Grape Pomace in Ewes Diet Affects Metagenomic Profile, Volatile Compounds and Biogenic Amines Contents of Ripened Cheese
Abstract
1. Introduction
2. Methods and Materials
2.1. Cheese Manufacturing
2.2. DNA Extraction, 16S rRNA Gene Amplicon Library Preparation and Sequencing
2.3. Determination of Volatile Compound in Cheese
2.4. Determination of Biogenic Amines in Cheese
2.5. Bioinformatic and Statistical Analysis
3. Results
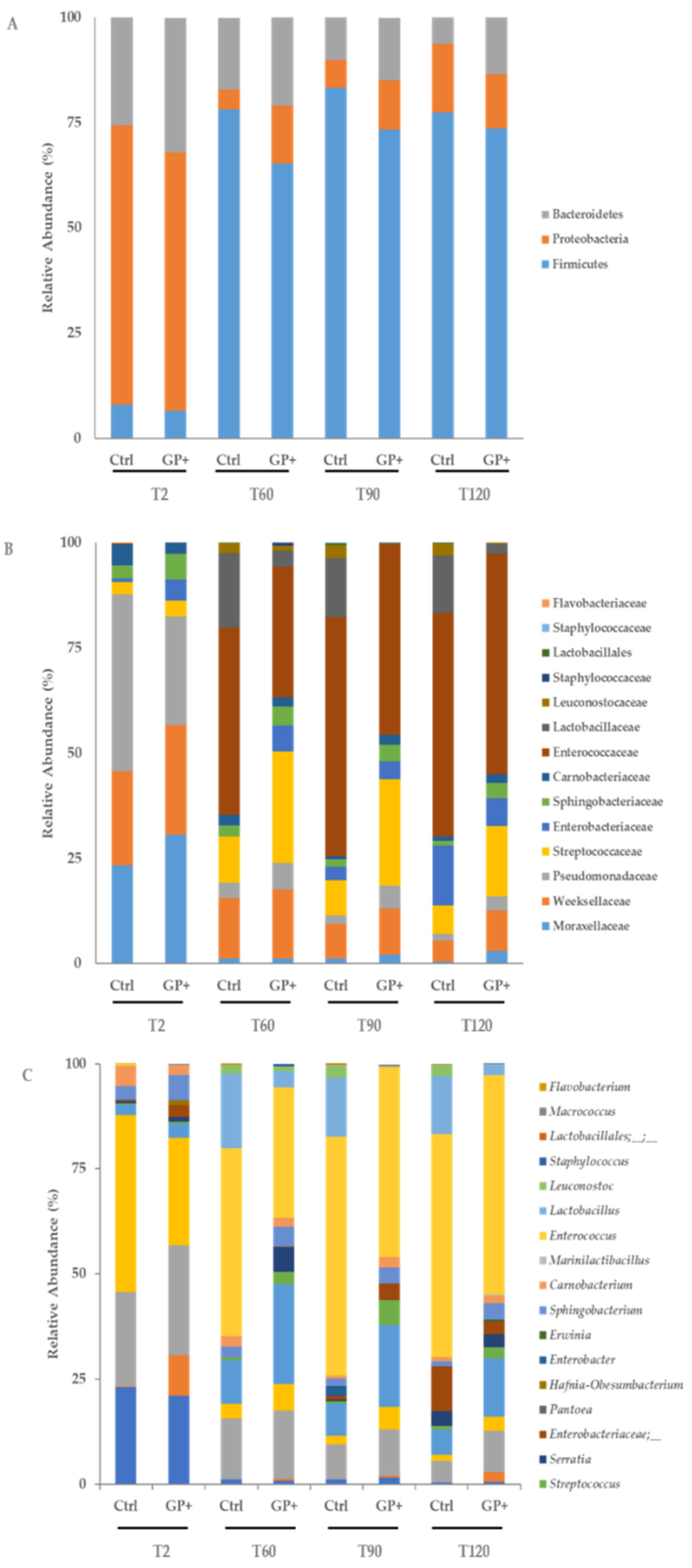
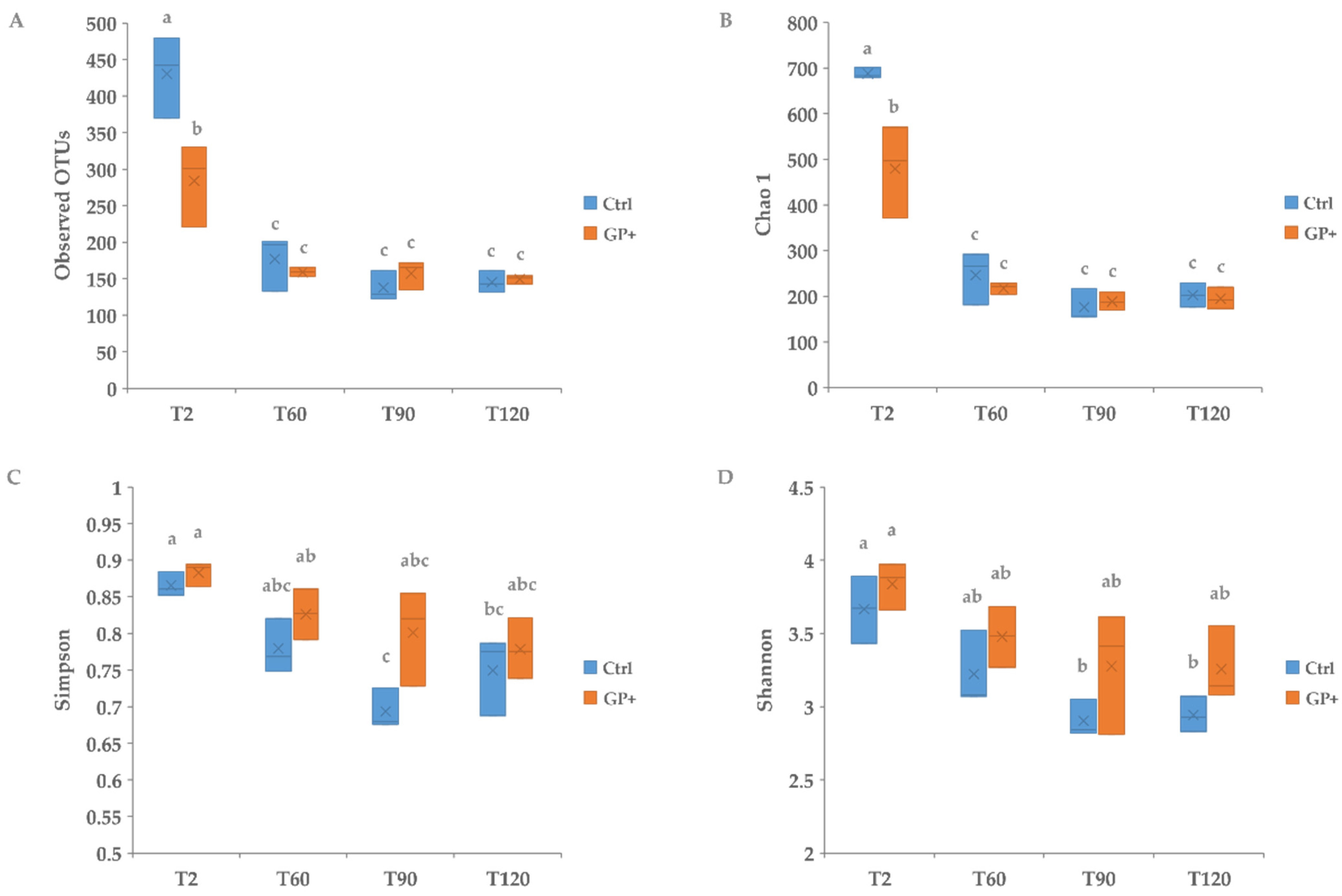
3.1. Microbiota Cheese Characterization
3.2. Volatile Compounds in Cheese
3.3. Biogenic Amines in Cheese
3.4. Microbioma, Volatile Profile and Biogenic Amines Correlation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed]
- Montel, M.-C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef]
- Afshari, R.; Pillidge, C.J.; Dias, D.A.; Osborn, A.M.; Gill, H. Cheesomics: The future pathway to understanding cheese flavour and quality. Crit. Rev. Food Sci. Nutr. 2018, 60, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Zepeda, A.; Sanchez-Flores, A.; Quirasco Baruch, M. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016, 57, 116–127. [Google Scholar] [CrossRef]
- Gezginc, Y.; Karabekmez-Erdem, T.; Tatar, H.D.; Dağgeçen, E.C.; Ayman, S.; Akyol, I. Metagenomics and volatile profile of Turkish artisanal Tulum cheese microbiota. Food Biosci. 2021, 45, 101497. [Google Scholar] [CrossRef]
- Lopez, C.C.; Serio, A.; Rossi, C.; Mazzarrino, G.; Marchetti, S.; Castellani, F.; Grotta, L.; Fiorentino, F.P.; Paparella, A.; Martino, G. Effect of diet supplementation with Ascophyllum nodosum on cow milk composition and microbiota. J. Dairy Sci. 2016, 99, 6285–6297. [Google Scholar] [CrossRef]
- Ianni, A.; Di Domenico, M.; Bennato, F.; Peserico, A.; Martino, C.; Rinaldi, A.; Candeloro, L.; Grotta, L.; Cammà, C.; Pomilio, F. Metagenomic and volatile profiles of ripened cheese obtained from dairy ewes fed a dietary hemp seed supplementation. J. Dairy Sci. 2020, 103, 5882–5892. [Google Scholar] [CrossRef]
- Jing-Wei, Z.; Yi-Yuan, S.; Xin, L.; Hua, Z.; Hui, N.; Luo-Yun, F.; Ben-Hai, X.; Jin-Jin, T.; Lin-Shu, J. Microbiome and Metabolic Changes of Milk in Response to Dietary Supplementation With Bamboo Leaf Extract in Dairy Cows. Front. Nutr. 2021, 8, 723446. [Google Scholar] [CrossRef]
- Correddu, F.; Nudda, A.; Battacone, G.; Boe, R.; Francesconi, A.H.D.; Pulina, G. Effects of grape seed supplementation, alone or associated with linseed, on ruminal metabolism in Sarda dairy sheep. Anim. Feed Sci. Technol. 2014, 199, 61–72. [Google Scholar] [CrossRef]
- Moate, P.J.; Williams, S.R.O.; Torok, V.A.; Hannah, M.C.; Ribaux, B.E.; Tavendale, M.H.; Eckard, R.J.; Jacobs, J.L.; Auldist, M.J.; Wales, W.J. Grape marc reduces methane emissions when fed to dairy cows. J. Dairy Sci. 2014, 97, 5073–5087. [Google Scholar] [CrossRef]
- Iannaccone, M.; Elgendy, R.; Giantin, M.; Martino, C.; Giansante, D.; Ianni, A.; Dacasto, M.; Martino, G. RNA Sequencing-Based Whole-Transcriptome Analysis of Friesian Cattle Fed with Grape Pomace-Supplemented Diet. Animals 2018, 8, 188. [Google Scholar] [CrossRef]
- Buffa, M.; Guamis, B.; Pavia, M.; Trujillo, A.J. Lipolysis in cheese made from raw, pasteurized or high-pressure-treated goats’ milk. Int. Dairy J. 2001, 11, 175–179. [Google Scholar] [CrossRef]
- McSweeney, P.L.H.; Sousa, M.J. Biochemical pathways for the production of flavour compounds in cheeses during ripening: A review. Lait 2000, 80, 293–324. [Google Scholar] [CrossRef]
- McSweeney, P.L.H. Biochemistry of cheese ripening. Int. J. Dairy Technol. 2004, 57, 127–144. [Google Scholar] [CrossRef]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Microbiology of cheese ripening. In Fundamentals of Cheese Science. In Fundamentals of Cheese Science, 2nd ed.; Fox, P.F., Guinee, T.P., Cogan, T.M., McSweeney, P.L.H., Eds.; Springer: New York, NY, USA, 2017; pp. 333–390. [Google Scholar] [CrossRef]
- Bertuzzi, A.S.; McSweeney, P.L.H.; Rea, M.C.; Kilcawley, K.N. Detection of Volatile Compounds of Cheese and Their Contribution to the Flavor Profile of Surface-Ripened Cheese. Compr. Rev. Food Sci. Food Saf. 2018, 17, 371–390. [Google Scholar] [CrossRef]
- Drake, M.A.; Delahunty, C.M. Sensory character of cheese and its evaluation. In Cheese: Chemistry, Physics, and Microbiology, 4th ed.; McSweeney, P.L.H., Fox, P.F., Cotter, P.D., Everett, D.W., Eds.; Academic Press: San Diego, CA, USA, 2017; Volume 1, pp. 455–487. [Google Scholar]
- Schirone, M.; Tofalo, R.; Mazzone, G.; Corsetti, A.; Suzzi, G. Biogenic amine content and microbiological profile of Pecorino di Farindola cheese. Food Microbiol. 2011, 28, 128–136. [Google Scholar] [CrossRef]
- Galgano, F.; Suzzi, G.; Favati, F.; Caruso, M.C.; Martuscelli, M.; Gardini, F.; Salzano, G. Biogenic amines during ripening in ‘Semicotto Caprino’ cheese: Role of enterococci. Int. J. Food Sci. Technol. 2001, 36, 153–160. [Google Scholar] [CrossRef]
- Bennato, F.; Ianni, A.; Florio, M.; Grotta, L.; Pomilio, F.; Saletti, M.A.; Martino, G. Nutritional Properties of Milk from Dairy Ewes Fed with a Diet Containing Grape Pomace. Foods 2022, 11, 1878. [Google Scholar] [CrossRef]
- Illumina. 16S Metagenomic Sequencing Library Preparation Protocol: Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System; Part No. 15044223 Rev. B.; Illumina: San Diego, CA, USA, 2013; Available online: https://www.illumina.com/content/dam/illuminasupport/documents/documentation/chemistrydocumentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 12 April 2022).
- Bennato, F.; Ianni, A.; Innosa, D.; Grotta, L.; D’Onofrio, A.; Martino, G. Chemical-nutritional characteristics and aromatic profile of milk and related dairy products obtained from goats fed with extruded linseed. Asian-Australas. J. Anim. Sci. 2019, 33, 148–156. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Cardinali, F.; Osimani, A.; Taccari, M.; Milanović, V.; Garofalo, C.; Clementi, F.; Polverigiani, S.; Zitti, S.; Raffaelli, N.; Mozzon, M.; et al. Impact of thistle rennet from Carlina acanthifolia All. subsp. acanthifolia on bacterial diversity and dynamics of a specialty Italian raw ewes’ milk cheese. Int. J. Food Microbiol. 2017, 255, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, M.; Hosseini, S.M.; Ferrocino, I.; Amoozegar, M.A.; Cocolin, L. Molecular investigation of bacterial communities during the manufacturing and ripening of semi-hard Iranian Liqvan cheese. Food Microbiol. 2017, 66, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Santamarina-García, G.; Hernández, I.; Amores, G.; Virto, M. Characterization of Microbial Shifts during the Production and Ripening of Raw Ewe Milk-Derived Idiazabal Cheese by High-Throughput Sequencing. Biology 2022, 11, 769. [Google Scholar] [CrossRef]
- Hassan, Y.I.; Kosir, V.; Yin, X.; Ross, K.; Diarra, M.S. Grape Pomace as a Promising Antimicrobial Alternative in Feed: A Critical Review. J. Agric. Food Chem. 2019, 67, 9705–9718. [Google Scholar] [CrossRef]
- De Pasquale, I.; Calasso, M.; Mancini, L.; Ercolini, D.; La Storia, A.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Causal Relationship between Microbial Ecology Dynamics and Proteolysis during Manufacture and Ripening of Protected Designation of Origin (PDO) Cheese Canestrato Pugliese. Appl. Environ. Microbiol. 2014, 80, 4085–4094. [Google Scholar] [CrossRef]
- Endres, C.M.; Castro, M.S.; Trevisol, L.D.; Severo, J.M.; Mann, M.B.; Varela, A.P.M.; Frazzon, A.P.G.; Mayer, F.Q.; Frazzon, J. Molecular characterization of the bacterial communities present in sheep’s milk and cheese produced in South Brazilian Region via 16S rRNA gene metabarcoding sequencing. LWT 2021, 147, 111579. [Google Scholar] [CrossRef]
- Esteban-Blanco, C.; Gutiérrez-Gil, B.; Puente-Sánchez, F.; Marina, H.; Tamames, J.; Acedo, A.; Arranz, J.J. Microbiota characterization of sheep milk and its association with somatic cell count using 16s rRNA gene sequencing. J. Anim. Breed. Genet. 2020, 137, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Han, S.; Zhang, S.; Xue, Y.; Zhang, Y.; Lu, H.; Wang, S. Microbial Properties of Raw Milk throughout the Year and Their Relationships to Quality Parameters. Foods 2022, 11, 3077. [Google Scholar] [CrossRef] [PubMed]
- Falardeau, J.; Keeney, K.; Trmčić, A.; Kitts, D.; Wang, S. Farm-to-fork profiling of bacterial communities associated with an artisan cheese production facility. Food Microbiol. 2019, 83, 48–58. [Google Scholar] [CrossRef]
- Bellassi, P.; Rocchetti, G.; Nocetti, M.; Lucini, L.; Masoero, F.; Morelli, L. A Combined Metabolomic and Metagenomic Approach to Discriminate Raw Milk for the Production of Hard Cheese. Foods 2021, 10, 109. [Google Scholar] [CrossRef]
- Jonnala, B.R.Y.; McSweeney, P.L.H.; Sheehan, J.J.; Cotter, P.D. Sequencing of the Cheese Microbiome and Its Relevance to Industry. Front. Microbiol. 2018, 9, 1020. [Google Scholar] [CrossRef] [PubMed]
- Delbès, C.; Ali-Mandjee, L.; Montel, M.-C. Monitoring Bacterial Communities in Raw Milk and Cheese by Culture-Dependent and -Independent 16S rRNA Gene-Based Analyses. Appl. Environ. Microbiol. 2007, 73, 1882–1891. [Google Scholar] [CrossRef]
- Ruegg, P.L. Investigation of mastitis problems on farms. Vet. Clin. North Am. Food Anim. Pract. 2003, 19, 47–73. [Google Scholar] [CrossRef]
- Raats, D.; Offek, M.; Minz, D.; Halpern, M. Molecular analysis of bacterial communities in raw cow milk and the impact of refrigeration on its structure and dynamics. Food Microbiol. 2011, 28, 465–471. [Google Scholar] [CrossRef]
- García-Cano, I.; Rocha-Mendoza, D.; Ortega-Anaya, J.; Wang, K.; Kosmerl, E.; Jiménez-Flores, R. Lactic acid bacteria isolated from dairy products as potential producers of lipolytic, proteolytic and antibacterial proteins. Appl. Microbiol. Biotechnol. 2019, 103, 5243–5257. [Google Scholar] [CrossRef]
- Liang, L.; Liu, G.; Zhang, F.; Li, Q.; Linhardt, R.J. Digestibility of squash polysaccharide under simulated salivary, gastric and intestinal conditions and its impact on short-chain fatty acid production in type-2 diabetic rats. Carbohydr. Polym. 2020, 235, 115904. [Google Scholar] [CrossRef]
- Ianni, A.; Innosa, D.; Martino, C.; Bennato, F.; Martino, G. Short communication: Compositional characteristics and aromatic profile of caciotta cheese obtained from Friesian cows fed with a dietary supplementation of dried grape pomace. J. Dairy Sci. 2019, 102, 1025–1032. [Google Scholar] [CrossRef] [PubMed]
- Padilla, B.; Belloch, C.; López-Díez, J.J.; Flores, M.; Manzanares, P. Potential impact of dairy yeasts on the typical flavour of traditional ewes’ and goats’ cheeses. Int. Dairy J. 2014, 35, 122–129. [Google Scholar] [CrossRef]
- Dopieralska, P.; Barłowska, J.; Teter, A.; Król, J.; Brodziak, A.; Domaradzki, P. Changes in Fatty Acid and Volatile Compound Profiles During Storage of Smoked Cheese Made from the Milk of Native Polish Cow Breeds Raised in the Low Beskids. Animals 2020, 10, 2103. [Google Scholar] [CrossRef] [PubMed]
- Bontinis, T.G.; Mallatou, H.; Pappa, E.C.; Massouras, T.; Alichanidis, E. Study of proteolysis, lipolysis and volatile profile of a traditional Greek goat cheese (Xinotyri) during ripening. Small Rumin. Res. 2012, 105, 193–201. [Google Scholar] [CrossRef]
- Sablé, S.; Cottenceau, G. Current Knowledge of Soft Cheeses Flavor and Related Compounds. J. Agric. Food Chem. 1999, 47, 4825–4836. [Google Scholar] [CrossRef]
- Moio, L.; Piombino, P.; Addeo, F. Odour-impact compounds of Gorgonzola cheese. J. Dairy Res. 2000, 67, 273–285. [Google Scholar] [CrossRef]
- Novella-Rodríguez, S.; Veciana-Nogués, M.; Izquierdo-Pulido, M.; Vidal-Carou, M. Distribution of Biogenic Amines and Polyamines in Cheese. J. Food Sci. 2003, 68, 750–756. [Google Scholar] [CrossRef]
- Roig-Sagués, A.X.; Molina, A.P.; Hernández-Herrero, M.M. Histamine and tyramine-forming microorganisms in Spanish traditional cheeses. Eur. Food Res. Technol. 2002, 215, 96–100. [Google Scholar] [CrossRef]
- Martuscelli, M.; Gardini, F.; Torriani, S.; Mastrocola, D.; Serio, A.; Chaves-López, C.; Schirone, M.; Suzzi, G. Production of biogenic amines during the ripening of Pecorino Abruzzese cheese. Int. Dairy J. 2005, 15, 571–578. [Google Scholar] [CrossRef]
- Pintado, A.I.E.; Pinho, O.; Ferreira, I.M.P.M.V.O.; Pintado, M.M.E.; Gomes, A.M.P.; Malcata, F.X. Microbiological, biochemical and biogenic amine profiles of Terrincho cheese manufactured in several dairy farms. Int. Dairy J. 2008, 18, 631–640. [Google Scholar] [CrossRef]
- Ladero, V.; Fernández, M.; Calles-Enríquez, M.; Sánchez-Llana, E.; Cañedo, E.; Martín, M.C.; Alvarez, M.A. Is the production of the biogenic amines tyramine and putrescine a species-level trait in enterococci? Food Microbiol. 2012, 30, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Kučerová, K.; Svobodova, H.; Tůma, S.; Ondráčková, I.; Plocková, M. Production of biogenic amines by Enterococci. Czech J. Food Sci. 2010, 27, 50–55. [Google Scholar] [CrossRef]
- Suzzi, G.; Caruso, M.; Gardini, F.; Lombardi, A.; Vannini, L.; Guerzoni, M.E.; Andrighetto, C.; Lanorte, M.T. A survey of the enterococci isolated from an artisanal Italian goat’s cheese (semicotto caprino). J. Appl. Microbiol. 2000, 89, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Marcobal, A.; Rivas, B.D.L.; Muã±Oz, R. First genetic characterization of a bacterial β-phenylethylamine biosynthetic enzyme in Enterococcus faecium RM58. FEMS Microbiol. Lett. 2006, 258, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Xu, W.; Du, L.; Wang, D.; Zhu, Y.; Geng, Z.; Zhang, M.; Xu, W. Heterologous Expression and Characterization of Tyrosine Decarboxylase from Enterococcus faecalis R612Z1 and Enterococcus faecium R615Z1. J. Food Prot. 2014, 77, 592–598. [Google Scholar] [CrossRef]
- Linares, D.M.; Martín, M.; Ladero, V.; Alvarez, M.A.; Fernández, M. Biogenic Amines in Dairy Products. Crit. Rev. Food Sci. Nutr. 2011, 51, 691–703. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J.M.; Chicón, R.; Cabezas, L. Biogenic amine content and proteolysis in Manchego cheese manufactured with Lactobacillus paracasei subsp. paracasei as adjunct and other autochthonous strains as starters. Int. Dairy J. 2015, 47, 94–101. [Google Scholar] [CrossRef]
- Alvarez, M.A.; Moreno-Arribas, M.V. The problem of biogenic amines in fermented foods and the use of potential biogenic amine-degrading microorganisms as a solution. Trends Food Sci. Technol. 2014, 39, 146–155. [Google Scholar] [CrossRef]
- Brink, B.T.; Damink, C.; Joosten, H.M.; Veld, J.H.I. Occurrence and formation of biologically active amines in foods. Int. J. Food Microbiol. 1990, 11, 73–84. [Google Scholar] [CrossRef]
- Pircher, A.; Bauer, F.; Paulsen, P. Formation of cadaverine, histamine, putrescine and tyramine by bacteria isolated from meat, fermented sausages and cheeses. Eur. Food Res. Technol. 2007, 226, 225–231. [Google Scholar] [CrossRef]
- Linares, D.M.; Del Río, B.; Ladero, V.; Martínez, N.; Fernández, M.; Martín, M.C.; Álvarez, M.A. Factors Influencing Biogenic Amines Accumulation in Dairy Products. Front. Microbiol. 2012, 3, 180. [Google Scholar] [CrossRef] [PubMed]
- Ladero, V.; Rattray, F.P.; Mayo, B.; Martin, M.C.; Fernandez, M.; Alvarez, M.A. Sequencing and Transcriptional Analysis of the Biosynthesis Gene Cluster of Putrescine-Producing Lactococcus lactis. Appl. Environ. Microbiol. 2011, 77, 6409–6418. [Google Scholar] [CrossRef] [PubMed]

| T2 | T60 | T90 | T120 | SEM | P | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ctrl | GP+ | Ctrl | GP+ | Ctrl | GP+ | Ctrl | GP+ | D | R | D × R | ||
| Alcohols | ||||||||||||
| 1-Pentanol, 3-methyl- | nd | nd | nd | nd | nd | nd | nd b | 0.13 a | <0.01 | 0.0219 | 0.0033 | 0.0033 |
| 2-Nonanol | nd | nd | nd | nd | nd | nd | 0.15 | 0.13 | 0.02 | 0.8928 | 0.0432 | 0.9953 |
| 1-Decanol | nd | nd | nd | nd | nd | nd | 0.10 | 0.08 | <0.01 | 0.0977 | <0.0001 | 0.041 |
| Aldeydes | ||||||||||||
| Hexanal | nd | nd | nd | nd | nd | nd | 1.85 | 1.14 | 0.31 | 0.2482 | <0.0001 | 0.2161 |
| Octanal | 0.20 | 0.27 | nd | nd | nd | nd | nd | nd | 0.02 | 0.6346 | 0.0005 | 0.8407 |
| Nonanal | 3.46 | 1.96 | 0.15 | 0.06 | nd | nd | nd | nd | 1.34 | 0.3090 | 0.0002 | 0.3727 |
| Carboxylic Acids | ||||||||||||
| Butanoic acid | nd | nd | 1.92 a,b | 1.21 a,b | 0.95 a,b | 0.48 b | 3.26 a | 1.92 a,b | 0.61 | 0.0518 | 0.0001 | 0.3756 |
| Hexanoic acid | 5.89 d | 5.68 d | 20.23 a | 14.88 b,c | 12.22 c | 10.62 c | 18.56 a,b | 10.71 c | 1.88 | <0.0001 | <0.0001 | 0.0021 |
| Octanoic Acid | 24.50 c,d | 27.97 b,c | 28.87 b,c | 44.47 a | 32.59 a,b,c | 39.11 a,b | 16.49 d | 42.95 a | 13.63 | <0.0001 | 0.0022 | 0.0009 |
| Nonanoic acid | nd | nd | 0.16 b | 0.37 c | nd c | 0.31 a,b | nd c | 0.30 a | 0.00 | <0.0001 | <0.0001 | < 0.0001 |
| n-Decanoic acid | 53.31 a | 46.10 a,b | 21.13 b | 32.78 a,b | 27.47 a,b | 41.52 a,b | 26.93 b | 26.94 b | 61.52 | 0.2211 | 0.0011 | 0.1698 |
| Undecanoic acid | 3.6 a | 2.48 a,b | nd b,c | 0.05 b,c | nd b,c | 0.09 b,c | nd c | 0.04 c | 0.61 | 0.5170 | <0.0001 | 0.515 |
| Dodecanoic acid | nd | nd | 0.71 b | 1.08 a,b | 1.03 a,b | 1.36 a | 0.77 b | 1.13 a,b | 0.02 | 0.0003 | <0.0001 | 0.0574 |
| Tetradecanoic acid | nd | nd | nd | nd | nd | nd | 0.08 a | 0.06 a | <0.01 | 0.6132 | <0.0001 | 0.8149 |
| Ketones | ||||||||||||
| 2-Pentanone | nd | nd | 2.84 a | 1.08 b,c | 0.65 c | 0.99 b,c | 2.28 a,b | 1.04 b,c | 0.18 | 0.0012 | <0.0001 | 0.0019 |
| 2-Heptanon | 1.81 b | 5.07 a,b | 9.79 a | 1.47 b | 7.76 a | 1.49 b | 5.68 a,b | 2.00 b | 2.13 | 0.0001 | 0.1611 | 0.0002 |
| 4,6 Octadiyn-3-one, 2-methyl- | nd | nd | nd | nd | nd | nd | 0.43 a | 0.08 b | 0.01 | 0.0083 | <0.0001 | 0.0007 |
| 2-Nonanone | 2.39 c | 8.44 a,b,c | 11.51 a,b | 1.74 b,c | 12.89 a | 2.74 b,c | 13.39 a | 5.9 a,b,c | 7.42 | 0.0010 | 0.1036 | 0.001 |
| 8-Nonen-2-one | nd | nd | 0.64 a | 0.05 b | 0.35 a,b | 0.09 b | 0.62 a | 0.3 b | 0.01 | <0.0001 | <0.0001 | 0.0002 |
| 2-Decanone | nd | nd | 0.17 a | 0.06 a | nd | nd | nd | nd | <0.01 | 0.0553 | 0.0001 | 0.0345 |
| 2-Undecanone | nd | nd | nd | nd | 0.44 a | 0.17 a | 0.64 a | 0.56 a | 0.05 | 0.2712 | 0.0001 | 0.6036 |
| Esters | ||||||||||||
| Butanoic acid, ethyl ester | nd | nd | nd | nd | nd | nd | 0.5 a | 0.39 a | 0.01 | 0.3833 | <0.0001 | 0.4451 |
| Hexanoic acid, ethyl ester | nd | nd | 0.26 b | 0.11 b | 1.36 a,b | 0.07 b | 2.81 a | 1.02 a,b | 0.52 | 0.0113 | 0.0006 | 0.0758 |
| Octanoic acid, ethyl ester | nd | nd | 0.12 b | 0.24 b | 0.89 b | 0.23 b | 2.80 a | 1.19 b | 0.12 | 0.0012 | <0.0001 | 0.0009 |
| Decanoic acid, ethyl ester | 1.05 b,c | 0.26 c | nd | nd | 1.01 b,c | 0.52 b,c | 2.75 a | 2.02 a,b | 0.27 | 0.0363 | <0.0001 | 0.5851 |
| Dodecanoic acid-, ethyl ester | nd | nd | nd | nd | nd | nd | 0.07 a | 0.10 a | <0.01 | 0.5967 | <0.0001 | 0.7936 |
| Valeric acid, 3-tridecyl ester | nd | nd | nd | nd | nd | nd | nd b | 0.01 a | <0.01 | 0.0080 | 0.0007 | 0.0007 |
| Lactones | ||||||||||||
| Pantolactone | nd | nd | nd | nd | 0.09 a | nd a | 1.05 a | 0.48 a | 0.19 | 0.2639 | 0.0028 | 0.3892 |
| δ-Decalactone | nd | nd | 0.37 a | 0.13 c | 0.22 b,c | 0.14 c | 0.35 a,b | 0.20 c | <0.01 | <0.0001 | <0.0001 | 0.0004 |
| γ-Dodecalactone | nd | nd | nd | nd | nd | nd | 0.08 a | 0.06 a | <0.01 | 0.5646 | 0.0005 | 0.7491 |
| δ-Dodecalactone | nd | nd | nd | nd | nd | nd | 0.09 a | 0.07 a | <0.01 | 0.5288 | <0.0001 | 0.6952 |
| Others | ||||||||||||
| Ethylbenzene | 0.8 a | 0.24 b | 0.52 a,b | 0.1 b | 0.02 b | 0.01 b | 0.12 b | 0.05 b | 0.03 | 0.0031 | 0.0008 | 0.0471 |
| P-xylene | 1.13 a | 0.4 b | 0.61 a,b | 0.13 b | 0.01 b | 0.01 b | nd | nd | 0.04 | 0.0018 | <0.0001 | 0.0082 |
| 1,1-Dodecanediol, diacetate | nd | nd | nd | nd | 0.06 a | 0.06 a | 0.04 a | 0.11 a | 0.01 | 0.6148 | 0.3213 | 0.8223 |
| T2 | T60 | T90 | T120 | SEM | P | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ctrl | GP+ | Ctrl | GP+ | Ctrl | GP+ | Ctrl | GP+ | D | R | D × R | ||
| Triptamine | nd | nd | nd | nd | nd | nd | nd | nd | ||||
| 2-phenylethylamine | nd | nd | 38.11 c | 13.35 d | 46.28 c | 15.04 d | 85.37 a | 63.51 b | 12.12 | <0.001 | <0.001 | 0.1523 |
| Putrescein | nd | nd | nd | 7.63 b | nd | 5.34 b | nd | 14.11 a | 3.22 | 0.0144 | ||
| Cadaverine | nd | nd | nd | 18.33 | nd | 10.67 | 15.74 | 20.00 | 8.62 | |||
| Hystamine | nd | nd | nd | nd | nd | nd | nd | nd | ||||
| Serotonine | nd | nd | nd | nd | nd | nd | nd | nd | ||||
| Tyramine | nd | nd | 32.53 b | 42.72 b | 40.18 b | 34.55 b | 101.23 a | 103.03 a | 86.31 | 0.6861 | <0.0001 | 0.4532 |
| Spermidine | nd | nd | nd | nd | nd | nd | nd | nd | ||||
| Spermine | nd | nd | nd | nd | nd | nd | nd | nd | ||||
| Alcohols | Aldeydes | Carboxylic Acids | Ketones | Esters | Lactones | 2-phenylethylamine | Putrescein | Cadaverine | Tyramine | |
|---|---|---|---|---|---|---|---|---|---|---|
| Moraxellaceae | −0.254 | 0.811 | 0.139 | −0.272 | −0.400 | −0.417 | −0.568 | −0.371 | −0.464 | −0.615 |
| Weeksellaceae | −0.452 | 0.706 | 0.338 | −0.393 | −0.647 | −0.616 | −0.604 | −0.280 | −0.353 | −0.613 |
| Pseudomonadaceae | −0.321 | 0.918 | 0.259 | −0.430 | −0.463 | −0.478 | −0.629 | −0.340 | −0.443 | −0.655 |
| Streptococcaceae | 0.030 | −0.548 | 0.563 | −0.419 | −0.169 | −0.097 | 0.176 | 0.459 | 0.647 | 0.401 |
| Enterobacteriaceae | 0.487 | −0.236 | −0.255 | 0.186 | 0.606 | 0.498 | 0.188 | 0.282 | 0.402 | 0.319 |
| Sphingobacteriaceae | −0.226 | 0.143 | 0.589 | −0.537 | −0.555 | −0.500 | −0.378 | 0.091 | 0.104 | −0.236 |
| Carnobacteriaceae | −0.240 | 0.549 | 0.464 | −0.562 | −0.508 | −0.418 | −0.476 | −0.172 | −0.166 | −0.436 |
| Enterococcaceae | 0.364 | −0.819 | −0.299 | 0.423 | 0.529 | 0.518 | 0.703 | 0.309 | 0.363 | 0.685 |
| Lactobacillaceae | 0.036 | −0.436 | −0.710 | 0.799 | 0.401 | 0.442 | 0.320 | −0.165 | −0.248 | 0.127 |
| Leuconostocaceae | −0.005 | −0.390 | −0.550 | 0.601 | 0.415 | 0.358 | 0.325 | −0.171 | −0.140 | 0.122 |
| Staphylococcaceae | −0.240 | −0.118 | 0.345 | −0.265 | −0.250 | −0.265 | −0.239 | 0.258 | 0.361 | −0.037 |
| Lactobacillales | 0.173 | −0.197 | −0.522 | 0.529 | 0.411 | 0.320 | 0.076 | −0.071 | −0.193 | 0.005 |
| Flavobacteriaceae | −0.158 | 0.294 | −0.003 | −0.053 | −0.149 | −0.160 | −0.143 | −0.209 | −0.270 | −0.233 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bennato, F.; Di Domenico, M.; Ianni, A.; Di Gialleonardo, L.; Cammà, C.; Martino, G. Grape Pomace in Ewes Diet Affects Metagenomic Profile, Volatile Compounds and Biogenic Amines Contents of Ripened Cheese. Fermentation 2022, 8, 598. https://doi.org/10.3390/fermentation8110598
Bennato F, Di Domenico M, Ianni A, Di Gialleonardo L, Cammà C, Martino G. Grape Pomace in Ewes Diet Affects Metagenomic Profile, Volatile Compounds and Biogenic Amines Contents of Ripened Cheese. Fermentation. 2022; 8(11):598. https://doi.org/10.3390/fermentation8110598
Chicago/Turabian StyleBennato, Francesca, Marco Di Domenico, Andrea Ianni, Luigina Di Gialleonardo, Cesare Cammà, and Giuseppe Martino. 2022. "Grape Pomace in Ewes Diet Affects Metagenomic Profile, Volatile Compounds and Biogenic Amines Contents of Ripened Cheese" Fermentation 8, no. 11: 598. https://doi.org/10.3390/fermentation8110598
APA StyleBennato, F., Di Domenico, M., Ianni, A., Di Gialleonardo, L., Cammà, C., & Martino, G. (2022). Grape Pomace in Ewes Diet Affects Metagenomic Profile, Volatile Compounds and Biogenic Amines Contents of Ripened Cheese. Fermentation, 8(11), 598. https://doi.org/10.3390/fermentation8110598





