In Vitro Assessment of Probiotic and Technological Properties of Lactic Acid Bacteria Isolated from Indigenously Fermented Cereal-Based Food Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation and Purification of LAB Isolates
2.3. Evaluation of Probiotic Properties (In Vitro)
2.3.1. Acid Tolerance
2.3.2. Bile Tolerance
2.3.3. Antagonistic Activity
2.3.4. Antifungal Activity
2.3.5. Resistance to Phenol
2.3.6. Tolerance to Lysozyme
2.3.7. Cell Autoaggregation
2.3.8. Cell-Surface Hydrophobicity
2.3.9. Survivability in Gastric and Pancreatic Juices
2.3.10. Antioxidative Potential of LAB Isolates
2.3.11. Hydrogen Peroxide Production
2.3.12. Bile Salt Hydrolase (BSH) Activity
2.4. Safety Assessment
2.4.1. Antibiotic Sensitivity Test
2.4.2. Hemolytic Activity
2.4.3. DNase Activity
2.4.4. Qualitative Evaluation for Biogenic Amine (BA) Formation
2.5. Technological Properties of the LAB Isolates
2.5.1. Amylolytic Activity
2.5.2. Screening of Exopolysaccharides
2.5.3. Milk Fermenting Ability
2.6. Phenotypic and Biochemical Characterization
2.7. Molecular Identification of Probiotic LAB Isolates Using 16s rDNA Gene Sequencing
2.8. Statistical Analysis
3. Results
3.1. Acid and Bile Tolerance
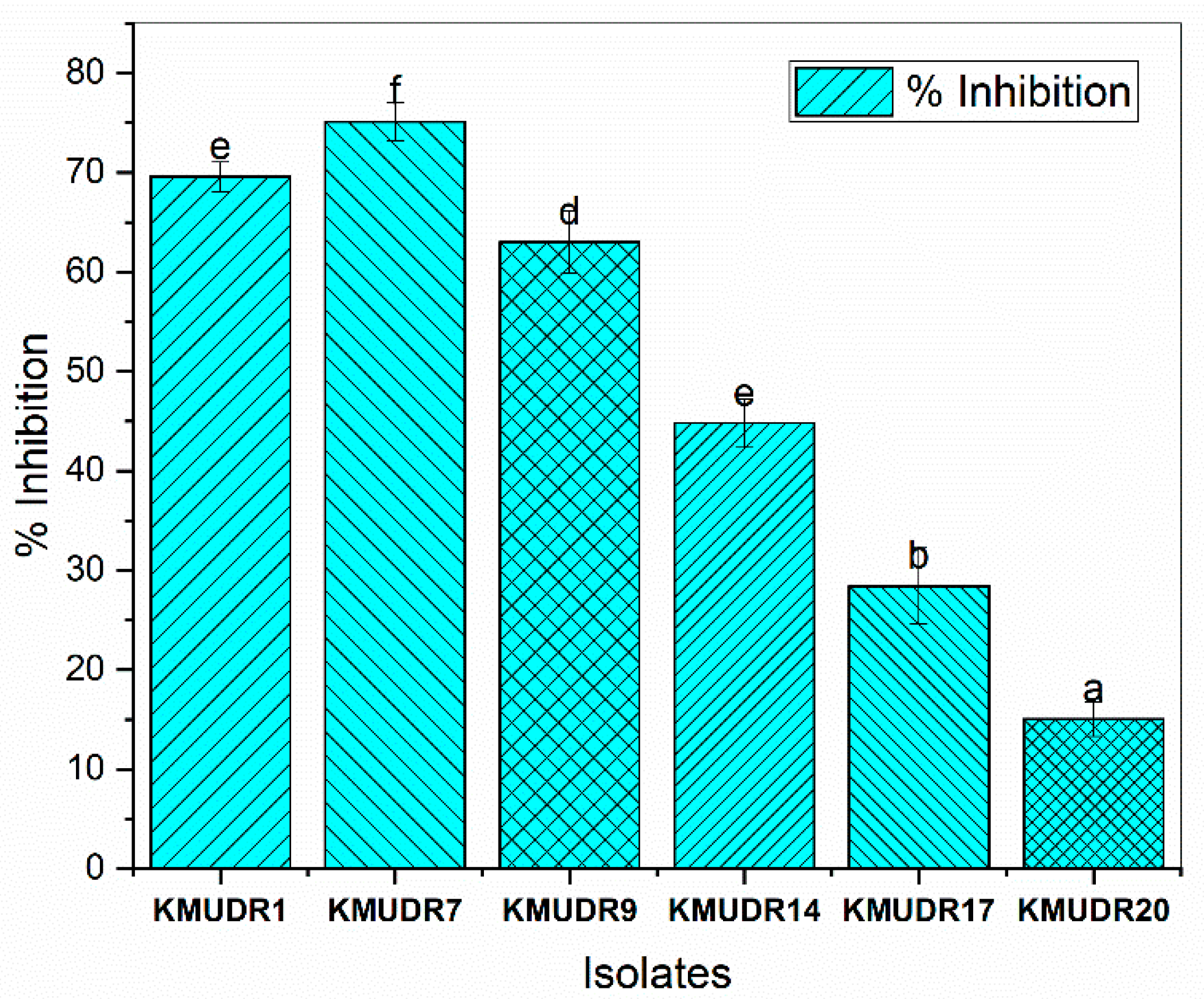
3.2. Antagonistic and Antifungal Activity
3.3. Resistance to Phenol
3.4. Lysozyme Tolerance and Cell-Surface Properties
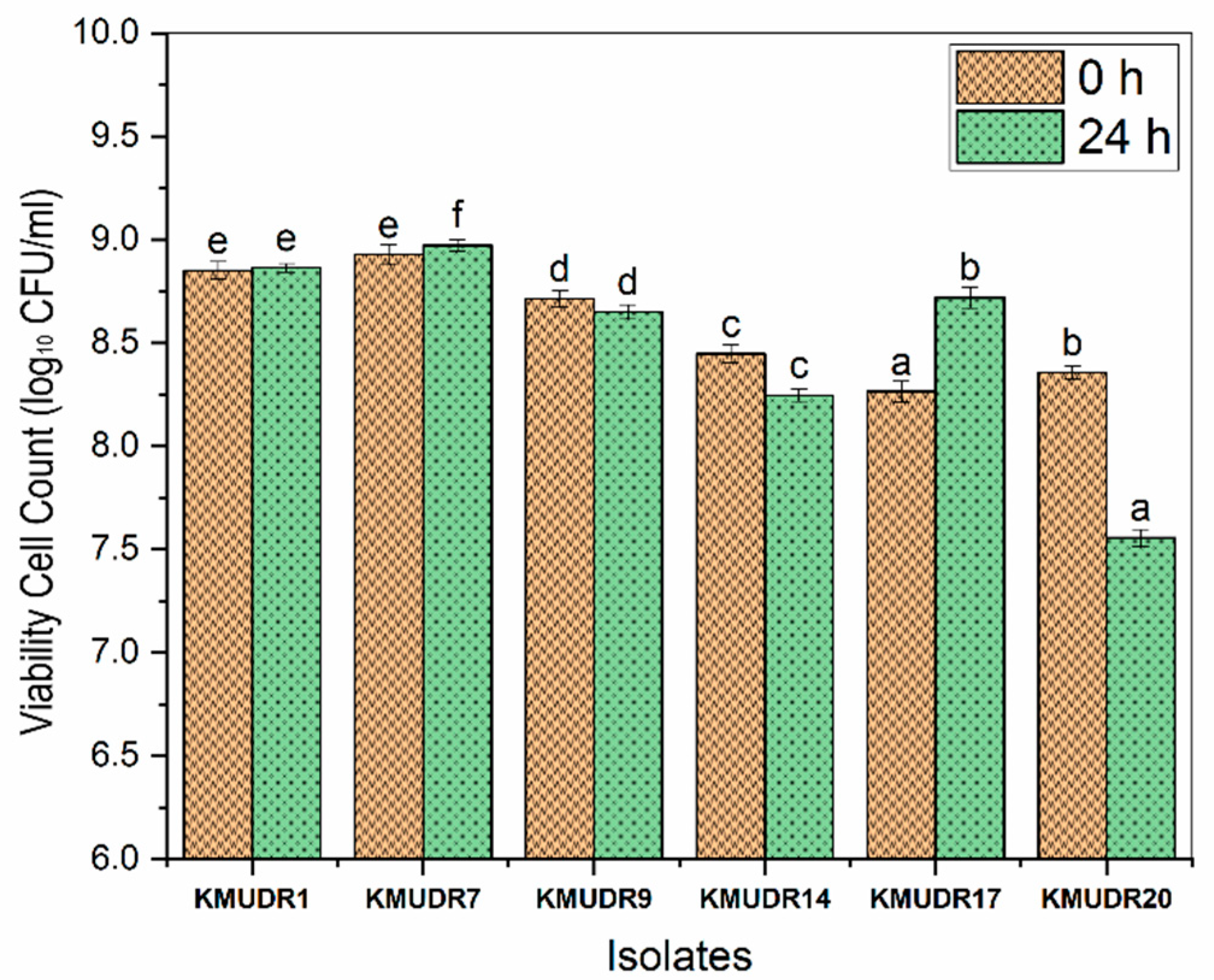
3.5. Survivability in Gastric and Pancreatic Juices
3.6. Antioxidative Potential
3.7. H2O2 Production and BSH Activity
3.8. Safety Assessment
3.8.1. Antibiotic Sensitivity Test
3.8.2. Hemolytic Activity, DNase Activity, and Biogenic Amine Production
3.9. Technological Properties
3.9.1. Amylolytic Activity
3.9.2. EPS Production
3.9.3. Milk Fermenting Ability
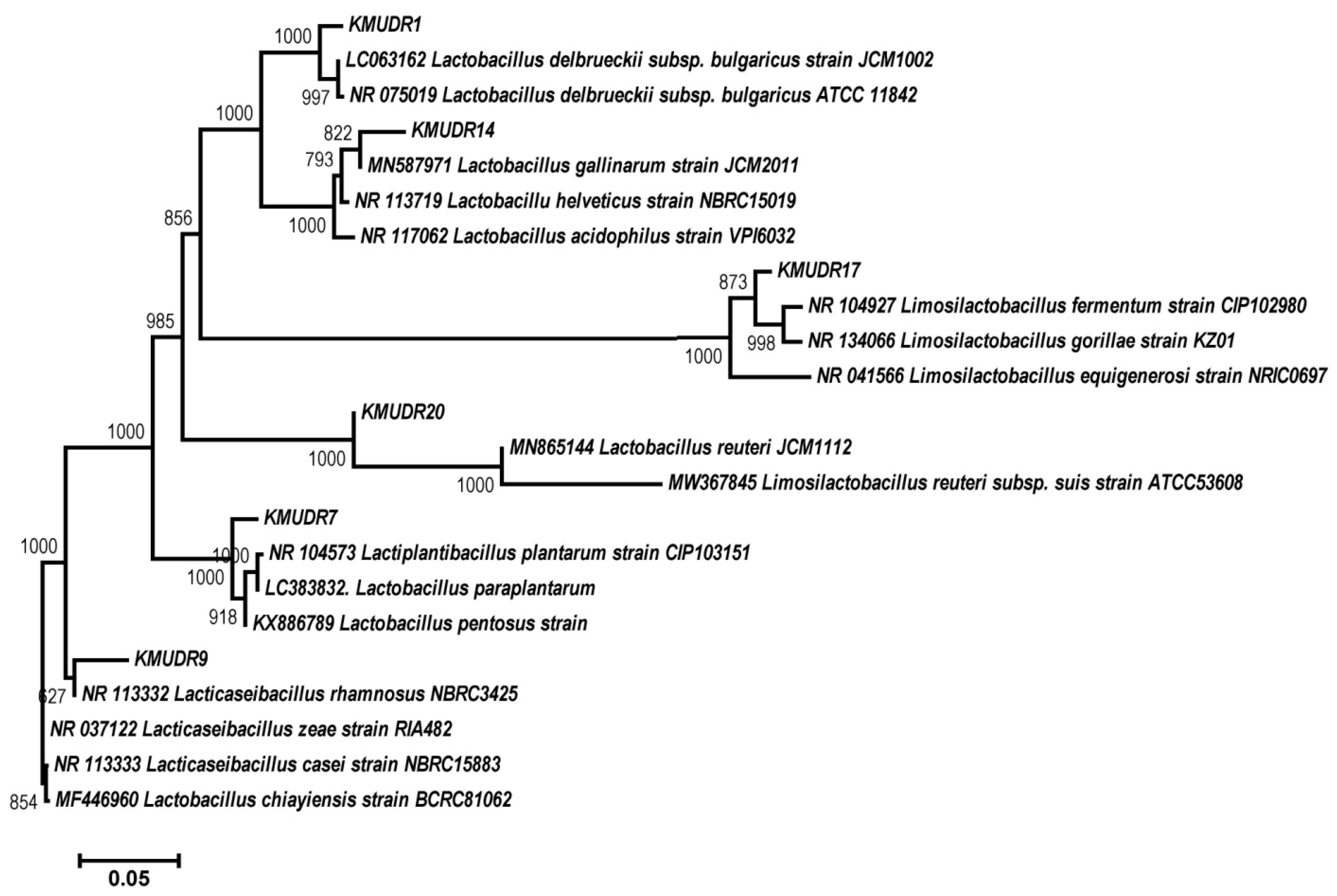
3.10. Phenotyping and Biochemical and Molecular Characterizations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mallappa, R.H.; Balasubramaniam, C.; Nataraj, B.H.; Ramesh, C.; Kadyan, S.; Pradhan, D.; Muniyappa, S.K.; Grover, S. Microbial Diversity and Functionality of Traditional Fermented Milk Products of India: Current Scenario and Future Perspectives. Int. Dairy J. 2021, 114, 104941. [Google Scholar] [CrossRef]
- Saeed, Z.K.; Abbas, B.A.; Othman, R.M. Molecular identification and phylogenetic analysis of lactic acid bacteria isolated from goat raw milk. Iraqi J. Vet. Sci. 2020, 34, 259–263. [Google Scholar] [CrossRef]
- Setta, M.C.; Matemu, A.; Mbega, E.R. Potential of probiotics from fermented cereal-based beverages in improving health of poor people in Africa. J. Food Sci. Technol. 2020, 57, 3935–3946. [Google Scholar] [CrossRef]
- Cunningham, M.; Azcarate-Peril, M.A.; Barnard, A.; Benoit, V.; Grimaldi, R.; Guyonnet, D.; Holscher, H.D.; Hunter, K.; Manurung, S.; Obis, D.; et al. Shaping the Future of Probiotics and Prebiotics. Trends Microbiol. 2021, 29, 667–685. [Google Scholar] [CrossRef]
- Abarquero, D.; Renes, E.; Fresno, J.M.; Tornadijo, M.E. Study of Exopolysaccharides from Lactic Acid Bacteria and Their Industrial Applications: A Review. Int. J. Food Sci. Technol. 2022, 57, 16–26. [Google Scholar] [CrossRef]
- Endo, A.; Tanizawa, Y.; Arita, M. Isolation and Identification of Lactic Acid Bacteria from Environmental Samples. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; Volume 1887. [Google Scholar]
- Verni, M.; Rizzello, C.G.; Coda, R. Fermentation Biotechnology Applied to Cereal Industry By-Products: Nutritional and Functional Insights. Front. Nutr. 2019, 6, 42. [Google Scholar] [CrossRef] [Green Version]
- Rezac, S.; Kok, C.R.; Heermann, M.; Hutkins, R. Fermented Foods as a Dietary Source of Live Organisms. Front. Microbiol. 2018, 9, 1785. [Google Scholar] [CrossRef]
- Sethi, S.; Anurag, R.K. Probiotic and Prebiotic Plant Milk Dairy Foods. In Probiotics and Prebiotics in Foods; Academic Press: Cambridge, MA, USA, 2021. [Google Scholar] [CrossRef]
- Vinderola, G.; Reinheimer, J.; Salminen, S. The Enumeration of Probiotic Issues: From Unavailable Standardised Culture Media to a Recommended Procedure? Int. Dairy J. 2019, 96, 58–65. [Google Scholar] [CrossRef]
- Rodríguez, L.G.R.; Mohamed, F.; Bleckwedel, J.; Medina, R.B.; De Vuyst, L.; Hebert, E.M.; Mozzi, F. Diversity and Functional Properties of Lactic Acid Bacteria Isolated From Wild Fruits and Flowers Present in Northern Argentina. Front. Microbiol. 2019, 10, 1091. [Google Scholar] [CrossRef]
- Paiva, I.H.R.; Duarte-Silva, E.; Peixoto, C.A. The role of prebiotics in cognition, anxiety, and depression. Eur. Neuropsychopharmacol. 2020, 34, 1–18. [Google Scholar] [CrossRef]
- Górska, A.; Przystupski, D.; Niemczura, M.J.; Kulbacka, J. Probiotic Bacteria: A Promising Tool in Cancer Prevention and Therapy. Curr. Microbiol. 2019, 76, 939–949. [Google Scholar] [CrossRef] [Green Version]
- Ofosu, F.K.; Elahi, F.; Daliri, E.B.-M.; Han, S.-I.; Oh, D.-H. Impact of thermal treatment and fermentation by lactic acid bacteria on sorghum metabolite changes, their antioxidant and antidiabetic activities. Food Biosci. 2021, 45, 101502. [Google Scholar] [CrossRef]
- Lei, W.; Liu, C.; Pan, L.; Peng, C.; Wang, J.; Zhou, H. Screening of probiotic Lactobacilli with potential anti-allergic activity based on hyaluronidase inhibition and degranulation of RBL-2H3 cells in vitro. LWT 2020, 140, 110707. [Google Scholar] [CrossRef]
- Gil-Rodríguez, A.M.; Beresford, T. Bile Salt Hydrolase and Lipase Inhibitory Activity in Reconstituted Skim Milk Fermented with Lactic Acid Bacteria. J. Funct. Foods 2021, 77, 104342. [Google Scholar] [CrossRef]
- Barcenilla, C.; Ducic, M.; López, M.; Prieto, M.; Álvarez-Ordóñez, A. Application of lactic acid bacteria for the biopreservation of meat products: A systematic review. Meat Sci. 2021, 183, 108661. [Google Scholar] [CrossRef]
- Shahbazi, R.; Sharifzad, F.; Bagheri, R.; Alsadi, N.; Yasavoli-Sharahi, H.; Matar, C. Anti-Inflammatory and Immunomodulatory Properties of Fermented Plant Foods. Nutrients 2021, 13, 1516. [Google Scholar] [CrossRef]
- Meena, K.K.; Kapila, S.; Chand, R.; Kumawat, D.K.; Verma, G. Specific Immune Response of Oral Administration of Dahi and Milk Fermented with Lactobacillus helveticus against Salmonella enteritidis in Mice. Indian J. Dairy Sci. 2017, 70, 404–410. [Google Scholar]
- Minj, J.; Chandra, P.; Paul, C.; Sharma, R.K. Bio-functional properties of probiotic Lactobacillus: Current applications and research perspectives. Crit. Rev. Food Sci. Nutr. 2020, 61, 2207–2224. [Google Scholar] [CrossRef]
- Mulaw, G.; Tessema, T.S.; Muleta, D.; Tesfaye, A. In Vitro Evaluation of Probiotic Properties of Lactic Acid Bacteria Isolated from Some Traditionally Fermented Ethiopian Food Products. Int. J. Microbiol. 2019, 2019, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilango, S.; Antony, U. Probiotic microorganisms from non-dairy traditional fermented foods. Trends Food Sci. Technol. 2021, 118, 617–638. [Google Scholar] [CrossRef]
- Tamang, J.P.; Cotter, P.D.; Endo, A.; Han, N.S.; Kort, R.; Liu, S.Q.; Mayo, B.; Westerik, N.; Hutkins, R. Fermented Foods in a Global Age: East meets West. Compr. Rev. Food Sci. Food Saf. 2020, 19, 184–217. [Google Scholar] [CrossRef] [Green Version]
- Kohajdová, Z. Fermented Cereal Products; Elsevier, B.V.: Amsterdam, The Netherlands, 2017; ISBN 9780444636775. [Google Scholar]
- Aspri, M.; Papademas, P.; Tsaltas, D. Review on Non-Dairy Probiotics and Their Use in Non-Dairy Based Products. Fermentation 2020, 6, 30. [Google Scholar] [CrossRef] [Green Version]
- Chaudhari, A.; Dwivedi, M.K. The Concept of Probiotics, Prebiotics, Postbiotics, Synbiotics, Nutribiotics, and Pharmabiotics. In Probiotics in the Prevention and Management of Human Diseases; Academic Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Ray, M.; Ghosh, K.; Singh, S.; Mondal, K.C. Folk to functional: An explorative overview of rice-based fermented foods and beverages in India. J. Ethn. Foods 2016, 3, 5–18. [Google Scholar] [CrossRef] [Green Version]
- Nath, S.; Roy, M.; Sikidar, J.; Deb, B.; Sharma, I.; Guha, A. Characterization and in-vitro screening of probiotic potential of novel Weissella confusa strain GCC_19R1 isolated from fermented sour rice. Curr. Res. Biotechnol. 2021, 3, 99–108. [Google Scholar] [CrossRef]
- Sreeja, V.; Prajapati, J.B. Ethnic Fermented Foods and Beverages of Gujarat and Rajasthan. In Ethnic Fermented Foods and Beverages of India: Science History and Culture; Springer: New Delhi, India, 2020. [Google Scholar]
- Pintu, R.; Verma, B. Optimization of Rabadi-like Sorghum Based Fermented Milk Beverage. J. Agrisearch 2019, 6, 194–198. [Google Scholar] [CrossRef]
- McFarland, L.V.; Evans, C.T.; Goldstein, E.J.C. Strain-Specificity and Disease-Specificity of Probiotic Efficacy: A Systematic Review and Meta-Analysis. Front. Med. 2018, 5, 124. [Google Scholar] [CrossRef] [Green Version]
- Ahire, J.J.; Jakkamsetty, C.; Kashikar, M.S.; Lakshmi, S.G.; Madempudi, R.S. In Vitro Evaluation of Probiotic Properties of Lactobacillus plantarum UBLP40 Isolated from Traditional Indigenous Fermented Food. Probiotics Antimicrob. Proteins 2021, 13, 1413–1424. [Google Scholar] [CrossRef]
- Yadav, R.; Puniya, A.K.; Shukla, P. Probiotic Properties of Lactobacillus Plantarum RYPR1 from an Indigenous Fermented Beverage Raabadi. Front. Microbiol. 2016, 7, 1683. [Google Scholar] [CrossRef] [Green Version]
- Aladeboyeje, O.T.; Sanli, N.O.; Buyuk, U. Evaluation of the Antimicrobial Efficacy of Some Fermented Traditional Turkish Beverages with Probiotic Potentials. Johns. Matthey Technol. Rev. 2021, 66, 337–350. [Google Scholar] [CrossRef]
- Heperkan, Z.D.; Bolluk, M.; Bülbül, S.S. Structural analysis and properties of dextran produced by Weissella confusa and the effect of different cereals on its rheological characteristics. Int. J. Biol. Macromol. 2019, 143, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Şentürk, D.Z.; Dertli, E.; Erten, H.; Şimşek, Ö. Structural and technological characterization of ropy exopolysaccharides produced by Lactobacillus plantarum strains isolated from Tarhana. Food Sci. Biotechnol. 2019, 29, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Fekri, A.; Torbati, M.; Khosrowshahi, A.Y.; Shamloo, H.B.; Azadmard-Damirchi, S. Functional effects of phytate-degrading, probiotic lactic acid bacteria and yeast strains isolated from Iranian traditional sourdough on the technological and nutritional properties of whole wheat bread. Food Chem. 2019, 306, 125620. [Google Scholar] [CrossRef]
- Somashekaraiah, R.; Shruthi, B.; Deepthi, B.V.; Sreenivasa, M.Y. Probiotic Properties of Lactic Acid Bacteria Isolated From Neera: A Naturally Fermenting Coconut Palm Nectar. Front. Microbiol. 2019, 10, 1382. [Google Scholar] [CrossRef]
- Zago, M.; Fornasari, M.E.; Carminati, D.; Burns, P.; Suàrez, V.; Vinderola, G.; Reinheimer, J.; Giraffa, G. Characterization and probiotic potential of Lactobacillus plantarum strains isolated from cheeses. Food Microbiol. 2011, 28, 1033–1040. [Google Scholar] [CrossRef]
- Bhushan, B.; Sakhare, S.M.; Narayan, K.S.; Kumari, M.; Mishra, V.; Dicks, L.M.T. Characterization of Riboflavin-Producing Strains of Lactobacillus plantarum as Potential Probiotic Candidate through in vitro Assessment and Principal Component Analysis. Probiotics Antimicrob. Proteins 2020, 13, 453–467. [Google Scholar] [CrossRef]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th Ed. CLSI M100 ED30:2020; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- Matuschek, E.; Brown, D.F.J.; Kahlmeter, G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014, 20, O255–O266. [Google Scholar] [CrossRef] [Green Version]
- Sangprapai, W.; Prasitpuriprecha, C.; Jantama, K.; Jantama, S.S. Probiotics Isolated from Thai Fermented Foods for Potential Uses against Foodborne Pathogens. Asia-Pac. J. Sci. Technol. 2022, 27, 1–11. [Google Scholar] [CrossRef]
- Adesulu-Dahunsi, A.T.; Jeyaram, K.; Sanni, A.I.; Banwo, K. Production of exopolysaccharide by strains of Lactobacillus plantarum YO175 and OF101 isolated from traditional fermented cereal beverage. PeerJ 2018, 6, e5326. [Google Scholar] [CrossRef] [Green Version]
- Ruas-Madiedo, P.; Reyes-Gavilán, C.D.L. Invited Review: Methods for the Screening, Isolation, and Characterization of Exopolysaccharides Produced by Lactic Acid Bacteria. J. Dairy Sci. 2005, 88, 843–856. [Google Scholar] [CrossRef] [Green Version]
- Samedi, L.; Charles, A.L. Isolation and characterization of potential probiotic Lactobacilli from leaves of food plants for possible additives in pellet feeding. Ann. Agric. Sci. 2019, 64, 55–62. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.R.; Sardi, J.D.C.O.; Pitangui, N.D.S.; Roque, S.M.; da Silva, A.C.B.; Rosalen, P.L. Probiotics as an alternative antimicrobial therapy: Current reality and future directions. J. Funct. Foods 2020, 73, 104080. [Google Scholar] [CrossRef]
- Jung, J.; Kim, S.; Lee, J.Y.; Yoon, S.; You, S.; Kim, S.H. Multifunctional properties of Lactobacillus plantarum strains WiKim83 and WiKim87 as a starter culture for fermented food. Food Sci. Nutr. 2019, 7, 2505–2516. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Tiwari, S.K. Probiotic Potential of Lactobacillus plantarum LD1 Isolated from Batter of Dosa, a South Indian Fermented Food. Probiotics Antimicrob. Proteins 2014, 6, 73–81. [Google Scholar] [CrossRef]
- Das, D.; Goyal, A. Potential Probiotic Attributes and Antagonistic Activity of an Indigenous Isolate Lactobacillus plantarum DM5 from an Ethnic Fermented Beverage “Marcha” of North Eastern Himalayas. Int. J. Food Sci. Nutr. 2014, 65, 335–344. [Google Scholar] [CrossRef]
- Mishra, B.K.; Hati, S.; Das, S.; Kumari, R. Evaluation of Probiotic Potentials of Lactobacillus Isolated from Traditional Fermented Foods of Garo Hills, Meghalaya, India. Rev. Med. Microbiol. 2018, 29, 120–128. [Google Scholar] [CrossRef]
- Sharma, K.; Attri, S.; Goel, G. Selection and Evaluation of Probiotic and Functional Characteristics of Autochthonous Lactic Acid Bacteria Isolated from Fermented Wheat Flour Dough Babroo. Probiotics Antimicrob. Proteins 2019, 11, 774–784. [Google Scholar] [CrossRef]
- Urdaneta, V.; Casadesús, J. Interactions between Bacteria and Bile Salts in the Gastrointestinal and Hepatobiliary Tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef]
- Prete, R.; Long, S.L.; Gallardo, A.L.; Gahan, C.G.; Corsetti, A.; Joyce, S.A. Beneficial bile acid metabolism from Lactobacillus plantarum of food origin. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayyash, M.M.; Abdalla, A.K.; AlKalbani, N.S.; Baig, M.A.; Turner, M.S.; Liu, S.Q.; Shah, N.P. Invited Review: Characterization of New Probiotics from Dairy and Nondairy Products—Insights into Acid Tolerance, Bile Metabolism and Tolerance, and Adhesion Capability. J. Dairy Sci. 2021, 104, 8363–8379. [Google Scholar] [CrossRef] [PubMed]
- Pisoschi, A.M.; Pop, A.; Georgescu, C.; Turcuş, V.; Olah, N.K.; Mathe, E. An overview of natural antimicrobials role in food. Eur. J. Med. Chem. 2018, 143, 922–935. [Google Scholar] [CrossRef] [PubMed]
- Kumariya, R.; Garsa, A.K.; Rajput, Y.; Sood, S.; Akhtar, N.; Patel, S. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 2019, 128, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Angmo, K.; Monika; Bhalla, T.C. Probiotic Attributes of Indigenous Lactobacillus Spp. Isolated from Traditional Fermented Foods and Beverages of North-Western Himalayas Using in Vitro Screening and Principal Component Analysis. J. Food Sci. Technol. 2016, 53, 2463–2475. [Google Scholar] [CrossRef] [Green Version]
- Angmo, K.; Kumari, A.; Savitri; Bhalla, T.C. Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. LWT 2016, 66, 428–435. [Google Scholar] [CrossRef]
- Dopazo, V.; Luz, C.; Quiles, J.M.; Calpe, J.; Romano, R.; Mañes, J.; Meca, G. Potential application of lactic acid bacteria in the biopreservation of red grape from mycotoxigenic fungi. J. Sci. Food Agric. 2021, 102, 898–907. [Google Scholar] [CrossRef]
- Schettino, R.; Pontonio, E.; Gobbetti, M.; Rizzello, C.G. Extension of the Shelf-Life of Fresh Pasta Using Chickpea Flour Fermented with Selected Lactic Acid Bacteria. Microorganisms 2020, 8, 1322. [Google Scholar] [CrossRef]
- Panebianco, F.; Caridi, A. New insights into the antifungal activity of lactic acid bacteria isolated from different food matrices. Grasas Aceites 2021, 72, e400. [Google Scholar] [CrossRef]
- Reuben, R.; Roy, P.C.; Sarkar, S.L.; Alam, R.-U.; Jahid, I.K. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiol. 2019, 19, 253. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, N.; Battista, N.; Prete, R.; Corsetti, A. Health-Promoting Role of Lactiplantibacillus plantarum Isolated from Fermented Foods. Microorganisms 2021, 9, 349. [Google Scholar] [CrossRef] [PubMed]
- Erttmann, S.F.; Gekara, N.O. Hydrogen peroxide release by bacteria suppresses inflammasome-dependent innate immunity. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Mokoena, M.P.; Mutanda, T.; Olaniran, A.O. Perspectives on the probiotic potential of lactic acid bacteria from African traditional fermented foods and beverages. Food Nutr. Res. 2016, 60, 29630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riane, K.; Sifour, M.; Ouled-Haddar, H.; Idoui, T.; Bounar, S.; Boussebt, S. Probiotic properties and antioxidant efficiency of lactobacillus plantarum 15 isolated from milk. J. Microbiol. Biotechnol. Food Sci. 2019, 9, 516–520. [Google Scholar] [CrossRef]
- Tsafrakidou, P.; Michaelidou, A.M.; Biliaderis, C.G. Fermented Cereal-based Products: Nutritional Aspects, Possible Impact on Gut Microbiota and Health Implications. Foods 2020, 9, 734. [Google Scholar] [CrossRef]
- Lee, H.; Gilliland, S.; Carter, S. Amylolytic Cultures of Lactobacillus acidophilus : Potential Probiotics to Improve Dietary Starch Utilization. J. Food Sci. 2001, 66, 338–344. [Google Scholar] [CrossRef]
- Xu, Y.; Zhou, T.; Tang, H.; Li, X.; Chen, Y.; Zhang, L.; Zhang, J. Probiotic potential and amylolytic properties of lactic acid bacteria isolated from Chinese fermented cereal foods. Food Control 2019, 111, 107057. [Google Scholar] [CrossRef]
- Chaves-López, C.; Rossi, C.; Maggio, F.; Paparella, A.; Serio, A. Changes Occurring in Spontaneous Maize Fermentation: An Overview. Fermentation 2020, 6, 36. [Google Scholar] [CrossRef] [Green Version]
- Angelin, J.; Kavitha, M. Exopolysaccharides from probiotic bacteria and their health potential. Int. J. Biol. Macromol. 2020, 162, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Heperkan, D.; Daskaya-Dikmen, C.; Bayram, B. Evaluation of lactic acid bacterial strains of boza for their exopolysaccharide and enzyme production as a potential adjunct culture. Process Biochem. 2014, 49, 1587–1594. [Google Scholar] [CrossRef]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm Matrixome: Extracellular Components in Structured Microbial Communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef]
- Korcz, E.; Varga, L. Exopolysaccharides from lactic acid bacteria: Techno-functional application in the food industry. Trends Food Sci. Technol. 2021, 110, 375–384. [Google Scholar] [CrossRef]
- Yerlikaya, O.; Ender, G.; Torunoglu, F.A.; Akbulut, N. Production of Probiotic Milk Drink Containing: Lactobacillus acidophilus, Bifidobacterium animalis Subsp. lactis and Lactobacilluscasei. Agro Food Ind. Hi. Tech 2013, 24, 49–52. [Google Scholar]
- Heidari, Z.; Ghasemi, M.F.; Modiri, L. Antimicrobial activity of bacteriocin produced by a new Latilactobacillus curvatus sp. LAB-3H isolated from traditional yogurt. Arch. Microbiol. 2021, 204, 1–12. [Google Scholar] [CrossRef]
- Afshari, A.; Hashemi, M.; Tavassoli, M.; Eraghi, V.; Noori, S.M.A. Probiotic bacteria from 10 different traditional Iranian cheeses: Isolation, characterization, and investigation of probiotic potential. Food Sci. Nutr. 2022, 10, 2009–2020. [Google Scholar] [CrossRef]


| Isolate | pH (2.0) | pH (3.0) | ||||
|---|---|---|---|---|---|---|
| 0 h | 2 h | 3 h | 0 h | 2 h | 3 h | |
| KMUDR1 | 9.35 ± 0.03 dC * | 9.07 ± 0.04 cB | 8.93 ± 0.04 cA | 9.53 ± 0.11 cD | 9.24 ± 0.09 cE | 9.1 ± 0.02 aF |
| KMUDR7 | 9.41 ± 0.02 eC | 9.33 ± 0.01 dB | 8.99 ± 0.03 dA | 9.62 ± 0.06 cD | 9.33 ± 0.07 aE | 9.36 ± 0.03 aF |
| KMUDR9 | 9.22 ± 0.03 cC | 8.8 ± 0.03 bB | 8.63 ± 0.03 bA | 9.38 ± 0.04 bD | 8.87 ± 0.05 bcE | 8.82 ± 0.11 aF |
| KMUDR14 | 9.15 ± 0.03 abC | 8.79 ± 0.08 bB | 8.6 ± 0.03 bA | 9.27 ± 0.06 abD | 8.84 ± 0.06 bE | 8.67 ± 0.13 bF |
| KMUDR17 | 9.12 ± 0.04 aC | 8.72 ± 0.08 bB | 8.55 ± 0.04 aA | 9.23 ± 0.05 aD | 8.72 ± 0.1 cE | 8.62 ± 0.08 cF |
| KMUDR20 | 9.19 ± 0.02 bcC | 8.67 ± 0.04 aB | 8.5 ± 0.03 aA | 9.17 ± 0.05 aD | 8.88 ± 0.08 cE | 8.66 ± 0.05 dF |
| Isolate | 0 h | 2 h | 3 h |
|---|---|---|---|
| KMUDR1 | 7.32 ± 0.07 cC * | 6.41 ± 0.04 eB | 6.13 ± 0.06 fA |
| KMUDR7 | 7.88 ± 0.04 dC | 6.68 ± 0.13 fB | 6.28 ± 0.04 eA |
| KMUDR9 | 7.28 ± 0.04 cC | 6.08 ± 0.05 dB | 5.93 ± 0.05 dA |
| KMUDR14 | 7.23 ± 0.05 bcC | 5.94 ± 0.04 cB | 5.38 ± 0.04 cA |
| KMUDR17 | 7.05 ± 0.03 aC | 5.74 ± 0.03 bB | 5.27 ± 0.06 bA |
| KMUDR20 | 7.16 ± 0.07 bC | 5.3 ± 0.06 aC | 5.05 ± 0.04 aA |
| Isolate | Tested Pathogens | |||||
|---|---|---|---|---|---|---|
| Staphylococcus aureus NCIM-5345 | Staph epidermis NCIM 2493 | Bacillus subtilis NCIM 2063 | E. coli NCIM 2065 | Enterococcus aerogenes NCIM 5139 | Pseudomonas auruginosa NCIM 3471 | |
| KMUDR1 | 15 ± 1 b * | 18 ± 1 b | 14.67 ± 0.58 c | 8.67 ± 0.58 b | 7.67 ± 0.58 b | 11 ± 1 c |
| KMUDR7 | 23.33 ± 1.15 c | 21.33 ± 1.53 c | 17 ± 1 d | 10.67 ± 0.58 c | 9 ± 1 d | 13.33 ± 1.53 d |
| KMUDR9 | 15.67 ± 1.15 b | 17 ± 1 b | 11.33 ± 0.58 b | 0 ± 0 a | 0 ± 0 a | 9 ± 1 b |
| KMUDR14 | 15 ± 1 b | 13.33 ± 1.53 a | 10.67 ± 1.15 b | 0 ± 0 a | 7.67 ± 0.58 b | 0 ± 0 a |
| KMUDR17 | 17 ± 1 b | 13.33 ± 1.53 a | 11.67 ± 1.15 b | 8.33 ± 0.58 b | 8 ± 1 b | 10 ± 1 bc |
| KMUDR20 | 12.33 ± 1.15 a | 12.33 ± 1.53 a | 9.67 ± 0.58 a | 0 ± 0 a | 0 ± 0 a | 0 ± 0 a |
| Isolate | Tested Fungal Strain | |||||
|---|---|---|---|---|---|---|
| Aspergillus brasiliensis NCIM 1196 | Alternaria solani MTCC 2101 | Candida albicans NCIM 3471 | ||||
| 2 Days Incubation | 7 Days Incubation | 2 Days Incubation | 7 Days Incubation | 2 Days Incubation | 7 Days Incubation | |
| KMUDR1 | 18.67 ± 1.7 d * | 16.33 ± 1.25 d | 16.17 ± 0.85 d | 13.5 ± 1.08 e | 14 ± 0.82 c | 11.33 ± 0.47 c |
| KMUDR7 | 22.67 ± 1.25 e | 20.67 ± 1.7 e | 22.33 ± 1.25 e | 18.17 ± 1.03 d | 21.33 ± 2.49 d | 18.67 ± 2.05 d |
| KMUDR9 | 15.33 ± 0.94 c | 11.33 ± 0.94 c | 13.17 ± 0.62 bc | 12.5 ± 1.08 cd | 14 ± 0.82 c | 13 ± 0.82 c |
| KMUDR14 | 8.33 ± 0.47 a | 7.33 ± 0.47 b | 15.17 ± 0.85 cd | 10.83 ± 0.62 bc | 0 ± 0 a | 0 ± 0 a |
| KMUDR17 | 9.33 ± 1.25 ab | 0 ± 0 a | 11.33 ± 0.62 ab | 10.5 ± 0.41 b | 12.67 ± 1.7 c | 10.67 ± 1.25 bc |
| KMUDR20 | 11 ± 0.82 b | 9 ± 0.82 b | 10 ± 1.78 a | 0 ± 0 a | 9.33 ± 1.25 b | 8.33 ± 1.25 b |
| Isolate | α-Amylase Activity | BSH Activity | H2O2 Production | |
|---|---|---|---|---|
| Sodium Taurodeoxycholate | Sodium Taurocholate | |||
| KMUDR1 | + a | + b | ++ b | + c |
| KMUDR7 | + a | ++ b | ++ b | ++ c |
| KMUDR9 | + a | + b | + b | - c |
| KMUDR14 | - a | + b | + b | - c |
| KMUDR17 | - a | - b | - b | - c |
| KMUDR20 | - a | + b | + b | - c |
| Isolates | |||||||
|---|---|---|---|---|---|---|---|
| Antibiotics | Concentration (µg/disc) | KMUDR1 | KMUDR7 | KMUDR9 | KMUDR14 | KMUDR17 | KMUDR20 |
| Gentamicin | (10 µg/disc) | MS * | R | S | MS | MS | S |
| Penicillin G | (10 µg/disc) | S | R | S | MS | MS | MS |
| Chloramphenicol | (30 µg/disc) | S | MS | S | R | MS | MS |
| Vancomycin | (15 µg/disc) | R | R | S | S | S | MS |
| Kanamycin | (30 µg/disc) | R | R | MS | S | MS | MS |
| Streptomycin | (30 µg/disc) | MS | R | MS | S | MS | MS |
| Tetracycline | (30 µg/disc) | R | R | R | R | R | R |
| Erythromycin | (15 μg/disc) | S | MS | MS | R | S | MS |
| Ciprofloxacin | (5 μg/disc) | R | R | R | R | R | R |
| Cefadroxil | (30 μg/disc) | R | R | S | S | MS | S |
| Isolate | Viability Count during Fermentation | Viability Count during Storage | ||||
|---|---|---|---|---|---|---|
| 0 h | 12 h | 24 h | 7 d | 14 d | 21 d | |
| KMUDR1 | 4.81 ± 0.09 dA * | 7.28 ± 0.07 eB | 8.81 ± 0.04 eC | 7.04 ± 0.08 dD | 7.34 ± 0.11 dE | 4.28 ± 0.16 dF |
| KMUDR7 | 5.1 ± 0.03 eA | 7.79 ± 0.1 fB | 8.94 ± 0.04 eC | 7.74 ± 0.07 eD | 7.99 ± 0.1 eE | 4.82 ± 0.06 eF |
| KMUDR9 | 4.67 ± 0.08 cA | 7.1 ± 0.05 dB | 8.55 ± 0.09 dC | 6.9 ± 0.05 cdD | 5.04 ± 0.08 cE | 3.56 ± 0.11 cF |
| KMUDR14 | 4.37 ± 0.1 bA | 6.81 ± 0.06 cB | 8.16 ± 0.12 cC | 6.72 ± 0.05 cD | 4.84 ± 0.12 bE | 2.78 ± 0.18 abF |
| KMUDR17 | 4.29 ± 0.07 abA | 8.72 ± 0.08 bB | 7.87 ± 0.12 bC | 6.49 ± 0.25 bD | 4.65 ± 0.11 aE | 2.68 ± 0.19 aF |
| KMUDR20 | 4.17 ± 0.05 aA | 5.91 ± 0.07 aB | 7.3 ± 0.1 aC | 5.9 ± 0.08 aD | 4.57 ± 0.12 aE | 2.96 ± 0.06 bF |
| Strain Designation | Strain Organism | Type of Cereal-Based Fermented Food | Closest Homolog | Similarity (%) | GenBank Accession Number (NCBI) |
|---|---|---|---|---|---|
| KMUDR1 | Lactobacillus delbrueckii subsp. bulgaricus | Raabadi | Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842 = JCM 1002 | 99.87 | ON954521 |
| KMUDR7 | Lactiplantibacillus plantarum | Makka Ki Raab | Lactiplantibacillus plantarum strain CIP 103151 | 96.51 | ON954522 |
| KMUDR9 | Lacticaseibacillus rhamnosus | Raabadi | Lacticaseibacillus rhamnosus strain NBRC 3425 | 99.49 | ON954523 |
| KMUDR14 | Lactobacillus helveticus | Oliya | Lactobacillus helveticus strain NBRC 15019 | 97.22 | ON954524 |
| KMUDR17 | Limosilactobacillus fermentum | Makka ki Raab | Limosilactobacillus fermentum strain CIP 102980 | 98.77 | ON954525 |
| KMUDR20 | Limosilactobacillus reuteri | Jalebi batter | Limosilactobacillus reuteri strain NBRC 15892 | 98.63 | ON954526 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meena, K.K.; Taneja, N.K.; Jain, D.; Ojha, A.; Kumawat, D.; Mishra, V. In Vitro Assessment of Probiotic and Technological Properties of Lactic Acid Bacteria Isolated from Indigenously Fermented Cereal-Based Food Products. Fermentation 2022, 8, 529. https://doi.org/10.3390/fermentation8100529
Meena KK, Taneja NK, Jain D, Ojha A, Kumawat D, Mishra V. In Vitro Assessment of Probiotic and Technological Properties of Lactic Acid Bacteria Isolated from Indigenously Fermented Cereal-Based Food Products. Fermentation. 2022; 8(10):529. https://doi.org/10.3390/fermentation8100529
Chicago/Turabian StyleMeena, Kamalesh Kumar, Neetu Kumra Taneja, Devendra Jain, Ankur Ojha, Dinesh Kumawat, and Vijendra Mishra. 2022. "In Vitro Assessment of Probiotic and Technological Properties of Lactic Acid Bacteria Isolated from Indigenously Fermented Cereal-Based Food Products" Fermentation 8, no. 10: 529. https://doi.org/10.3390/fermentation8100529
APA StyleMeena, K. K., Taneja, N. K., Jain, D., Ojha, A., Kumawat, D., & Mishra, V. (2022). In Vitro Assessment of Probiotic and Technological Properties of Lactic Acid Bacteria Isolated from Indigenously Fermented Cereal-Based Food Products. Fermentation, 8(10), 529. https://doi.org/10.3390/fermentation8100529







