Towards a Starter Culture for Cocoa Fermentation by the Selection of Acetic Acid Bacteria
Abstract
1. Introduction
2. Material and Methods
2.1. Cocoa Bean Sampling
2.2. Acetic Acid Bacteria Strains Isolation
2.3. Bacterial Species Identification
2.4. Biochemical Characterization of Acetobacter pasteurianus
2.4.1. Determination of the Growth Kinetics
2.4.2. Quantification of the Metabolite Consumption and Production
2.4.3. Statistical Analysis
2.5. Genomic Diversity Analysis of Acetobacter pasteurianus by Rep-PCR
3. Results
3.1. Identification of the Acetic Acid Bacteria Strains by 16S rRNA Gene Sequencing
3.2. Comparative Analysis of the A. pasteurianus Strains’ Metabolism in a Medium Stimulating Cocoa Pulp
3.2.1. Dendrogram and Heatmap Analysis
3.2.2. Comparison of the Molecule Consumption and Production between the Strain Clusters
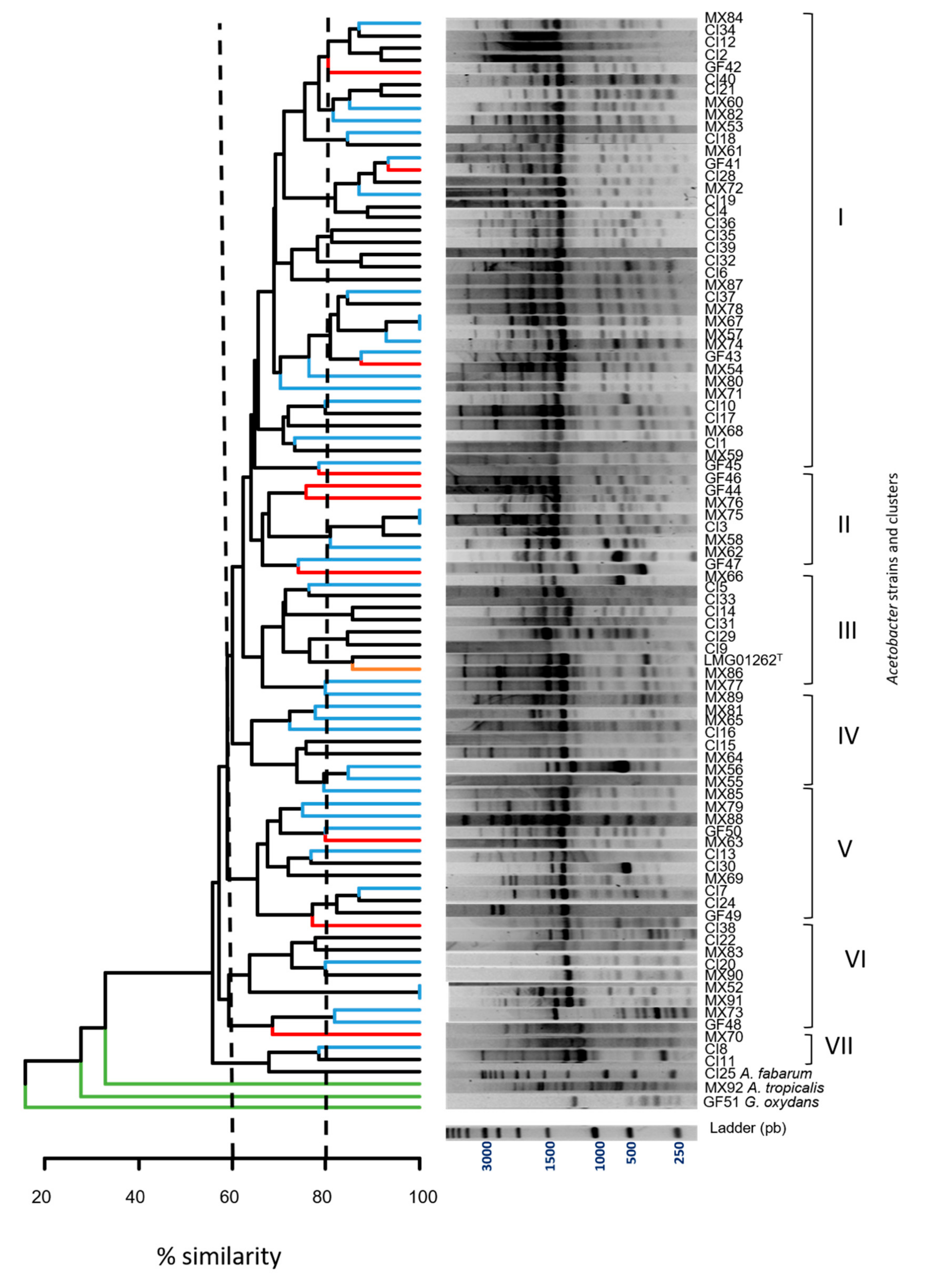
3.3. Genomic Polymorphism Analysis of the Acetic Acid Bacteria by (GTG)5 PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barel, M. Qualité du Cacao: L’impact du Traitement Post-Récolte; Quae Editions; Quae: Versailles, France, 2013; ISBN 978-2-7592-2052-6. [Google Scholar]
- Ozturk, G.; Young, G.M. Food Evolution: The impact of society and science on the fermentation of cocoa beans. Compr. Rev. Food Sci. Food Saf. 2017, 16, 431–455. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Weckx, S. The cocoa bean fermentation process: From ecosystem analysis to starter culture development. J. Appl. Microbiol. 2016, 121, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Schwan, R.F.; Wheals, A.E. The microbiology of cocoa fermentation and its role in chocolate quality. Crit. Rev. Food Sci. Nutr. 2004, 44, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, D.S.; Teniola, O.D.; Ban-Koffi, L.; Owusu, M.; Andersson, T.S.; Holzapfel, W.H. The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int. J. Food Microbiol. 2007, 114, 168–186. [Google Scholar] [CrossRef] [PubMed]
- Camu, N.; Gonzalez, A.; De Winter, T.; Van Schoor, A.; De Bruyne, K.; Vandamme, P.; Takrama, J.S.; Addo, S.K.; De Vuyst, L. Influence of turning and environmental contamination on the dynamics of populations of lactic acid and acetic acid bacteria involved in spontaneous cocoa bean heap fermentation in Ghana. Appl. Environ. Microbiol. 2008, 74, 86–98. [Google Scholar] [CrossRef]
- Camu, N.; De Winter, T.; Verbrugghe, K.; Cleenwerck, I.; Vandamme, P.; Takrama, J.S.; Vancanneyt, M.; De Vuyst, L. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl. Environ. Microbiol. 2007, 73, 1809–1824. [Google Scholar] [CrossRef]
- Daniel, H.M.; Vrancken, G.; Takrama, J.F.; Camu, N.; De Vos, P.; De Vuyst, L. Yeast diversity of Ghanaian cocoa bean heap fermentations. FEMS Yeast Res. 2009, 9, 774–783. [Google Scholar] [CrossRef]
- Garcia-Armisen, T.; Papalexandratou, Z.; Hendryckx, H.; Camu, N.; Vrancken, G.; De Vuyst, L.; Cornelis, P. Diversity of the total bacterial community associated with Ghanaian and Brazilian cocoa bean fermentation samples as revealed by a 16 S rRNA gene clone library. Appl. Microbiol. Biotechnol. 2010, 87, 2281–2292. [Google Scholar] [CrossRef]
- Lefeber, T.; Papalexandratou, Z.; Gobert, W.; Camu, N.; De Vuyst, L. On-farm implementation of a starter culture for improved cocoa bean fermentation and its influence on the flavour of chocolates produced thereof. Food Microbiol. 2012, 30, 379–392. [Google Scholar] [CrossRef]
- Meersman, E.; Steensels, J.; Mathawan, M.; Wittocx, P.J.; Saels, V.; Struyf, N.; Bernaert, H.; Vrancken, G.; Verstrepen, K.J. Detailed analysis of the microbial population in Malaysian Spontaneous Cocoa Pulp Fermentations Reveals a Core and Variable Microbiota. PLoS ONE 2013, 8, e81559. [Google Scholar] [CrossRef]
- Pettipher, G.L. Analysis of cocoa pulp and the formulation of a standardised artificial cocoa pulp medium. J. Sci. Food Agric. 1986, 37, 297–309. [Google Scholar] [CrossRef]
- Papalexandratou, Z.; Lefeber, T.; Bahrim, B.; Lee, O.S.; Daniel, H.M.; De Vuyst, L. Hanseniaspora opuntiae, Saccharomyces cerevisiae, Lactobacillus fermentum, and Acetobacter pasteurianus predominate during well-performed Malaysian cocoa bean box fermentations, underlining the importance of these microbial species for a successful cocoa bean fermentation process. Food Microbiol. 2013, 35, 73–85. [Google Scholar] [CrossRef]
- Papalexandratou, Z.; Vrancken, G.; De Bruyne, K.; Vandamme, P.; De Vuyst, L. Spontaneous organic cocoa bean box fermentations in Brazil are characterized by a restricted species diversity of lactic acid bacteria and acetic acid bacteria. Food Microbiol. 2011, 28, 1326–1338. [Google Scholar] [CrossRef] [PubMed]
- Papalexandratou, Z.; Falony, G.; Romanens, E.; Jimenez, J.C.; Amores, F.; Daniel, H.M.; De Vuyst, L. Species diversity, community dynamics, and metabolite kinetics of the microbiota associated with traditional Ecuadorian spontaneous cocoa bean fermentations. Appl. Environ. Microbiol. 2011, 77, 7698–7714. [Google Scholar] [CrossRef]
- Papalexandratou, Z.; Camu, N.; Falony, G.; De Vuyst, L. Comparison of the bacterial species diversity of spontaneous cocoa bean fermentations carried out at selected farms in Ivory Coast and Brazil. Food Microbiol. 2011, 28, 964–973. [Google Scholar] [CrossRef]
- Ardhana, M. The microbial ecology of cocoa bean fermentations in Indonesia. Int. J. Food Microbiol. 2003, 86, 87–99. [Google Scholar] [CrossRef]
- Crafack, M.; Mikkelsen, M.B.; Saerens, S.; Knudsen, M.; Blennow, A.; Lowor, S.; Takrama, J.; Swiegers, J.H.; Petersen, G.B.; Heimdal, H.; et al. Influencing cocoa flavour using Pichia kluyveri and Kluyveromyces marxianus in a defined mixed starter culture for cocoa fermentation. Int. J. Food Microbiol. 2013, 167, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Holm, C.S.; Aston, J.W.; Douglas, K. The effects of the organic acids in cocoa on the flavour of chocolate. J. Sci. Food Agric. 1993, 61, 65–71. [Google Scholar] [CrossRef]
- John, W.A.; Kumari, N.; Böttcher, N.L.; Koffi, K.J.; Grimbs, S.; Vrancken, G.; D’Souza, R.N.; Kuhnert, N.; Ullrich, M.S. Aseptic artificial fermentation of cocoa beans can be fashioned to replicate the peptide profile of commercial cocoa bean fermentations. Food Res. Int. 2016, 89, 764–772. [Google Scholar] [CrossRef]
- De Taeye, C.; Eyamo Evina, V.J.; Caullet, G.; Niemenak, N.; Collin, S. Fate of Anthocyanins through Cocoa Fermentation. Emergence of New Polyphenolic Dimers. J. Agric. Food Chem. 2016, 64, 8876–8885. [Google Scholar] [CrossRef] [PubMed]
- Urbańska, B.; Derewiaka, D.; Lenart, A.; Kowalska, J. Changes in the composition and content of polyphenols in chocolate resulting from pre-treatment method of cocoa beans and technological process. Eur. Food Res. Technol. 2019, 245, 2101–2112. [Google Scholar] [CrossRef]
- Hansen, C.E.; del Olmo, M.; Burri, C. Enzyme activities in cocoa beans during fermentation. J. Sci. Food Agric. 1998, 77, 273–281. [Google Scholar] [CrossRef]
- Teyssier, C.; Hamdouche, Y. Acetic Acid Bacteria. In Fermented Foods. Part I: Biochemistry and Biotechnology; CRC Press: Boca Raton, FL, USA, 2016; p. 97. [Google Scholar]
- Sengun, I.Y.; Karabiyikli, S. Importance of acetic acid bacteria in food industry. Food Control 2011, 22, 647–656. [Google Scholar] [CrossRef]
- De Roos, J.; De Vuyst, L. Acetic acid bacteria in fermented foods and beverages. Curr. Opin. Biotechnol. 2018, 49, 115–119. [Google Scholar] [CrossRef]
- Bortolini, C.; Patrone, V.; Puglisi, E.; Morelli, L. Detailed analyses of the bacterial populations in processed cocoa beans of different geographic origin, subject to varied fermentation conditions. Int. J. Food Microbiol. 2016, 236, 98–106. [Google Scholar] [CrossRef]
- De Melo Pereira, G.V.; Magalhães, K.T.; de Almeida, E.G.; da Silva Coelho, I.; Schwan, R.F. Spontaneous cocoa bean fermentation carried out in a novel-design stainless steel tank: Influence on the dynamics of microbial populations and physical–chemical properties. Int. J. Food Microbiol. 2013, 161, 121–133. [Google Scholar] [CrossRef]
- De Melo Pereira, G.V.; Da Cruz Pedrozo Miguel, M.G.; Ramos Lacerda, C.; Schwan, R.F. Microbiological and physicochemical characterization of small-scale cocoa fermentations and screening of yeast and bacterial strains to develop a defined starter culture. Appl. Environ. Microbiol. 2012, 78, 5395–5405. [Google Scholar] [CrossRef]
- De Melo Pereira, G.V.; Magalhães, K.T.; Schwan, R.F. rDNA-based DGGE analysis and electron microscopic observation of cocoa beans to monitor microbial diversity and distribution during the fermentation process. Food Res. Int. 2013, 53, 482–486. [Google Scholar] [CrossRef]
- Hamdouche, Y.; Guehi, T.; Durand, N.; Kedjebo, K.B.D.; Montet, D.; Meile, J.C. Dynamics of microbial ecology during cocoa fermentation and drying: Towards the identification of molecular markers. Food Control 2015, 48, 117–122. [Google Scholar] [CrossRef]
- De Vuyst, L.; Camu, N.; De Winter, T.; Vandemeulebroecke, K.; Van de Perre, V.; Vancanneyt, M.; De Vos, P.; Cleenwerck, I. Validation of the (GTG)5-rep-PCR fingerprinting technique for rapid classification and identification of acetic acid bacteria, with a focus on isolates from Ghanaian fermented cocoa beans. Int. J. Food Microbiol. 2008, 125, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Visintin, S.; Alessandria, V.; Valente, A.; Dolci, P.; Cocolin, L. Molecular identification and physiological characterization of yeasts, lactic acid bacteria and acetic acid bacteria isolated from heap and box cocoa bean fermentations in West Africa. Int. J. Food Microbiol. 2016, 216, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Illeghems, K.; De Vuyst, L.; Weckx, S. Complete genome sequence and comparative analysis of Acetobacter Pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem. BMC Genom. 2013, 14, 526. [Google Scholar] [CrossRef]
- Sato, J.; Wakayama, M.; Takagi, K. Lactate dehydrogenase involved in lactate metabolism of Acetobacter pasteurianus. Procedia Environ. Sci. 2015, 28, 67–71. [Google Scholar] [CrossRef][Green Version]
- Lefeber, T.; Janssens, M.; Camu, N.; De Vuyst, L. Kinetic analysis of strains of lactic acid bacteria and acetic acid bacteria in cocoa pulp simulation media toward development of a starter culture for cocoa bean fermentation. Appl. Environ. Microbiol. 2010, 76, 7708–7716. [Google Scholar] [CrossRef]
- Adler, P.; Frey, L.J.; Berger, A.; Bolten, C.J.; Hansen, C.E.; Wittmann, C. The key to acetate: Metabolic fluxes of acetic acid bacteria under cocoa pulp fermentation-simulating conditions. Appl. Environ. Microbiol. 2014, 80, 4702–4716. [Google Scholar] [CrossRef] [PubMed]
- Crafack, M.; Keul, H.; Eskildsen, C.E.; Petersen, M.A.; Saerens, S.; Blennow, A.; Skovmand-Larsen, M.; Swiegers, J.H.; Petersen, G.B.; Heimdal, H.; et al. Impact of starter cultures and fermentation techniques on the volatile aroma and sensory profile of chocolate. Food Res. Int. 2014, 63, 306–316. [Google Scholar] [CrossRef]
- Lefeber, T.; Janssens, M.; Moens, F.; Gobert, W.; De Vuyst, L. Interesting Starter Culture Strains for Controlled Cocoa Bean Fermentation Revealed by Simulated Cocoa Pulp Fermentations of Cocoa-Specific Lactic Acid Bacteria. Appl. Environ. Microbiol. 2011, 77, 6694–6698. [Google Scholar] [CrossRef] [PubMed]
- Magalhães da Veiga Moreira, I.; de Figueiredo Vilela, L.; da Cruz Pedroso Miguel, M.; Santos, C.; Lima, N.; Freitas Schwan, R. Impact of a Microbial Cocktail Used as a Starter Culture on Cocoa Fermentation and Chocolate Flavor. Molecules 2017, 22, 766. [Google Scholar] [CrossRef]
- Organisme International de la Vigne et du vin: Recueil des Méthodes Internationales D’analyses, Analyse Microbiologique des vins et des Moûts: Détection, Différenciation et Dénombrement des Microorganismes 2010. Available online: https://www.oiv.int/fr/technical-standards-and-documents/methods-of-analysis/compendium-of-international-methods-of-analysis-of-wines-and-musts-2-vol (accessed on 24 February 2021).
- Moens, F.; Lefeber, T.; De Vuyst, L. Oxidation of metabolites highlights the microbial interactions and Role of Acetobacter pasteurianus during cocoa bean fermentation. Appl. Environ. Microbiol. 2014, 80, 1848–1857. [Google Scholar] [CrossRef]
- Teyssier, C.; Marchandin, H.; Simeon De Buochberg, M.; Ramuz, M.; Jumas-Bilak, E. Atypical 16S rRNA gene copies in Ochrobactrum intermedium strains reveal a large genomic rearrangement by recombination between rrn copies. J. Bacteriol. 2003, 185, 2901–2909. [Google Scholar] [CrossRef]
- Tulini, F.L.; Winkelströter, L.K.; De Martinis, E.C.P. Identification and evaluation of the probiotic potential of Lactobacillus paraplantarum FT259, a bacteriocinogenic strain isolated from Brazilian semi-hard artisanal cheese. Anaerobe 2013, 22, 57–63. [Google Scholar] [CrossRef]
- Altschul, S. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approch using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Alfenore, S.; Molina-Jouve, C.; Guillouet, S.; Uribelarrea, J.-L.; Goma, G.; Benbadis, L. Improving ethanol production and viability of Saccharomyces cerevisiae by a vitamin feeding strategy during fed-batch process. Appl. Microbiol. Biotechnol. 2002, 60, 67–72. [Google Scholar] [CrossRef]
- Bailey, J.E.; Ollis, D.F. Biochemical Engineering Fundamentals, 2nd ed.; McGraw-Hill Chemical Engineering Series; McGraw-Hill: New York, NY, USA, 1986; ISBN 978-0-07-003212-5. [Google Scholar]
- Steensels, J.; Meersman, E.; Snoek, T.; Saels, V.; Verstrepen, K.J. Large-Scale Selection and Breeding to generate industrial yeasts with superior aroma production. Appl. Environ. Microbiol. 2014, 80, 6965–6975. [Google Scholar] [CrossRef]
- Brase, C.H. Understanding Basic Statistics, 8th ed; Cengage Learning, Inc.: Boston, MA, USA, 2017; ISBN 978-1-337-55807-5. [Google Scholar]
- Versalovic, J.; Koeuth, T.; Lupski, R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef]
- Versalovic, J.; Schneider, M.; de Bruijn, F.; Lupski, J. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 1994, 5, 25–40. [Google Scholar]
- Gonzalez, A.; Hierro, N.; Poblet, M.; Rozes, N.; Mas, A.; Guillamon, J.M. Application of molecular methods for the differentiation of acetic acid bacteria in a red wine fermentation. J. Appl. Microbiol. 2004, 96, 853–860. [Google Scholar] [CrossRef]
- Valera, M.J.; Torija, M.J.; Mas, A.; Mateo, E. Acetobacter malorum and Acetobacter cerevisiae identification and quantification by Real-Time PCR with TaqMan-MGB probes. Food Microbiol. 2013, 36, 30–39. [Google Scholar] [CrossRef]
- Ramasamy, D.; Mishra, A.K.; Lagier, J.-C.; Padhmanabhan, R.; Rossi, M.; Sentausa, E.; Raoult, D.; Fournier, P.-E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int. J. Syst. Evol. Microbiol. 2014, 64, 384–391. [Google Scholar] [CrossRef]
- Cleenwerck, I.; Camu, N.; Engelbeen, K.; De Winter, T.; Vandemeulebroecke, K.; De Vos, P.; De Vuyst, L. Acetobacter ghanensis sp. nov., a novel acetic acid bacterium isolated from traditional heap fermentations of Ghanaian cocoa beans. Int. J. Syst. Evol. Microbiol. 2007, 57, 1647–1652. [Google Scholar] [CrossRef]
- Cleenwerck, I.; Gonzalez, A.; Camu, N.; Engelbeen, K.; De Vos, P.; De Vuyst, L. Acetobacter fabarum sp. nov., an acetic acid bacterium from a Ghanaian cocoa bean heap fermentation. Int. J. Syst. Evol. Microbiol. 2008, 58, 2180–2185. [Google Scholar] [CrossRef]
- Sokollek, S.J.; Hertel, C.; Hammes, W.P. Description of Acetobacter oboediens sp. nov. and Acetobacter pomorum sp. nov., two new species isolated from industrial vinegar fermentations. Int. J. Syst. Bacteriol. 1998, 48, 935–940. [Google Scholar] [CrossRef]
- Baek, C.H.; Park, E.H.; Baek, S.Y.; Jeong, S.T.; Kim, M.D.; Kwon, J.-H.; Jeong, Y.J.; Yeo, S.H. Characterization of Acetobacter pomorum KJY8 Isolated from Korean Traditional Vinegar. J. Microbiol. Biotechnol. 2014, 24, 1679–1684. [Google Scholar] [CrossRef]
- Figueroa-Hernández, C.; Mota-Gutierrez, J.; Ferrocino, I.; Hernández-Estrada, Z.J.; González-Ríos, O.; Cocolin, L.; Suárez-Quiroz, M.L. The challenges and perspectives of the selection of starter cultures for fermented cocoa beans. Int. J. Food Microbiol. 2019, 301, 41–50. [Google Scholar] [CrossRef]
- Papalexandratou, Z.; Cleenwerck, I.; De Vos, P.; De Vuyst, L. (GTG)5 -PCR reference framework for acetic acid bacteria. FEMS Microbiol. Lett. 2009, 301, 44–49. [Google Scholar] [CrossRef]
- Azuma, Y.; Hosoyama, A.; Matsutani, M.; Furuya, N.; Horikawa, H.; Harada, T.; Hirakawa, H.; Kuhara, S.; Matsushita, K.; Fujita, N.; et al. Whole-genome analyses reveal genetic instability of Acetobacter pasteurianus. Nucleic Acids Res. 2009, 37, 5768–5783. [Google Scholar] [CrossRef]
- Wu, J.; Gullo, M.; Chen, F.; Giudici, P. Diversity of Acetobacter pasteurianus strains isolated from solid-state fermentation of cereal vinegars. Curr. Microbiol. 2010, 60, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, K.; Arai, H.; Ishii, M.; Igarashi, Y. Transcriptome response to different carbon sources in Acetobacter aceti. Microbiology 2011, 157, 899–910. [Google Scholar] [CrossRef]
- Saeki, A.; Matsushita, K.; Takeno, S.; Taniguchi, M.; Toyama, H.; Theeragool, G.; Lotong, N.; Adachi, O. Enzymes responsible for acetate oxidation by acetic acid bacteria. Biosci. Biotechnol. Biochem. 1999, 63, 2102–2109. [Google Scholar] [CrossRef][Green Version]
- Yang, H.; Yu, Y.; Fu, C.; Chen, F. Bacterial acid resistance toward organic weak acid revealed by RNA-Seq transcriptomic analysis in Acetobacter pasteurianus. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- De Ley, J.; Swings, J. Genus Gluconobacter. In Bergey’s Manual of Systematic Bacteriology; Williams and Wilkins: Philadelphia, PA, USA, 1984; pp. 275–278. [Google Scholar]
- De Muynck, C.; De Melo Pereira, C.S.S.; Naessens, M.; Parmentier, S.; Soetaert, W.; Vandamme, E.J. The Genus Gluconobacter Oxydans: Comprehensive overview of biochemistry and biotechnological applications. Crit. Rev. Biotechnol. 2007, 27, 147–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Shao, Y.; Chen, T.; Chen, W.; Chen, F. Global insights into acetic acid resistance mechanisms and genetic stability of Acetobacter pasteurianus strains by comparative genomics. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Beppu, T. Genetic organization of Acetobacter for acetic acid fermentation. Antonie Van Leeuwenhoek 1994, 64, 121–135. [Google Scholar] [CrossRef]
- Matsutani, M.; Nishikura, M.; Saichana, N.; Hatano, T.; Masud-Tippayasak, U.; Theergool, G.; Yakushi, T.; Matsushita, K. Adaptive mutation of Acetobacter pasteurianus SKU1108 enhances acetic acid fermentation ability at high temperature. J. Biotechnol. 2013, 165, 109–119. [Google Scholar] [CrossRef] [PubMed]


| Origin | Sampling Date | Geographical Area | Fermentation Type | Duration of Fermentation (Days) | Number of Fermentation Box/Heap |
|---|---|---|---|---|---|
| Ivory Coast | 2016 | Wagana | Box | 6 | 6 |
| Ivory Coast | 2018 | Abidjan | Heap | 7 | 1 |
| French Guiana | 2017 | Combi | Box | 7 | 1 |
| Mexico | 02/2018 | Comalcalco | Box | 5 | 2 |
| Mexico | 10/2018 | Comalcalco | Box | 4 | 2 |
| Bacterial Species Identification | Total Strains | Ivory Coast | French Guiana | Mexico | ||
|---|---|---|---|---|---|---|
| 2016 | 2018 | Main | Mid Crop | |||
| A. pasteurianus | 89 * | 24 * | 15 | 10 | 11 | 29 |
| A. tropicalis | 16 | 5 | 2 | 2 | 5 | 2 |
| A. fabarum | 2 | 1 | 1 | 0 | 0 | 0 |
| A. ghanensis | 7 | 5 | 2 | 0 | 0 | 0 |
| A. orientalis | 2 | 0 | 0 | 2 | 0 | 0 |
| A. malorum/A. cerevisiae | 6 | 0 | 0 | 0 | 0 | 6 |
| G. oxydans | 7 | 1 | 0 | 6 | 0 | 0 |
| G. nephelii | 1 | 1 | 0 | 0 | 0 | 0 |
| Sub-total | 37 | 20 | 16 | 37 | ||
| Total strains | 130 | 57 | 20 | 53 | ||
| Metabolism Group | Isolates Number | Isolates | Origin Country |
|---|---|---|---|
| I | 42 | LMG01262T; CI1; CI10; CI11; CI14; CI16; CI17; CI18; CI20; CI21;CI28; CI30; CI31; CI33; CI36; CI38; CI39; CI40; CI6; CI7; CI9; GF41; GF42; GF43; GF45; MX52; MX56; MX60; MX62; MX63; MX64; MX67; MX68; MX69; MX72; MX73; MX74; MX75; MX79; MX80; MX84; MX86; GF51 | Ivory Coast (20/36, 55.5% *), Mexico (17/40,42.5% *), and French Guiana (4/10, 40% *) |
| II | 44 | CI12; CI13; CI15; CI19; CI2; CI22; CI24;; CI29; CI3; CI32; CI34; CI35; CI37; CI4; CI5; CI8; GF44; GF46; GF47; GF48; GF49; GF50; MX53; MX54; MX55; MX57; MX58; MX59; MX65; MX66; MX70; MX71; MX76; MX77; MX78; MX81; MX82; MX83; MX85; MX87; MX88; MX89; MX90; MX91 | Mexico (22/40, 55% *), Ivory Coast (16/36, 44.5% *), French Guiana (6/10, 60% *) |
| (GTG)5 Grouping | Isolates Number | % Clustering | Isolates | Origin Country |
|---|---|---|---|---|
| I | 38 | 62.5% | MX84; CI34; CI12; CI2; GF42; CI40; CI21; MX60; MX82; MX53; CI18; MX61; GF41; CI28; MX72; CI19; CI4; CI36; CI35; CI39; CI32; CI6; MX87; CI37; MX78; MX67; MX57; MX74; GF43; MX54; MX80; MX71; CI10; CI17; MX68; CI1; MX59; GF45 | Mexico (40% *), Ivory Coast (50% *), French Guiana (30% *) |
| II | 8 | 65% | GF46; GF44; MX76; MX75; CI3; MX58; MX62; GF47 | Mexico (10% *), French Guiana (30% *), Ivory Coast (3% *) |
| III | 10 | 70% | MX66; CI5; CI33; CI14; CI31; CI29; CI9; LMG01262T; MX86; MX77 | Ivory Coast (17% *), Mexico (7.5% *) |
| IV | 8 | 65% | MX80; MX81; MX65; CI16; CI15; MX64; MX56; MX55 | Mexico (15% *), Ivory Coast (5.5% *) |
| V | 11 | 70% | MX85; MX79; MX88; GF50; MX63; CI13; CI30; MX69; CI7; CI24; GF49 | Mexico (12.5% *), Ivory Coast (11% *), French Guiana (20% *) |
| VI | 9 | 60% | CI38; CI22; MX83; CI20; MX90; MX52; MX91; MX73; GF48 | Mexico (12.5% *), Ivory Coast (8% *), French Guiana (10% *) |
| VII | 3 | 70% | MX70; CI8; CI11 | Mexico (2.5% *), Ivory Coast (5.5% *) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farrera, L.; Colas de la Noue, A.; Strub, C.; Guibert, B.; Kouame, C.; Grabulos, J.; Montet, D.; Teyssier, C. Towards a Starter Culture for Cocoa Fermentation by the Selection of Acetic Acid Bacteria. Fermentation 2021, 7, 42. https://doi.org/10.3390/fermentation7010042
Farrera L, Colas de la Noue A, Strub C, Guibert B, Kouame C, Grabulos J, Montet D, Teyssier C. Towards a Starter Culture for Cocoa Fermentation by the Selection of Acetic Acid Bacteria. Fermentation. 2021; 7(1):42. https://doi.org/10.3390/fermentation7010042
Chicago/Turabian StyleFarrera, Lucie, Alexandre Colas de la Noue, Caroline Strub, Benjamin Guibert, Christelle Kouame, Joël Grabulos, Didier Montet, and Corinne Teyssier. 2021. "Towards a Starter Culture for Cocoa Fermentation by the Selection of Acetic Acid Bacteria" Fermentation 7, no. 1: 42. https://doi.org/10.3390/fermentation7010042
APA StyleFarrera, L., Colas de la Noue, A., Strub, C., Guibert, B., Kouame, C., Grabulos, J., Montet, D., & Teyssier, C. (2021). Towards a Starter Culture for Cocoa Fermentation by the Selection of Acetic Acid Bacteria. Fermentation, 7(1), 42. https://doi.org/10.3390/fermentation7010042







