 2 |
| 3-methylhermidin (6) | 16.6 | 185 (M+, 100), 170 (65), 156 (68), 142 (30), 128 (20), 126 (15), 114 (13), 99 (17), 83 (40), 68 (74), 58 (64) |
| 3,4-dimethoxyphenol (11) | 17.0 | 154 (M+, 100), 139 (96), 125 (4), 111 (48), 96 (11), 93 (27), 81 (9), 69 (16), 65 (18), 55 (18) 1 |
| 3-methylhermidin quinone (7) | 18.7 | 183 (M+, 52), 154 (1), 137 (2), 126 (6), 112 (3), 98 (23), 83 (100), 70 (8), 55 (8) |
| hermidin quinone (3) | 20.9 | 169 (M+, 35), 142 (0.4), 112 (35), 84 (21), 69 (100), 56 (7), 53 (9) 1 |
| plicatanin B (bis-(3-methoxy-1N-methyl-maleimide); 8) | 35.8 | 280 (M+, 100), 265 (20), 251 (24), 237 (11), 208 (17), 195 (6), 180 (35), 178 (22), 166 (21), 152 (20), 123 (45), 109 (5), 95 (72), 80 (37), 72 (16), 67 (8), 53 (6) |
| 3-(3-methoxy-1N-methyl-malimido)-hermidin quinone (12) | 40.0 | 308 (M+, 67), 280 (54), 265 (14), 251 (27), 237 (13), 223 (11), 208 (72), 195 (34), 180 (76), 166 (13), 152 (30), 123 (62), 109 (8), 95 (100), 80 (57), 69 (10), 53 (11) |
| chrysohermidin (bis-(hermidin quinone); 4) | 43.9 | 336 (M+, 9), 321 (74), 308 (12), 280 (53), 265 (23), 251 (23), 236 (76), 208 (62), 195 (25), 180 (100), 167 (11), 152 (37), 123 (52), 111 (13), 95 (94), 80 (58), 68 (13), 53 (11) 1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lorenz, P.; Bunse, M.; Sauer, S.; Conrad, J.; Stintzing, F.C.; Kammerer, D.R. Conversion of Plant Secondary Metabolites upon Fermentation of Mercurialis perennis L. Extracts with two Lactobacteria Strains. Fermentation 2019, 5, 42. https://doi.org/10.3390/fermentation5020042
Lorenz P, Bunse M, Sauer S, Conrad J, Stintzing FC, Kammerer DR. Conversion of Plant Secondary Metabolites upon Fermentation of Mercurialis perennis L. Extracts with two Lactobacteria Strains. Fermentation. 2019; 5(2):42. https://doi.org/10.3390/fermentation5020042
Chicago/Turabian StyleLorenz, Peter, Marek Bunse, Simon Sauer, Jürgen Conrad, Florian C. Stintzing, and Dietmar R. Kammerer. 2019. "Conversion of Plant Secondary Metabolites upon Fermentation of Mercurialis perennis L. Extracts with two Lactobacteria Strains" Fermentation 5, no. 2: 42. https://doi.org/10.3390/fermentation5020042
APA StyleLorenz, P., Bunse, M., Sauer, S., Conrad, J., Stintzing, F. C., & Kammerer, D. R. (2019). Conversion of Plant Secondary Metabolites upon Fermentation of Mercurialis perennis L. Extracts with two Lactobacteria Strains. Fermentation, 5(2), 42. https://doi.org/10.3390/fermentation5020042