Effect of Different Inducer Sources on Cellulase Enzyme Production by White-Rot Basidiomycetes Pleurotus ostreatus and Phanerochaete chrysosporium under Submerged Fermentation
Abstract
1. Introduction
2. Materials and Methods
2.1. Organisms and Inoculum Preparation
2.2. Cultivation Conditions
2.2.1. Enzyme Activities with Carbon Inducer Sources
2.2.2. Enzyme Activities with Nitrogen Inducer Sources
2.2.3. Enzyme Activities with Agro-Biomass Inducer Sources
2.3. Enzyme Assay
2.4. Statistical Analysis
3. Results
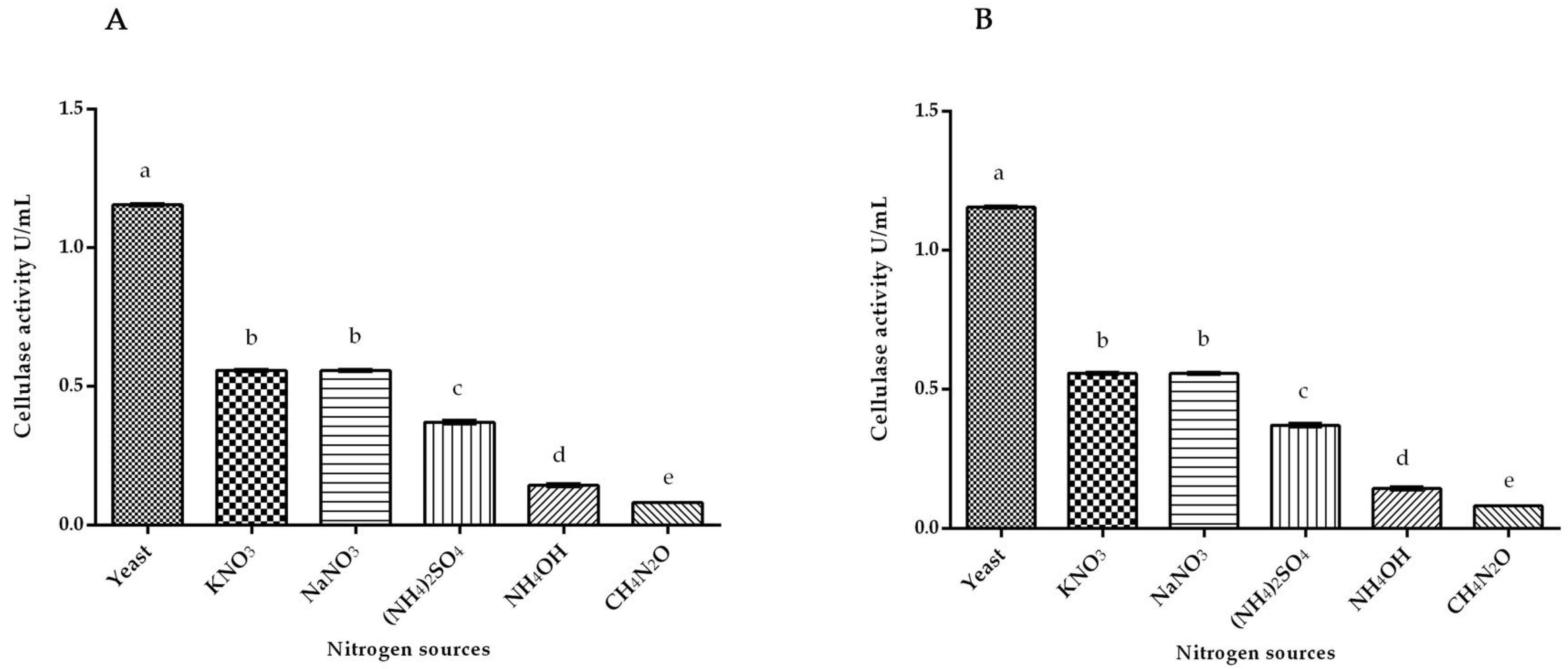
3.1. Cellulase Activity of White Rot Fungus P. ostreatus and P. chrysosporium under Submerged Fermentation on Different Nitrogen Inducer Sources
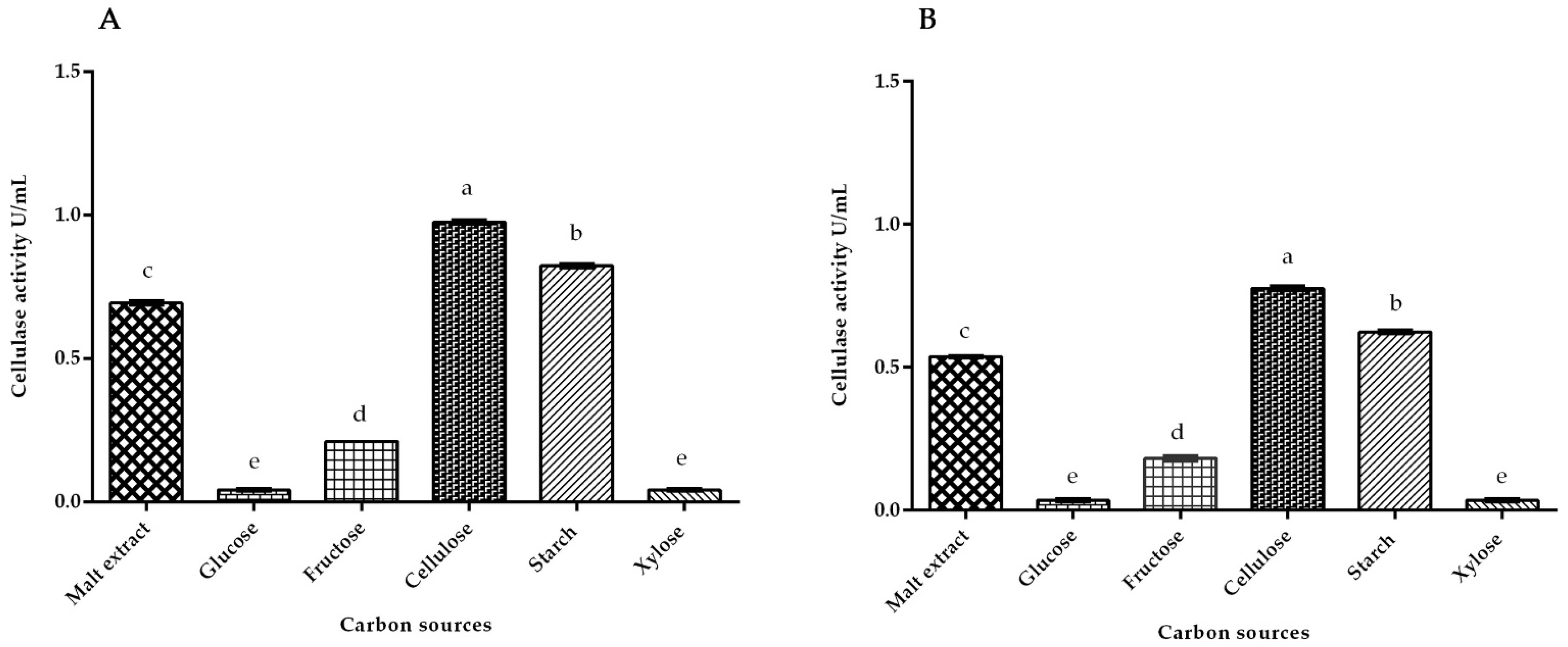
3.2. Cellulase Activity of White Rot Fungus P. ostreatus and P. chrysosporium under Submerged Fermentation on Different Carbon Inducer Sources
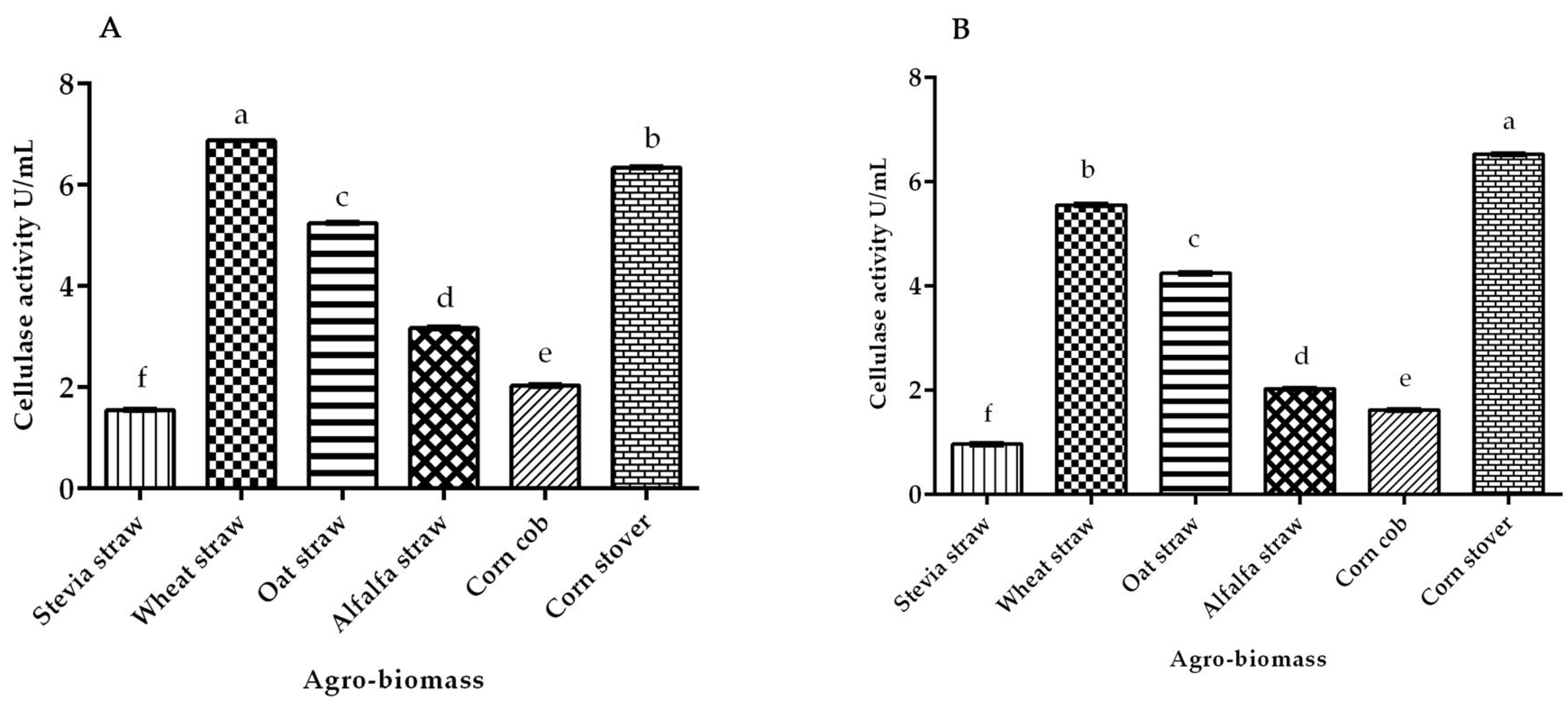
3.3. Cellulase Activity of White Rot Fungus P. ostreatus and P. chrysosporium under Submerged Fermentation on Different Agro-Biomass
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bhat, M. Cellulases and related enzymes in biotechnology. Biotechnol. Adv. 2000, 18, 355–383. [Google Scholar] [CrossRef]
- Lee, S.-M.; Koo, Y.-M. Pilot-scale production of cellulase using Trichoderma Reesei rut C-30 fed-batch mode. J. Microbiol. Biotechnol. 2001, 11, 229–233. [Google Scholar]
- Kirk, O.; Borchert, T.V.; Fuglsang, C.C. Industrial enzyme applications. Curr. Opin. Biotechnol. 2002, 13, 345–351. [Google Scholar] [CrossRef]
- Kuhad, R.; Gupta, R.; Khasa, Y. Bioethanol production from lignocellulosic biomass: An overview. In Wealth from Waste; Lal, B., Ed.; Teri Press: New Delhi, India, 2010. [Google Scholar]
- Bentil, J.A.; Thygesen, A.; Mensah, M.; Lange, L.; Meyer, A.S. Cellulase production by white-rot basidiomycetous fungi: Solid-state versus submerged cultivation. Appl. Microbiol. Biotechnol. 2018, 102, 5827–5839. [Google Scholar] [CrossRef] [PubMed]
- Okal, E.J.; Aslam, M.M.; Karanja, J.K.; Nyimbo, W.J. Mini review: Advances in understanding regulation of cellulase enzyme in white-rot basidiomycetes. Microb. Pathog. 2020, 147, 104410. [Google Scholar] [CrossRef]
- Elisashvili, V.; Penninckx, M.; Kachlishvili, E.; Tsiklauri, N.; Metreveli, E.; Kharziani, T.; Kvesitadze, G. Lentinus edodes and Pleurotus species lignocellulolytic enzymes activity in submerged and solid-state fermentation of lignocellulosic wastes of different composition. Bioresour. Technol. 2008, 99, 457–462. [Google Scholar] [CrossRef]
- Téllez-Téllez, M.; Diaz-Godinez, G. Omic tools to study enzyme production from fungi in the Pleurotus genus. BioResources 2019, 14, 2420–2457. [Google Scholar] [CrossRef]
- Persson, I.; Tjerneld, F.; Hahn-Hägerdal, B. Fungal cellulolytic enzyme production: A review. Process Biochem. 1991, 26, 65–74. [Google Scholar] [CrossRef]
- Barragán, L.P.; Figueroa, J.; Durán, L.R.; González, C.A.; Hennigs, C. Fermentative production methods. In Biotransformation of Agricultural Waste and By-Products, 1st ed.; Poltronieri, P., D’Urso, O., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 189–217. [Google Scholar]
- Elisashvili, V.; Kachlishvili, E.; Penninckx, M. Effect of growth substrate, method of fermentation, and nitrogen source on lignocellulose-degrading enzymes production by white-rot basidiomycetes. Ind. Microbiol. Biotechnol. 2008, 35, 1531–1538. [Google Scholar] [CrossRef]
- Kachlishvili, E.; Penninckx, M.J.; Tsiklauri, N.; Elisashvili, V. Effect of nitrogen source on lignocellulolytic enzyme production by white-rot basidiomycetes under solid-state cultivation. World J. Microbiol. Biotechnol. 2006, 22, 391–397. [Google Scholar] [CrossRef]
- Radhika, R.; Jebapriya, G.R.; Gnanadoss, J.J. Production of cellulase and laccase using Pleurotus sp. under submerged and solid-state fermentation. Int. J. Curr. Microbiol. 2013, 6, 7–13. [Google Scholar]
- Khan, M.H.; Ali, S.; Fakhru’l-Razi, A.; Alam, Z. Use of fungi for the bioconversion of rice straw into cellulase enzyme. J. Environ. Sci. Health B 2007, 42, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Uzcategui, E.; Raices, M.; Montesino, R.; Johansson, G.; Pettersson, G.; Eriksson, K. Pilot-scale production and purification of the cellulolytic enzyme system from the white-rot fungus Phanerochaete chrysosporium. Biotechnol. Appl. Biochem. 1991, 13, 323–334. [Google Scholar]
- Ghose, T. Measurement of cellulase activities. Pure Appl. Chem. 1987, 59, 257–268. [Google Scholar] [CrossRef]
- Chuwech, M.; Rakariyatham, N.; Chandet, N.; Tinoi, J. Utilization of pretreated corn cobs for cellulase production by Pycnoporus coccineus. Asia-Pac. J. Sci. Technol. 2016, 21, 310–318. [Google Scholar] [CrossRef]
- Jonathan, S.G.; Adeoyo, O.R. Evaluation of ten wild Nigerian mushrooms for amylase and cellulase activities. Mycobiology 2011, 39, 103–108. [Google Scholar] [CrossRef]
- Jonathan, S.; Fasidi, I. Effect of carbon, nitrogen and mineral sources on growth of Psathyerella atroumbonata (Pegler), a Nigerian edible mushroom. Food Chem. 2001, 72, 479–483. [Google Scholar] [CrossRef]
- Rajmane, S.; Korekar, S. Cellulase enzyme production of post-harvest fungi under the influence of carbon and nitrogen sources. Curr. Botany 2012, 3, 13–15. [Google Scholar]
- Wang, C. Cellulolytic enzymes of Volvariella volvacea. In Tropical Mushrooms; Chang, S.T., Quimio, M.H., Eds.; The Chinese Press: Kuala Lumpur, Malaysia, 1984; pp. 167–186. [Google Scholar]
- Hammad, A.; Swelim, M.; Gannam, R. Enhanced production of cellulase (S) by Aspergillus spp. isolated from agriculture wastes by solid state fermentation. Am.-Eurasian J. Agric. Environ. 2010, 8, 402–410. [Google Scholar]
- Elsebaay, H.; Shoukry, A.; Hassan, F.; El-Shall, H. Optimization of cellulase enzyme production from Pleurotus under submerged fermentation. Middle East J. Agric. Res. 2018, 7, 50–59. [Google Scholar]
- Sethi, S.; Gupta, S. Optimization of cultural parameters for cellulase enzyme production from fungi. BioLife 2014, 2, 989–996. [Google Scholar] [CrossRef]
- Gao, J.; Weng, H.; Zhu, D.; Yuan, M.; Guan, F.; Xi, Y. Production and characterization of cellulolytic enzymes from the thermoacidophilic fungal Aspergillus terreus M11 under solid-state cultivation of corn stover. Bioresour. Technol. 2008, 99, 7623–7629. [Google Scholar] [CrossRef] [PubMed]
- Niranjane, A.P.; Madhou, P.; Stevenson, T.W. The effect of carbohydrate carbon sources on the production of cellulase by Phlebia gigantea. Enzyme Microb. Technol. 2007, 40, 1464–1468. [Google Scholar] [CrossRef]
- Metreveli, E.; Khardziani, T.; Elisashvili, V. The Carbon Source Controls the Secretion and Yield of Polysaccharide-Hydrolyzing Enzymes of Basidiomycetes. Biomolecules 2021, 11, 1341. [Google Scholar] [CrossRef] [PubMed]
- Kobakhidze, A.; Asatiani, M.; Kachlishvili, E.; Elisashvili, V. Induction and catabolite repression of cellulase and xylanase synthesis in the selected white-rot basidiomycetes. Ann. Agrar. Sci. 2016, 14, 169–176. [Google Scholar] [CrossRef]
- Yoav, S.; Salame, T.M.; Feldman, D.; Levinson, D.; Ioelovich, M.; Morag, E.; Yarden, O.; Bayer, E.A.; Hadar, Y. Effects of cre1 modification in the white-rot fungus Pleurotus ostreatus PC9: Altering substrate preference during biological pretreatment. Biotechnol. Biofuels 2018, 11, 212. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Igarashi, K.; Samejima, M. Cellotriose and cellotetraose as inducers of the genes encoding cellobiohydrolases in the basidiomycete Phanerochaete chrysosporium. Appl. Environ. Microbiol. 2010, 76, 6164–6170. [Google Scholar] [CrossRef]
- Elisashvili, V.; Khardziani, T.S.; Tsiklauri, N.; Kachlishvili, E. Cellulase and xylanase activities in higher basidiomycetes. Biochemistry 1999, 64, 718–722. [Google Scholar]
- Wang, B.-T.; Hu, S.; Yu, X.-Y.; Jin, L.; Zhu, Y.-J.; Jin, F.-J. Studies of cellulose and starch utilization and the regulatory mechanisms of related enzymes in fungi. Polymers 2020, 12, 530. [Google Scholar] [CrossRef]
- Andlar, M.; Rezić, T.; Marđetko, N.; Kracher, D.; Ludwig, R.; Šantek, B. Lignocellulose degradation: An overview of fungi and fungal enzymes involved in lignocellulose degradation. Eng. Life Sci. 2018, 18, 768–778. [Google Scholar] [CrossRef]
- Vankuyk, P.A.; Benen, J.A.; Wösten, H.A.; Visser, J.; de Vries, R.P. A broader role for AmyR in Aspergillus niger: Regulation of the utilisation of D-glucose or D-galactose containing oligo-and polysaccharides. Appl. Microbiol. Biotechnol. 2012, 93, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Wu, V.W.; Lubbe, A.; Qin, L.; Deng, S.; Kennedy, M.; Bauer, D.; Singan, V.R.; Barry, K.; Northen, T.R. A fungal transcription factor essential for starch degradation affects integration of carbon and nitrogen metabolism. PLoS Genet. 2017, 13, e1006737. [Google Scholar] [CrossRef] [PubMed]
- Rytioja, J.; Hildén, K.; Yuzon, J.; Hatakka, A.; De Vries, R.P.; Mäkelä, M.R. Plant-polysaccharide-degrading enzymes from basidiomycetes. Microbiol. Mol. Biol. Rev. 2014, 78, 614–649. [Google Scholar] [CrossRef] [PubMed]
- Merck. Malt Extract. In Microbiology Manual, 12th ed.; Merck and Co., Inc.: Kenilworth, NJ, USA, 2010; p. 688. [Google Scholar]
- Kobakhidze, A.; Elisashvili, V.; Irbe, I.; Tsiklauri, N.; Andersone, I.; Andersons, B.; Isikhuemhen, O.S. Lignocellulolytic enzyme activity of new corticoid and poroid basidiomycetes isolated from latvian cultural monuments. J. Waste Convers. Bioprod. Biotechnol. 2012, 1, 16–21. [Google Scholar] [CrossRef]
- Cai, Y.; Gong, Y.; Liu, W.; Hu, Y.; Chen, L.; Yan, L.; Zhou, Y.; Bian, Y. Comparative secretomic analysis of lignocellulose degradation by Lentinula edodes grown on microcrystalline cellulose, lignosulfonate and glucose. J. Proteom. 2017, 163, 92–101. [Google Scholar] [CrossRef]
- Salmon, D.N.X.; Spier, M.R.; Soccol, C.R.; de Souza Vandenberghe, L.P.; Montibeller, V.W.; Bier, M.C.J.; Faraco, V. Analysis of inducers of xylanase and cellulase activities production by Ganoderma applanatum LPB MR-56. Fungal Biol. 2014, 118, 655–662. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; Van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Suzuki, H.; Igarashi, K.; Samejima, M. Real-time quantitative analysis of carbon catabolite derepression of cellulolytic genes expressed in the basidiomycete Phanerochaete chrysosporium. Appl. Microbiol. Biotechnol. 2008, 80, 99–106. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Messner, R.; Gruber, F.; Mandels, M.; Kubicek-Pranz, E.M. Triggering of cellulase biosynthesis by cellulose in Trichoderma reesei. Involvement of a constitutive, sophorose-inducible, glucose-inhibited beta-diglucoside permease. J. Biol. Chem. 1993, 268, 19364–19368. [Google Scholar] [CrossRef]
- Ilmen, M.; Saloheimo, A.; Onnela, M.-L.; Penttilä, M.E. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl. Environ. Microbiol. 1997, 63, 1298–1306. [Google Scholar] [CrossRef]
- Aslam, N.; Sheikh, M.; Ashraf, M.; Jalil, A. Expression pattern of Trichoderma cellulases under different carbon sources. Pak. J. Bot. 2010, 42, 2895–2902. [Google Scholar]
- Muthuvelayudham, R.; Viruthagiri, T. Fermentative production and kinetics of cellulase protein on Trichoderma reesei using sugarcane bagasse and rice straw. Afr. J. Biotechnol. 2006, 5, 1873–1881. [Google Scholar] [CrossRef]
- Mehboob, N.; Asad, M.J.; Asgher, M.; Gulfraz, M.; Mukhtar, T.; Mahmood, R.T. Exploring thermophilic cellulolytic enzyme production potential of Aspergillus fumigatus by the solid-state fermentation of wheat straw. Appl. Biochem. Biotechnol. 2014, 172, 3646–3655. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.; Machuca, A.; Milagres, A. Effect of cereal brans on Lentinula edodes growth and enzyme activities during cultivation on forestry waste. Lett. Appl. Microbiol. 2005, 40, 283–288. [Google Scholar] [CrossRef]
- Papinutti, V.; Forchiassin, F. Lignocellulolytic enzymes from Fomes sclerodermeus growing in solid-state fermentation. J. Food Eng. 2007, 81, 54–59. [Google Scholar] [CrossRef]
- Levin, L.; Herrmann, C.; Papinutti, V.L. Optimization of lignocellulolytic enzyme production by the white-rot fungus Trametes trogii in solid-state fermentation using response surface methodology. Biochem. Eng. J. 2008, 39, 207–214. [Google Scholar] [CrossRef]
- Haltrich, D.; Nidetzky, B.; Kulbe, K.D.; Steiner, W.; Župančič, S. Production of fungal xylanases. Bioresour. Technol. 1996, 58, 137–161. [Google Scholar] [CrossRef]
- Reddy, M.S.; Kanwal, H.K. Influence of carbon, nitrogen sources, inducers, and substrates on lignocellulolytic enzyme activities of Morchella spongiola. J. Agric. Food Res. 2022, 7, 100271. [Google Scholar] [CrossRef]
- Ortiz, G.E.; Guitart, M.E.; Cavalitto, S.F.; Albertó, E.O.; Fernández-Lahore, M.; Blasco, M. Characterization, optimization, and scale-up of cellulases production by Trichoderma reesei cbs 836.91 in solid-state fermentation using agro-industrial products. Bioprocess Biosyst. Eng. 2015, 38, 2117–2128. [Google Scholar] [CrossRef]
- Elisashvili, V.; Kachlishvili, E.; Tsiklauri, N.; Metreveli, E.; Khardziani, T.; Agathos, S.N. Lignocellulose-degrading enzyme production by white-rot Basidiomycetes isolated from the forests of Georgia. World J. Microbiol. Biotechnol. 2009, 25, 331–339. [Google Scholar] [CrossRef]
- Reddy, G.P.K.; Narasimha, G.; Kumar, K.D.; Ramanjaneyulu, G.; Ramya, A.; Kumari, B.S.; Reddy, B.R. Cellulase production by Aspergillus niger on different natural lignocellulosic substrates. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 835–845. [Google Scholar]
- Ganash, M.; Ghany, T.M.A.; Al Abboud, M.A.; Alawlaqi, M.M.; Qanash, H.; Amin, B.H. Lignocellulolytic Activity of Pleurotus ostreatus under Solid State Fermentation Using Silage, Stover, and Cobs of Maize. BioResources 2021, 16, 3797–3807. [Google Scholar] [CrossRef]
- Govumoni, S.P.; Jahnavi, G.; Sravanthi, K.; Haragopal, V.; Venkateshwar, S.; Rao, L.V. Extracellular lignocellulolytic enzymes by Phanerochaete chrysosporium (MTCC 787) under solid-state fermentation of agro wastes. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 700–710. [Google Scholar]
- Ji, G.; Gao, C.; Xiao, W.; Han, L. Mechanical fragmentation of corncob at different plant scales: Impact and mechanism on microstructure features and enzymatic hydrolysis. Bioresour. Technol. 2016, 205, 159–165. [Google Scholar] [CrossRef]
- Reyes, C.; Poulin, A.; Nyström, G.; Schwarze, F.W.; Ribera, J. Enzyme activities of five white-rot fungi in the presence of nanocellulose. J. Fungi 2021, 7, 222. [Google Scholar] [CrossRef]
- Lucho, S.R.; do Amaral, M.N.; Auler, P.A.; Bianchi, V.J.; Ferrer, M.Á.; Calderón, A.A.; Braga, E.J.B. Salt stress-induced changes in in vitro cultured Stevia rebaudiana Bertoni: Effect on metabolite contents, antioxidant capacity and expression of steviol glycosides-related biosynthetic genes. J. Plant Growth Regul. 2019, 38, 1341–1353. [Google Scholar] [CrossRef]
- Putnik, P.; Bezuk, I.; Barba, F.J.; Lorenzo, J.M.; Polunić, I.; Bursać, D.K. Sugar reduction: Stevia rebaudiana Bertoni as a natural sweetener. In Agri-Food Industry Strategies for Healthy Diets and Sustainability; Academic Press: Cambridge, MA, USA; Elsevier: Amsterdam, The Netherlands, 2020; pp. 123–152. [Google Scholar]
- Amore, A.; Giacobbe, S.; Faraco, V. Regulation of cellulase and hemicellulase gene expression in fungi. Curr. Genom. 2013, 14, 230–249. [Google Scholar] [CrossRef]
- Carle-Urioste, J.C.; Escobar-Vera, J.; El-Gogary, S.; Henrique-Silva, F.; Torigoi, E.; Crivellaro, O.; Herrera-Estrella, A.; El-Dorry, H. Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J. Biol. Chem. 1997, 272, 10169–10174. [Google Scholar] [CrossRef]
- Alfaro, M.; Majcherczyk, A.; Kües, U.; Ramírez, L.; Pisabarro, A.G. Glucose counteracts wood-dependent induction of lignocellulolytic enzyme secretion in monokaryon and dikaryon submerged cultures of the white-rot basidiomycete Pleurotus ostreatus. Sci. Rep. 2020, 10, 12421. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Datsomor, O.; Yan, Q.; Opoku-Mensah, L.; Zhao, G.; Miao, L. Effect of Different Inducer Sources on Cellulase Enzyme Production by White-Rot Basidiomycetes Pleurotus ostreatus and Phanerochaete chrysosporium under Submerged Fermentation. Fermentation 2022, 8, 561. https://doi.org/10.3390/fermentation8100561
Datsomor O, Yan Q, Opoku-Mensah L, Zhao G, Miao L. Effect of Different Inducer Sources on Cellulase Enzyme Production by White-Rot Basidiomycetes Pleurotus ostreatus and Phanerochaete chrysosporium under Submerged Fermentation. Fermentation. 2022; 8(10):561. https://doi.org/10.3390/fermentation8100561
Chicago/Turabian StyleDatsomor, Osmond, Qi Yan, Louis Opoku-Mensah, Guoqi Zhao, and Lin Miao. 2022. "Effect of Different Inducer Sources on Cellulase Enzyme Production by White-Rot Basidiomycetes Pleurotus ostreatus and Phanerochaete chrysosporium under Submerged Fermentation" Fermentation 8, no. 10: 561. https://doi.org/10.3390/fermentation8100561
APA StyleDatsomor, O., Yan, Q., Opoku-Mensah, L., Zhao, G., & Miao, L. (2022). Effect of Different Inducer Sources on Cellulase Enzyme Production by White-Rot Basidiomycetes Pleurotus ostreatus and Phanerochaete chrysosporium under Submerged Fermentation. Fermentation, 8(10), 561. https://doi.org/10.3390/fermentation8100561







