Robotic Cell Printing for Constructing Living Yeast Cell Microarrays in Microfluidic Chips
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Strain and Media
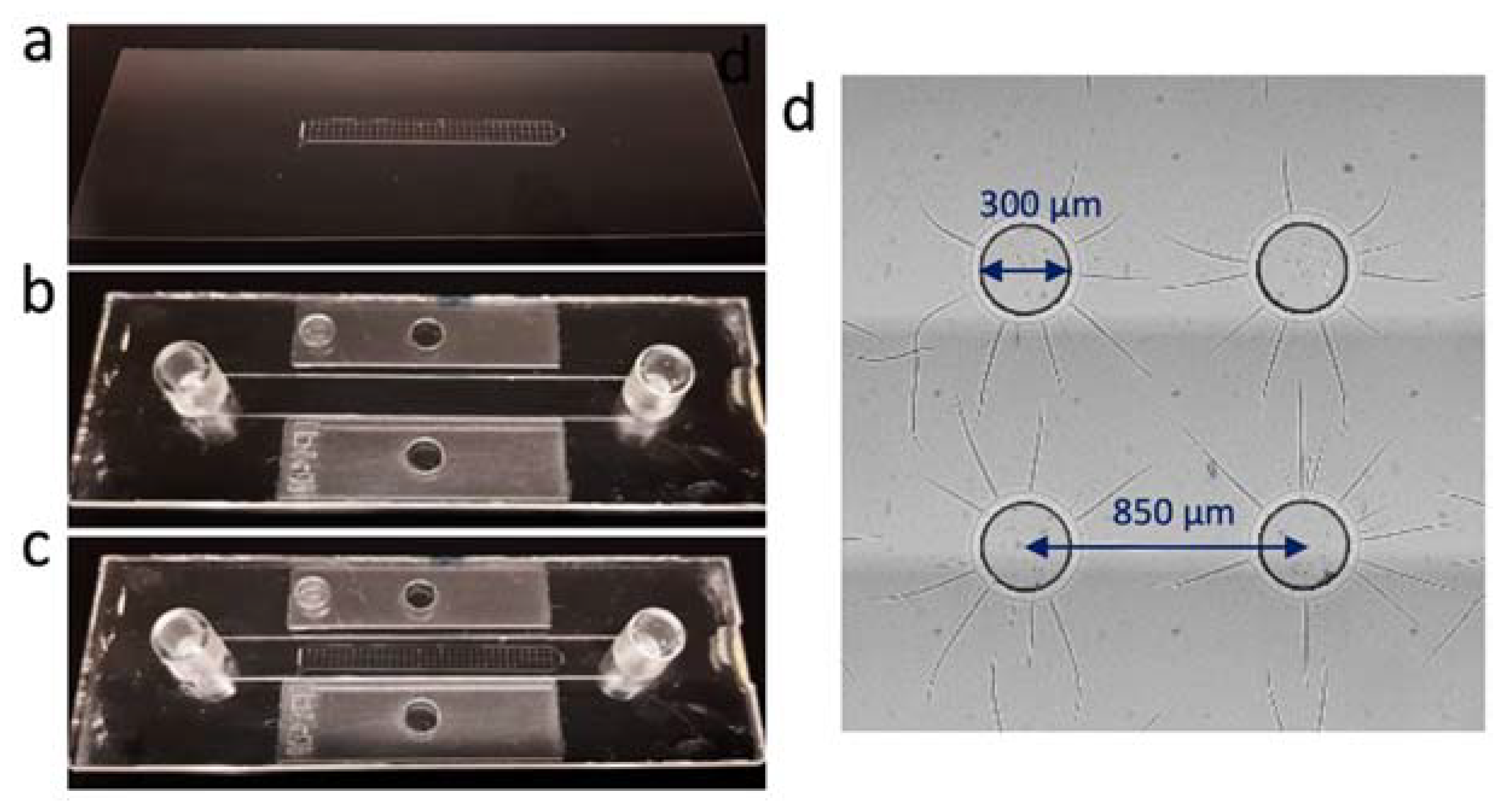
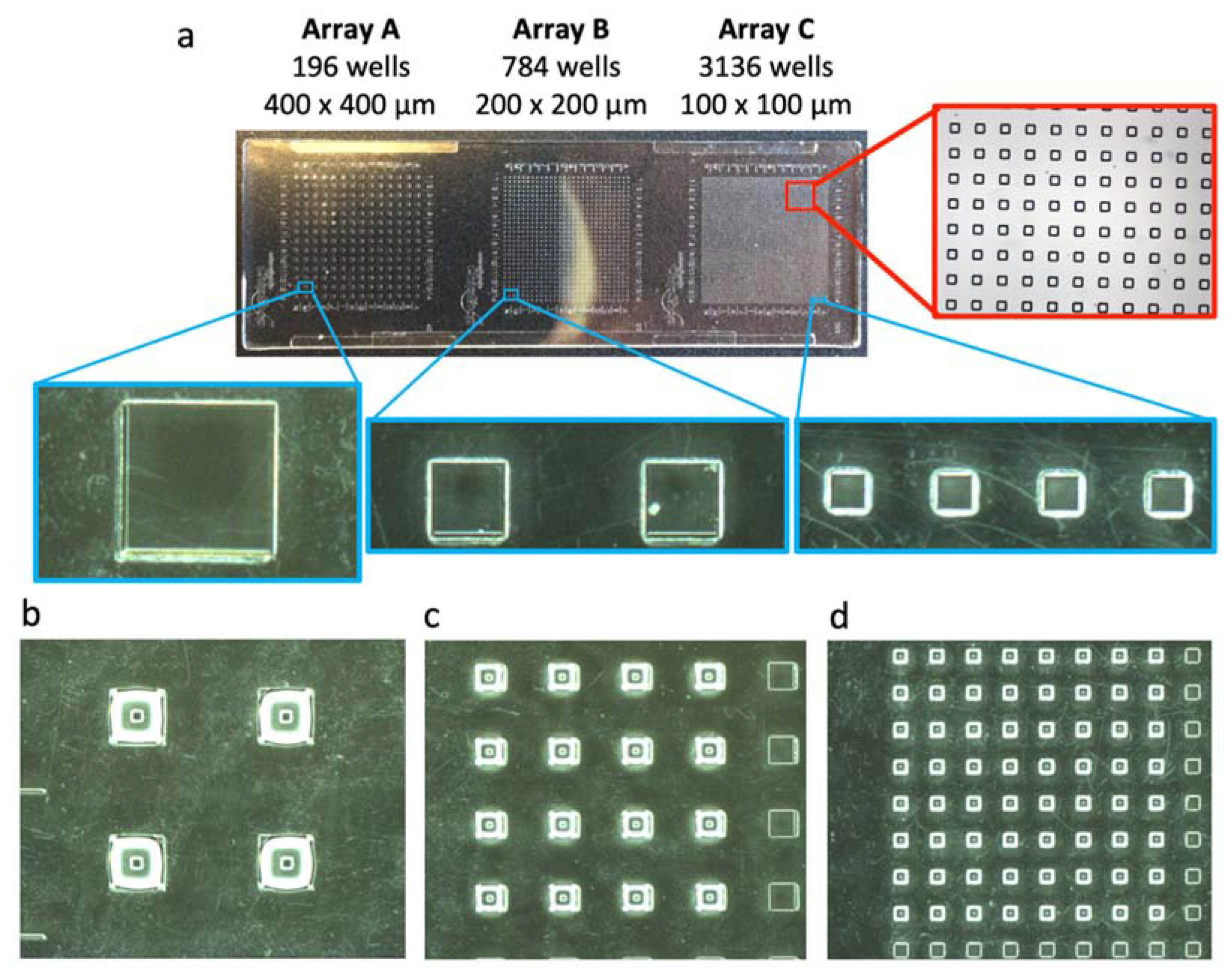
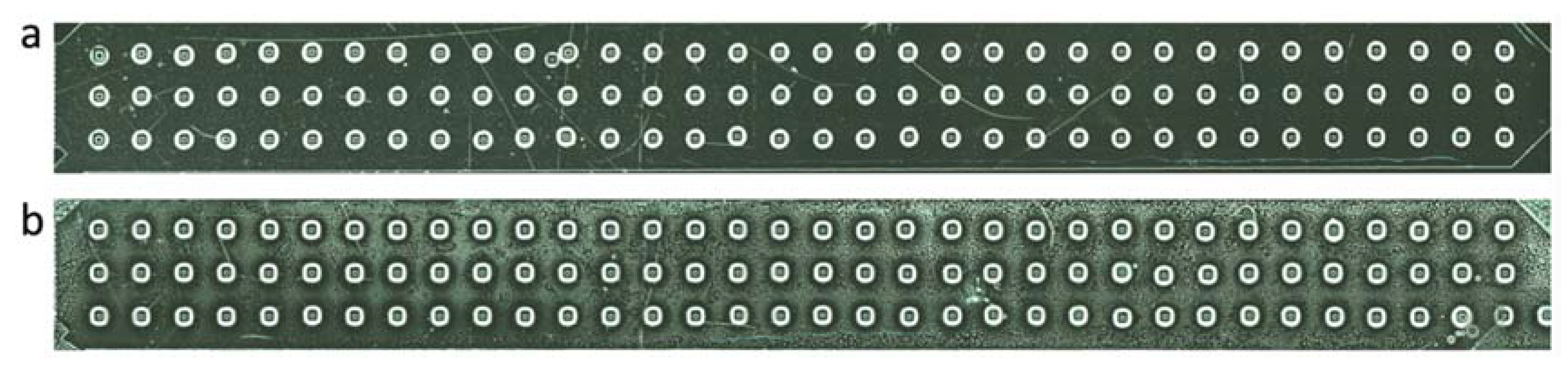
2.2. Fabrication of the Microwell Substrate and the Microfluidic Chip
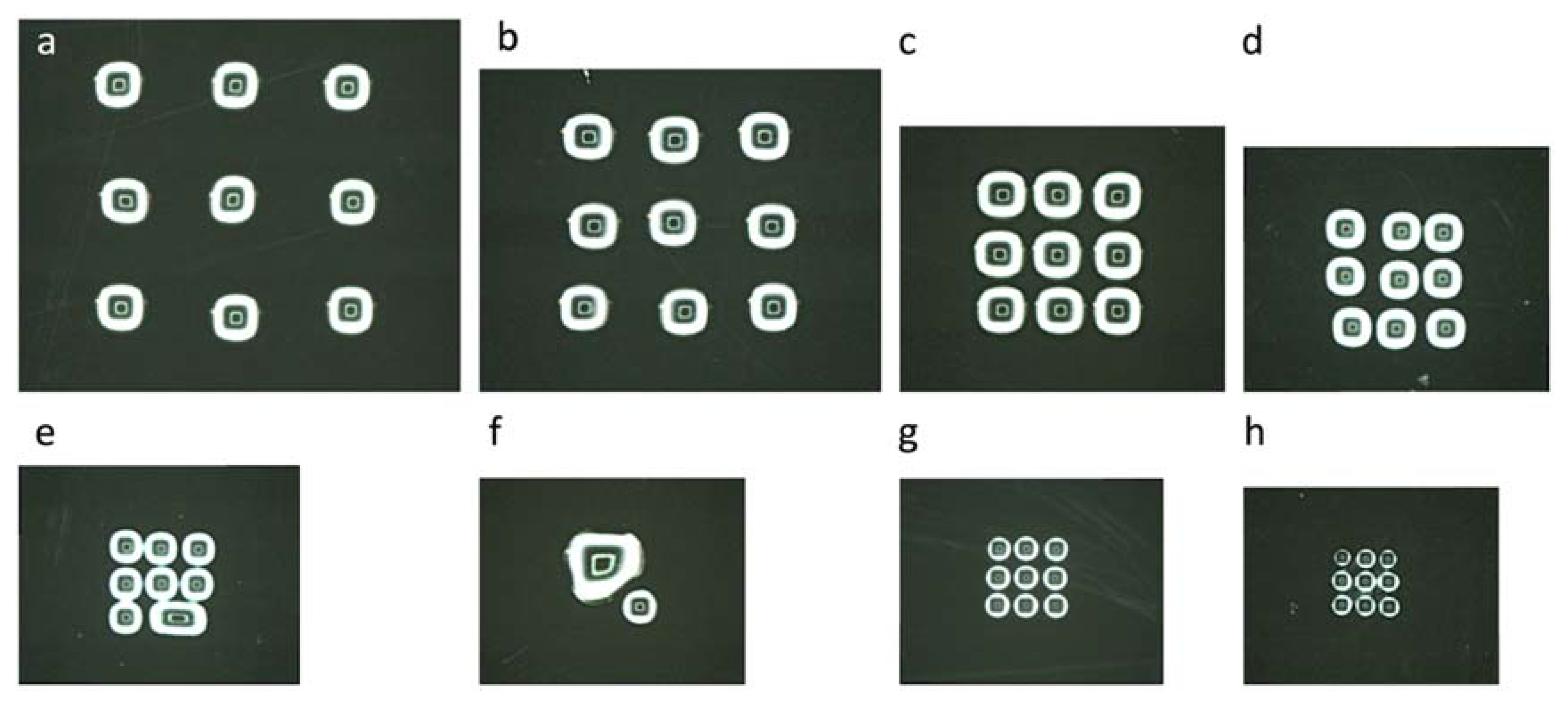
2.3. Cell Printing
2.4. Microscopy
2.5. Image Processing
3. Results
3.1. Construction of the Microfluidic Chip
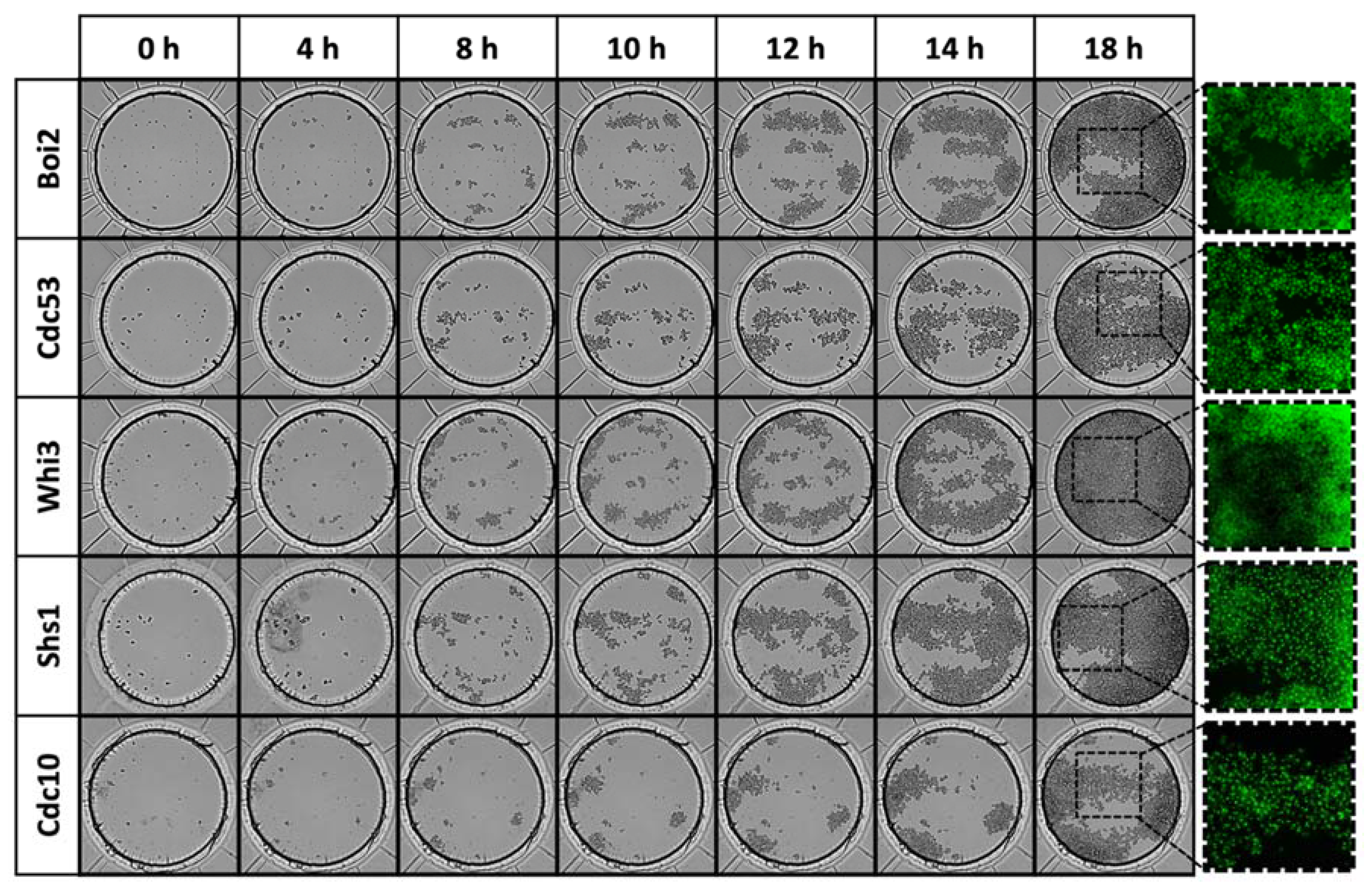
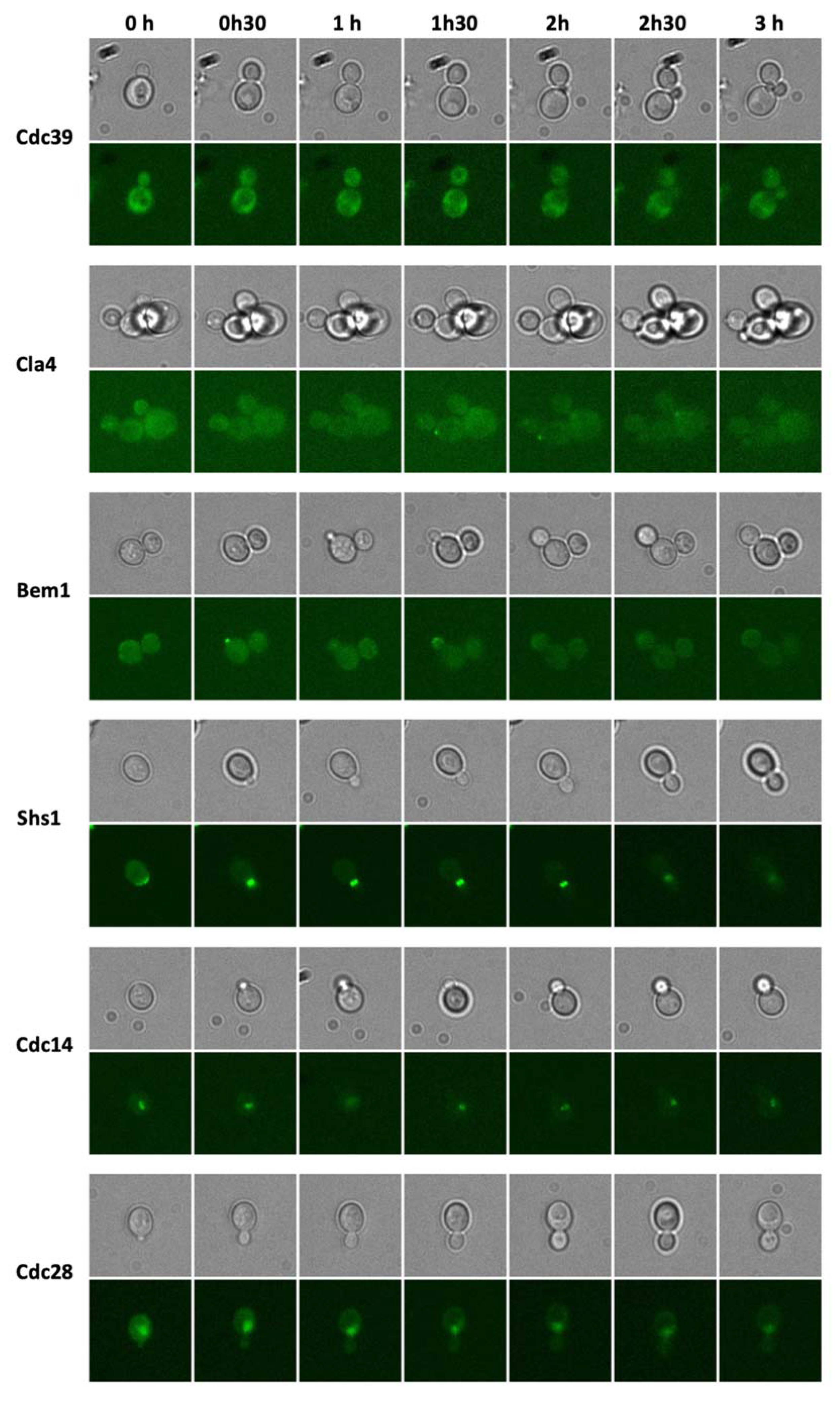
3.2. Living-Cell Microarray Development
3.3. Growth of Yeast Cells in the Microfluidic Chip
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Castel, D.; Pitaval, A.; Debily, M.A.; Gidrol, X. Cell microarrays in drug discovery. Drug Discov. Today 2006, 11, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Berthuy, O.I.; Muldur, S.K.; Rossi, F.; Colpo, P.; Blum, L.J.; Marquette, C.A. Multiplex cell microarrays for high-throughput screening. Lab Chip 2016, 16, 4248–4262. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Koom, W.; Koh, W.G. Cell Microarray Technologies for High-Throughput Cell-Based Biosensors. Sensors 2017, 17, 1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willaert, R.; Goossens, K. Microfluidic Bioreactors for Cellular Microarrays. Fermentation 2015, 1, 38–78. [Google Scholar] [CrossRef] [Green Version]
- Jonczyk, R.; Kurth, T.; Lavrentieva, A.; Walter, J.-G.; Scheper, T.; Stahl, F. Living Cell Microarrays: An Overview of Concepts. Microarrays 2016, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Willaert, R.; Sahli, H. On-Chip Living-Cell Microarrays for Network Biology. In Bioinformatics—Trends and Methodologies; InTechOpen: London, UK, 2011. [Google Scholar]
- Fernandes, T.G.; Diogo, M.M.; Clark, D.S.; Dordick, J.S.; Cabral, J.M.S. High-throughput cellular microarray platforms: Applications in drug discovery, toxicology and stem cell research. Trends Biotechnol. 2009, 27, 342–349. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Davis, M. Molecular and functional analysis using live cell microarrays. Curr. Opin. Chem. Biol. 2006, 10, 28–34. [Google Scholar] [CrossRef]
- Breker, M.; Gymrek, M.; Schuldiner, M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J. Cell Biol. 2013, 200, 839–850. [Google Scholar] [CrossRef] [Green Version]
- Dénervaud, N.; Becker, J.; Delgado-Gonzalo, R.; Damay, P.; Rajkumar, A.S.; Unser, M.; Shore, D.; Naef, F.; Maerkl, S.J. A chemostat array enables the spatio-temporal analysis of the yeast proteome. Proc. Natl. Acad. Sci. USA 2013, 110, 15842–15847. [Google Scholar] [CrossRef] [Green Version]
- Chong, Y.T.; Koh, J.L.Y.; Friesen, H.; Duffy, K.; Cox, M.J.; Moses, A.; Moffat, J.; Boone, C.; Andrews, B.J. Yeast proteome dynamics from single cell imaging and automated analysis. Cell 2015, 161, 1413–1424. [Google Scholar] [CrossRef] [Green Version]
- Yuan, H.; Zhang, R.; Shao, B.; Wang, X.; Ouyang, Q.; Hao, N.; Luo, C. Protein expression patterns of the yeast mating response. Integr. Biol. (UK) 2016, 8, 712–719. [Google Scholar] [CrossRef]
- Shao, B.; Yuan, H.; Zhang, R.; Wang, X.; Zhang, S.; Ouyang, Q.; Hao, N.; Luo, C. Reconstructing the regulatory circuit of cell fate determination in yeast mating response. PLoS Comput. Biol. 2017, 13, e1005671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarmush, M.L.; King, K.R. Living-Cell Microarrays. Annu. Rev. Biomed. Eng. 2009, 11, 235–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, M.C.; Hur, J.Y.; Cho, H.S.; Park, S.H.; Suh, K.Y. High-throughput single-cell quantification using simple microwell-based cell docking and programmable time-course live-cell imaging. Lab Chip 2011, 11, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Minc, N.; Boudaoud, A.; Chang, F. Mechanical Forces of Fission Yeast Growth. Curr. Biol. 2009, 19, 1096–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falconnet, D.; Niemistö, A.; Taylor, R.J.; Ricicova, M.; Galitski, T.; Shmulevich, I.; Hansen, C.L. High-throughput tracking of single yeast cells in a microfluidic imaging matrix. Lab Chip 2011, 11, 466–473. [Google Scholar] [CrossRef]
- Groisman, A.; Lobo, C.; Cho, H.; Campbell, J.K.; Dufour, Y.S.; Stevens, A.M.; Levchenko, A. A microfluidic chemostat for experiments with bacterial and yeast cells. Nat. Methods 2005, 2, 685–689. [Google Scholar] [CrossRef]
- Ryley, J.; Pereira-Smith, O.M. Microfluidics device for single cell gene expression analysis inSaccharomyces cerevisiae. Yeast 2006, 23, 1065–1073. [Google Scholar] [CrossRef]
- Bell, L.; Seshia, A.; Lando, D.; Laue, E.; Palayret, M.; Lee, S.F.; Klenerman, D. A microfluidic device for the hydrodynamic immobilisation of living fission yeast cells for super-resolution imaging. Sens. Actuators B Chem. 2014, 192, 36–41. [Google Scholar] [CrossRef] [Green Version]
- Carlo, D.D.; Wu, L.Y.; Lee, L.P. Dynamic single cell culture array. Lab Chip 2006, 6, 1445–1449. [Google Scholar] [CrossRef]
- Barbulovic-Nad, I.; Lucente, M.; Sun, Y.; Zhang, M.; Wheeler, A.R.; Bussmann, M. Bio-microarray fabrication techniques—A review. Crit. Rev. Biotechnol. 2006, 26, 237–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillemot, F.; Souquet, A.; Catros, S.; Guillotin, B.; Lopez, J.; Faucon, M.; Pippenger, B.; Bareille, R.; Rémy, M.; Bellance, S.; et al. High-throughput laser printing of cells and biomaterials for tissue engineering. Acta Biomater. 2010, 6, 2494–2500. [Google Scholar] [CrossRef] [PubMed]
- Bean, G.J.; Jaeger, P.A.; Bahr, S.; Ideker, T. Development of ultra-high-density screening tools for microbial “omics”. PLoS ONE 2014, 9, e85177. [Google Scholar] [CrossRef] [PubMed]
- Schaack, B.; Reboud, J.; Combe, S.; Fouqué, B.; Berger, F.; Boccard, S.; Odile Filhol-Cochet, F.C. A “DropChip” Cell Array for DNA and siRNA Transfection Combined with Drug Screening. Nanobiotechnology 2005, 1, 183–189. [Google Scholar] [CrossRef]
- Ringeisen, B.R.; Othon, C.M.; Barron, J.A.; Young, D.; Spargo, B.J. Jet-based methods to print living cells. Biotechnol. J. 2006, 1, 930–948. [Google Scholar] [CrossRef]
- Roth, E.A.; Xu, T.; Das, M.; Gregory, C.; Hickman, J.J.; Boland, T. Inkjet printing for high-throughput cell patterning. Biomaterials 2004, 25, 3707–3715. [Google Scholar] [CrossRef]
- Ferris, C.J.; Gilmore, K.G.; Wallace, G.G.; In Het Panhuis, M. Biofabrication: An overview of the approaches used for printing of living cells. Appl. Microbiol. Biotechnol. 2013, 97, 4243–4258. [Google Scholar] [CrossRef]
- Demirci, U.; Montesano, G. Cell encapsulating droplet vitrification. Lab Chip 2007, 7, 1428–1433. [Google Scholar] [CrossRef]
- Lee, W.; Debasitis, J.C.; Lee, V.K.; Lee, J.H.; Fischer, K.; Edminster, K.; Park, J.K.; Yoo, S.S. Multi-layered culture of human skin fibroblasts and keratinocytes through three-dimensional freeform fabrication. Biomaterials 2009, 30, 1587–1595. [Google Scholar] [CrossRef]
- Gonzalez-Macia, L.; Morrin, A.; Smyth, M.R.; Killard, A.J. Advanced printing and deposition methodologies for the fabrication of biosensors and biodevices. Analyst 2010, 135, 845–867. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Rossignol, F.; Macdonald, J. Inkjet printing for biosensor fabrication: Combining chemistry and technology for advanced manufacturing. Lab Chip 2015, 15, 2538–2558. [Google Scholar] [CrossRef] [PubMed]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preibisch, S.; Saalfeld, S.; Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 2009, 25, 1463–1465. [Google Scholar] [CrossRef]
- Pemberton, L.F. Preparation of yeast cells for live-cell imaging and indirect immunofluorescence. Methods Mol. Biol. 2014, 1205, 79–90. [Google Scholar]
- Zhang, R.; Yuan, H.; Wang, S.; Ouyang, Q.; Chen, Y.; Hao, N.; Luo, C. High-throughput single-cell analysis for the proteomic dynamics study of the yeast osmotic stress response. Sci. Rep. 2017, 7, 42200. [Google Scholar] [CrossRef]
- Mirzaei, M.; Pla-Roca, M.; Safavieh, R.; Nazarova, E.; Safavieh, M.; Li, H.; Vogel, J.; Juncker, D. Microfluidic perfusion system for culturing and imaging yeast cell microarrays and rapidly exchanging media. Lab Chip 2010, 10, 2449–2457. [Google Scholar] [CrossRef]
- Kang, D.H.; Han, W.B.; Choi, N.; Kim, Y.J.; Kim, T.S. Tightly Sealed 3D Lipid Structure Monolithically Generated on Transparent SU-8 Microwell Arrays for Biosensor Applications. ACS Appl. Mater. Interfaces 2018, 10, 40401–40410. [Google Scholar] [CrossRef]
- Wu, Z.Z.; Zhao, Y.; Kisaalita, W.S. Interfacing SH-SY5Y human neuroblastoma cells with SU-8 microstructures. Colloids Surf. B Biointerfaces 2006, 52, 14–21. [Google Scholar] [CrossRef]
- Calvert, P. Inkjet printing for materials and devices. Chem. Mater. 2001, 13, 3299–3305. [Google Scholar] [CrossRef]
- Derby, B. Bioprinting: Inkjet printing proteins and hybrid cell-containing materials and structures. J. Mater. Chem. 2008, 18, 5717–5721. [Google Scholar] [CrossRef]
- Newman, J.D.; Turner, A.P.F.; Marrazza, G. Ink-jet printing for the fabrication of amperometric glucose biosensors. Anal. Chim. Acta 1992, 262, 13–17. [Google Scholar] [CrossRef]
- Montenegro-Nicolini, M.; Miranda, V.; Morales, J.O. Inkjet Printing of Proteins: An Experimental Approach. AAPS J. 2017, 19, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Matsugaki, A.; Yamazaki, D.; Nakano, T. Selective patterning of netrin-1 as a novel guiding cue for anisotropic dendrogenesis in osteocytes. Mater. Sci. Eng. C 2020, 108, 110391. [Google Scholar] [CrossRef]
- Xu, T.; Petridou, S.; Lee, E.H.; Roth, E.A.; Vyavahare, N.R.; Hickman, J.J.; Boland, T. Construction of high-density bacterial colony arrays and patterns by the ink-jet method. Biotechnol. Bioeng. 2004, 85, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Lu, J.; Chen, H.; Huang, L.; Cai, J.; Xu, Z. Application of inkjet printing technique for biological material delivery and antimicrobial assays. Anal. Biochem. 2011, 410, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, G.; Maloney, N.; Hegner, M. Ink-jet printing: Perfect tool for cantilever array sensor preparation for microbial growth detection. J. Sensors 2012, 2012. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Chen, F.; He, Z.; Ma, Y.; Uchiyama, K.; Lin, J.M. A novel approach for precisely controlled multiple cell patterning in microfluidic chips by inkjet printing and the detection of drug metabolism and diffusion. Analyst 2016, 141, 2940–2947. [Google Scholar] [CrossRef]
- Collart, M.A.; Struhl, K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994, 8, 525–537. [Google Scholar] [CrossRef] [Green Version]
- Kruk, J.A.; Dutta, A.; Fu, J.; Gilmour, D.S.; Reese, J.C. The multifunctional Ccr4-Not complex directly promotes transcription elongation. Genes Dev. 2011, 25, 581–593. [Google Scholar] [CrossRef] [Green Version]
- Ghaemmaghami, S.; Huh, W.K.; Bower, K.; Howson, R.W.; Belle, A.; Dephoure, N.; O’Shea, E.K.; Weissman, J.S. Global analysis of protein expression in yeast. Nature 2003, 425, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Benton, B.K.; Tinkelenberg, A.; Gonzalez, I.; Cross, F.R. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol. Cell. Biol. 1997, 17, 5067–5076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madden, K.; Snyder, M. Cell Polarity and Morphogenesis in Budding Yeast. Annu. Rev. Microbiol. 1998, 52, 687–744. [Google Scholar] [CrossRef]
- Mino, A.; Tanaka, K.; Kamei, T.; Umikawa, M.; Fujiwara, T.; Takai, Y. Shs1p: A novel member of septin that interacts with Spa2p, involved in polarized growth in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1998, 251, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Visintin, R.; Craig, K.; Hwang, E.S.; Prinz, S.; Tyers, M.; Amon, A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell 1998, 2, 709–718. [Google Scholar] [CrossRef]
- Mendenhall, M.D.; Hodge, A.E. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998, 62, 1191–1243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chymkowitch, P.; Eldholm, V.; Lorenz, S.; Zimmermann, C.; Lindvall, J.M.; Bjørås, M.; Meza-Zepeda, L.A.; Enserink, J.M. Cdc28 kinase activity regulates the basal transcription machinery at a subset of genes. Proc. Natl. Acad. Sci. USA 2012, 109, 10450–10455. [Google Scholar] [CrossRef] [Green Version]
- Weig, M.; Brown, A.J.P. Genomics and the development of new diagnostics and anti-Candida drugs. Trends Microbiol. 2007, 15, 310–317. [Google Scholar] [CrossRef]
- Howson, R.; Huh, W.K.; Ghaemmaghami, S.; Falvo, J.V.; Bower, K.; Belle, A.; Dephoure, N.; Wykoff, D.D.; Weissman, J.S.; O’Shea, E.K. Construction, verification and experimental use of two epitope-tagged collections of budding yeast strains. Comp. Funct. Genom. 2005, 6, 2–16. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.A.; Geva-Zatorsky, N.; Eden, E.; Frenkel-Morgenstern, M.; Issaeva, I.; Sigal, A.; Milo, R.; Cohen-Saidon, C.; Liron, Y.; Kam, Z.; et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 2008, 322, 1511–1516. [Google Scholar] [CrossRef] [Green Version]
- Michael, C.A.; Dominey-Howes, D.; Labbate, M. The Antimicrobial Resistance Crisis: Causes, Consequences, and Management. Front. Public Health 2014, 2, 145. [Google Scholar] [CrossRef] [PubMed]
- Willaert, R. Micro- and Nanoscale Approaches in Antifungal Drug Discovery. Fermentation 2018, 4, 43. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yvanoff, C.; Torino, S.; Willaert, R.G. Robotic Cell Printing for Constructing Living Yeast Cell Microarrays in Microfluidic Chips. Fermentation 2020, 6, 26. https://doi.org/10.3390/fermentation6010026
Yvanoff C, Torino S, Willaert RG. Robotic Cell Printing for Constructing Living Yeast Cell Microarrays in Microfluidic Chips. Fermentation. 2020; 6(1):26. https://doi.org/10.3390/fermentation6010026
Chicago/Turabian StyleYvanoff, Charlotte, Stefania Torino, and Ronnie G. Willaert. 2020. "Robotic Cell Printing for Constructing Living Yeast Cell Microarrays in Microfluidic Chips" Fermentation 6, no. 1: 26. https://doi.org/10.3390/fermentation6010026
APA StyleYvanoff, C., Torino, S., & Willaert, R. G. (2020). Robotic Cell Printing for Constructing Living Yeast Cell Microarrays in Microfluidic Chips. Fermentation, 6(1), 26. https://doi.org/10.3390/fermentation6010026





