New Avoparcin-like Molecules from the Avoparcin Producer Amycolatopsis coloradensis ATCC 53629
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Cultivation Conditions and Identification of Morphological Variants
2.2. Avoparcin Complex Extraction, Analysis and Purification
3. Results
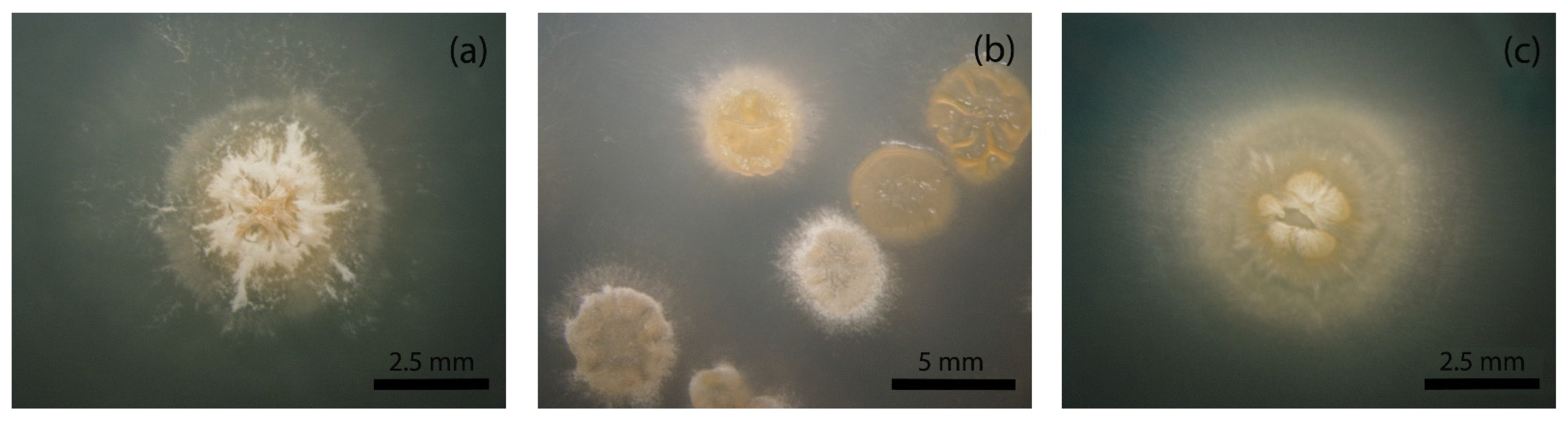
3.1. Identification of a Heterogeneous Population in A. coloradensis ATCC 53629
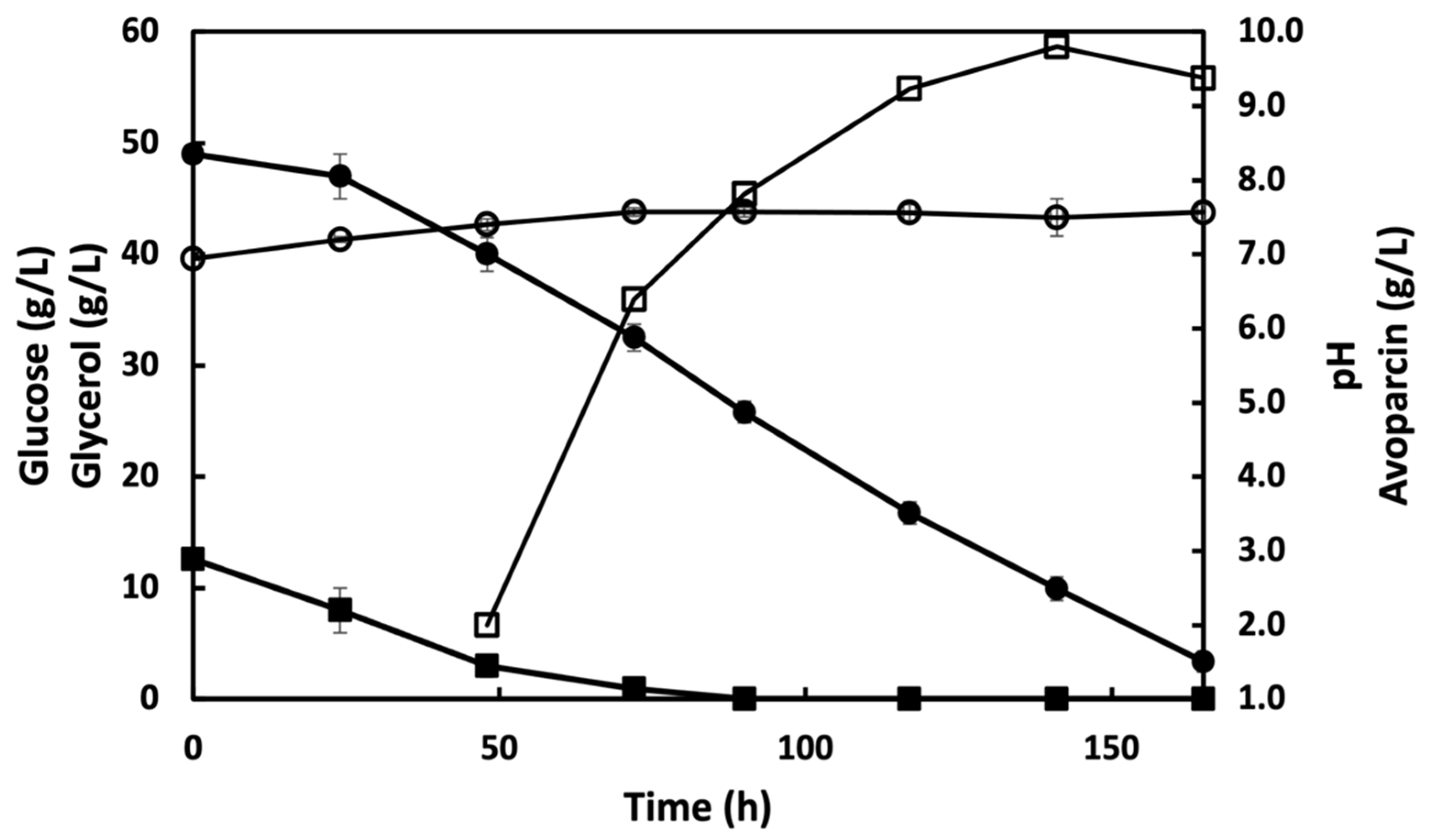
3.2. Fermentation of A. coloradensis ATCC 53629 for the Production of Avoparcin
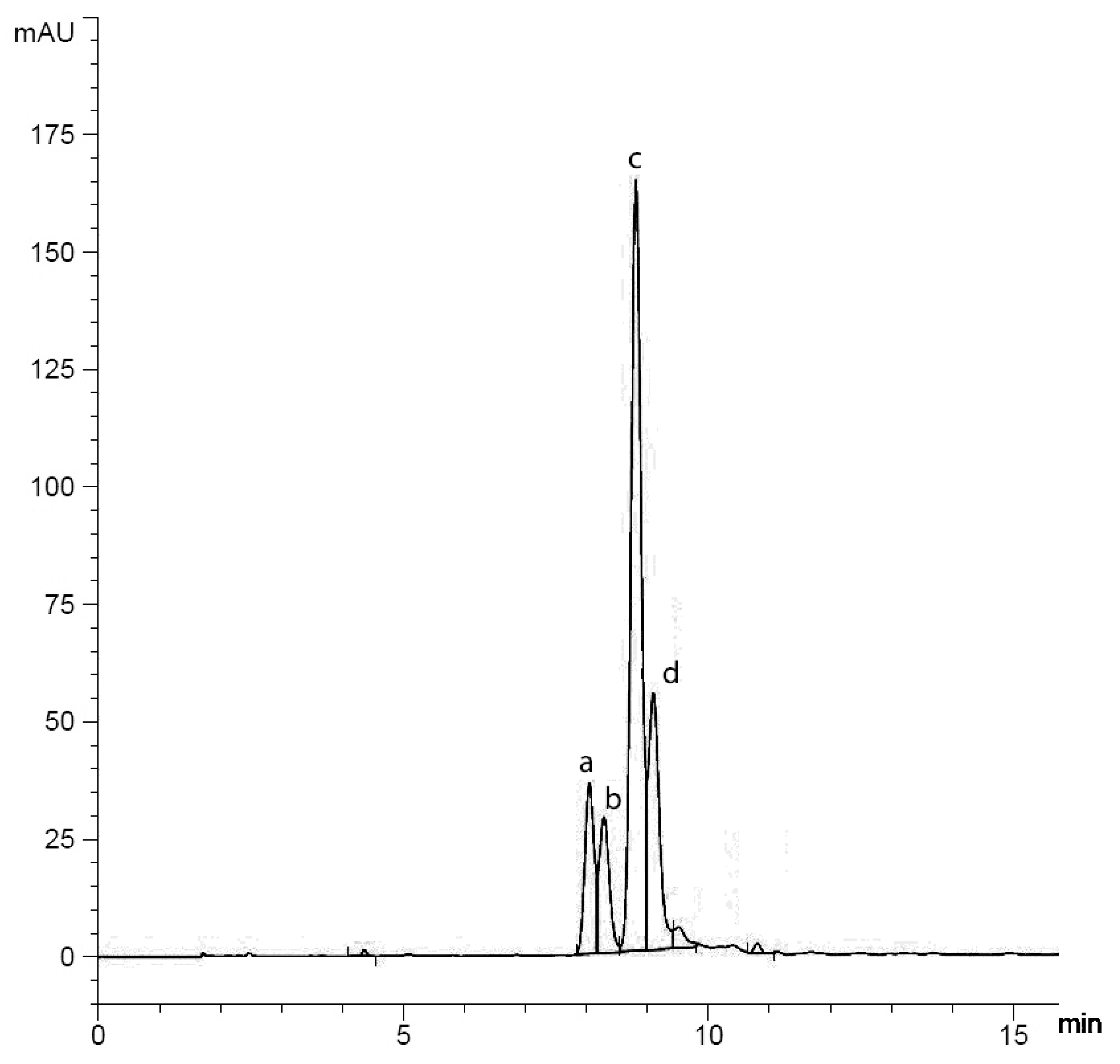
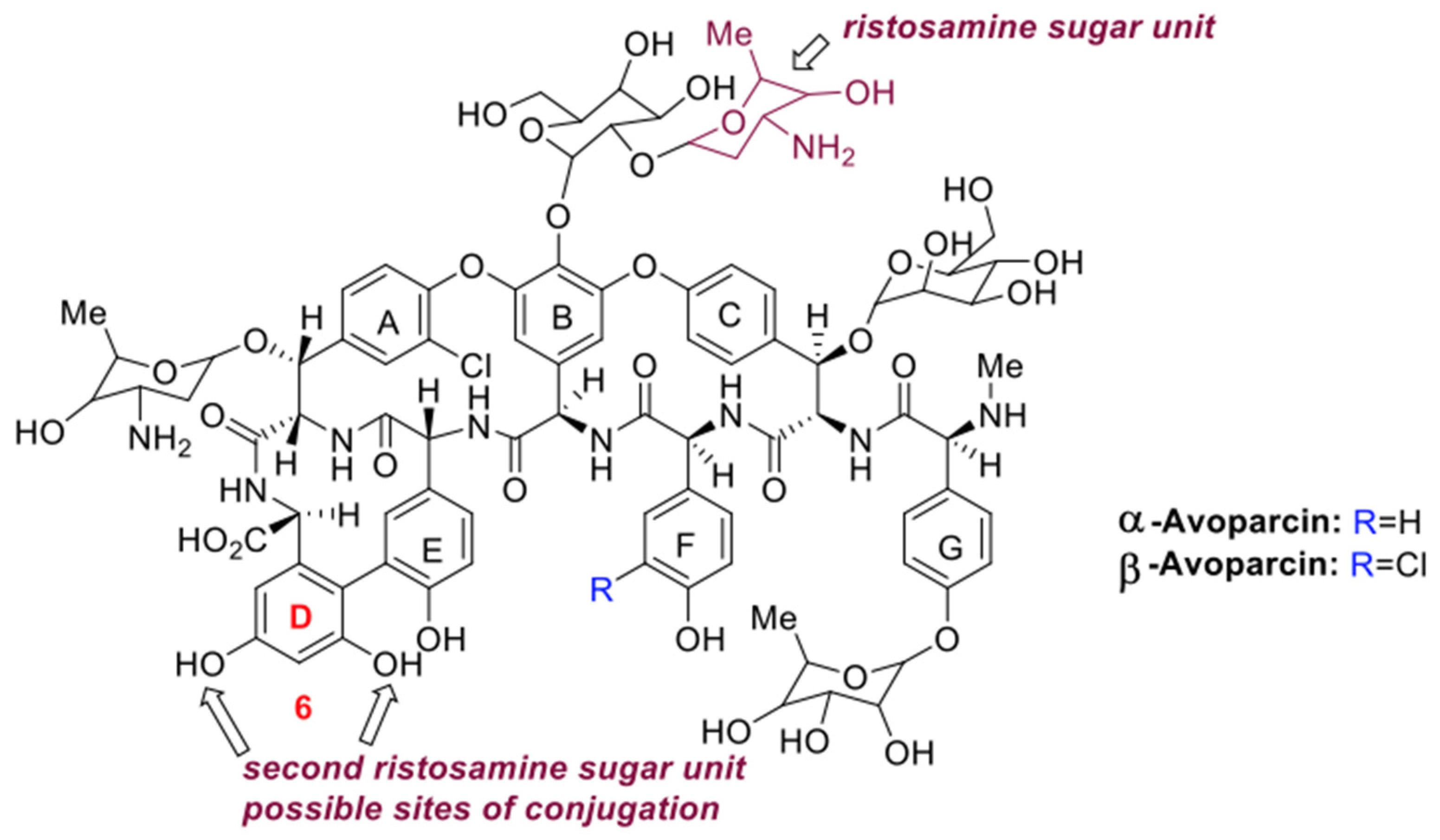
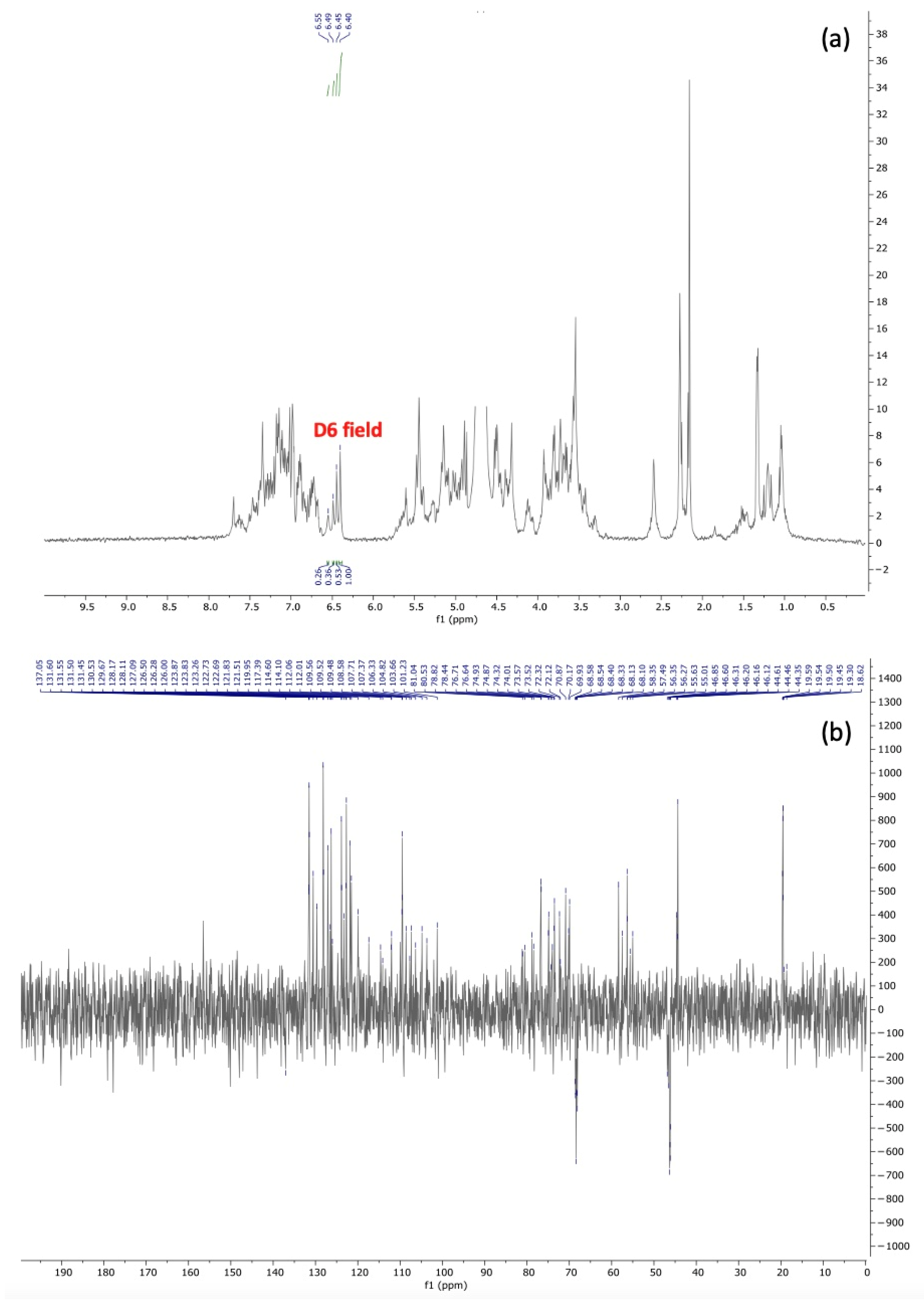
3.3. Purification and Chemical Characterization of the Avoparcin Complex
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bérdy, J. Thoughts and Facts about Antibiotics: Where We Are Now and Where We Are Heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Katz, L.; Baltz, R.H. Natural Product Discovery: Past, Present, and Future. J. Ind. Microbiol. Biotechnol. 2016, 43, 155–176. [Google Scholar] [CrossRef]
- Hopwood, D.A. Streptomyces in Nature and Medicine: The Antibiotic Makers; Oxford University Press: Oxford, UK, 2007; ISBN 978-0-19-515066-7. [Google Scholar]
- Zhang, Z.; Du, C.; de Barsy, F.; Liem, M.; Liakopoulos, A.; van Wezel, G.P.; Choi, Y.H.; Claessen, D.; Rozen, D.E. Antibiotic Production in Streptomyces Is Organized by a Division of Labor through Terminal Genomic Differentiation. Sci. Adv. 2020, 6, eaay5781. [Google Scholar] [CrossRef] [Green Version]
- West, S.A.; Cooper, G.A. Division of Labour in Microorganisms: An Evolutionary Perspective. Nat. Rev. Microbiol. 2016, 14, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Wahl, L.M. The Division of Labor: Genotypic versus Phenotypic Specialization. Am. Nat. 2002, 160, 135–145. [Google Scholar] [CrossRef]
- Kumari, R.; Singh, P.; Lal, R. Genetics and Genomics of the Genus Amycolatopsis. Indian J. Microbiol. 2016, 56, 233–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.; Xu, T.; Wang, J.; Hou, Y.; Liu, C.; Liu, S.; Wu, S. Secondary Metabolites of the Genus Amycolatopsis: Structures, Bioactivities and Biosynthesis. Molecules 2021, 26, 1884. [Google Scholar] [CrossRef]
- Kunstmann, M.; Mitscher, L.; Porter, J.; Shay, A.; Darken, M. LL-AV290, a New Antibiotic. I. Fermentation, Isolation, and Characterization. Antimicrob. Agents Chemother. 1968, 8, 242–245. [Google Scholar] [PubMed]
- Redin, G.; Dornbush, A. LL-AV290, a New Antibiotic. II. Antibacterial Efficacy in Mice and in Vitro. Antimicrob. Agents Chemother. 1968, 8, 246–248. [Google Scholar]
- Ping, S.; Murray, D. Antibiotic AV290-Syntan complexes and Animal Feed Supplements. U.S. Patent 3,832,462, 27 August 1974. [Google Scholar]
- Witte, W.; Klare, I. Glycopeptide-Resistant Enterococcus Faecium Outside Hospitals: A Commentary. Microb. Drug Resist. 1995, 1, 259–263. [Google Scholar] [CrossRef]
- Wegener, H.C. Antibiotics in Animal Feed and Their Role in Resistance Development. Curr. Opin. Microbiol. 2003, 6, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Wegener, H.C.; Aarestrup, F.M.; Jensen, L.B.; Hammerum, A.M.; Bager, F. Use of Antimicrobial Growth Promoters in Food Animals and Enterococcus Faecium Resistance to Therapeutic Antimicrobial Drugs in Europe. Emerg. Infect. Dis. 1999, 5, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Marcone, G.L.; Carrano, L.; Marinelli, F.; Beltrametti, F. Protoplast Preparation and Reversion to the Normal Filamentous Growth in Antibiotic-Producing Uncommon Actinomycetes. J. Antibiot. 2010, 63, 83–88. [Google Scholar] [CrossRef] [Green Version]
- Beltrametti, F.; Lazzarini, A.; Brunati, C.; Selva, E.; Marinelli, F. Production of Demannosyl-A40926 by a Nonomuraea Sp. ATCC 39727 Mutant Strain. J. Antibiot. 2003, 56, 310–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binda, E.; Berini, F.; Marinelli, F.; Bava, A.; Beltrametti, F. Heterogeneous A40926 Self-Resistance Profile in Nonomuraea Gerenzanensis Population Informs Strain Improvement. Fermentation 2021, 7, 140. [Google Scholar] [CrossRef]
- Beltrametti, F.; Consolandi, A.; Carrano, L.; Bagatin, F.; Rossi, R.; Leoni, L.; Zennaro, E.; Selva, E.; Marinelli, F. Resistance to Glycopeptide Antibiotics in the Teicoplanin Producer Is Mediated by van Gene Homologue Expression Directing the Synthesis of a Modified Cell Wall Peptidoglycan. Antimicrob. Agents Chemother. 2007, 51, 1135–1141. [Google Scholar] [CrossRef] [Green Version]
- Murray, D.; Korshalla, J.D.; Ping, S. Method for the Production and Isolation of Antibiot AV290 Sulfate. U.S. Patent 3855410, 17 December 1974. [Google Scholar]
- Wong, S.K.; Yao, W.Y. Purity Assessment for Avoparcin. Accredit. Qual. Assur. 2013, 18, 411–420. [Google Scholar] [CrossRef]
- Corti, A.; Cassani, G. Synthesis and Characterization of D-Alanyl-D-Alanine-Agarose. Appl. Biochem. Biotechnol. 1985, 11, 101–109. [Google Scholar] [CrossRef]
- Volff, J.-N.; Altenbuchner, J. Genetic Instability of the Streptomyces Chromosome. Mol. Microbiol. 1998, 27, 239–246. [Google Scholar] [CrossRef]
- Kadota, M.; Imanaka, M.; Ogawa, N.; Kumashiro, K.; Mori, T.; Oka, H.; Ikai, Y.; Horie, M.; Nakazawa, H. Determination of the Avoparcin(AV) in Glycopeptide-Type Antibiotics by High-Performance Liquid Chromatography. (1). Isolation and Purification of α-AV and β-AV from the Avoparcin Complex. Annu. Rep. Okayama Prefect Instig. Environ. Sci. Public Health 1991, 15, 7–12. [Google Scholar]
- Ellestad, G.A.; Leese, R.A.; Morton, G.O.; Barbatschi, F.; Gore, W.; McGahren, W.J.; Armitage, I.M. Avoparcin and Epiavoparcin. J. Am. Chem. Soc. 1981, 103, 6522–6524. [Google Scholar] [CrossRef]
- McGahren, W.J.; Martin, J.H.; Morton, G.O.; Hargreaves, R.T.; Leese, R.A.; Lovell, F.M.; Ellestad, G.A.; O’Brien, E.; Holker, J.S.E. Structure of Avoparcin Components. J. Am. Chem. Soc. 1980, 102, 1671–1684. [Google Scholar] [CrossRef]
- Mcgahren, W.J.; Leese, R.A.; Barbatschi, F.; Morton, G.O.; Kuck, N.A.; Ellestad, G.A. Components and Degradation Compounds of the Avoparcin Complex. J. Antibiot. 1983, 36, 1671–1682. [Google Scholar] [CrossRef]
- Ellestad, G.A.; Swenson, W.; Mcgahren, W.J. Epimerization and Stereochemistry of Avoparcin. J. Antibiot. 1983, 36, 1683–1690. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, M.; Takahashi, S.; Inukai, M.; Nakamura, T.; Kinoshita, T. Helvecardins A and B, Novel Glycopeptide Antibiotics. II. Structural Elucidation. J. Antibiot. 1991, 44, 271–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inoue, K.; Mizuno, Y.; Yoshimi, Y.; Nunome, M.; Hino, T.; Tsutsumiuchi, K.; Oka, H. Determination of Avoparcin in Animal Tissues and Milk Using LC-ESI-MS/MS and Tandem-SPE. J. Sep. Sci. 2008, 31, 3871–3878. [Google Scholar] [CrossRef]
- Xu, L.; Huang, H.; Wei, W.; Zhong, Y.; Tang, B.; Yuan, H.; Zhu, L.; Huang, W.; Ge, M.; Yang, S.; et al. Complete Genome Sequence and Comparative Genomic Analyses of the Vancomycin-Producing Amycolatopsis Orientalis. BMC Genom. 2014, 15, 363. [Google Scholar] [CrossRef] [Green Version]
- Doroghazi, J.R.; Metcalf, W.W. Comparative Genomics of Actinomycetes with a Focus on Natural Product Biosynthetic Genes. BMC Genom. 2013, 14, 611. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Wu, Q.; Shen, Q.; Wang, H. Progress in Understanding the Genetic Information and Biosynthetic Pathways behind Amycolatopsis Antibiotics, with Implications for the Continued Discovery of Novel Drugs. ChemBioChem 2016, 17, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto-Shinohara, Y.; Imaizumi, T.; Sukenobe, J.; Murakami, Y.; Kawamura, S.; Komatsu, Y. Survival Rate of Microbes after Freeze-Drying and Long-Term Storage. Cryobiology 2000, 41, 251–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jałowiecki, Ł.; Krzymińska, I.; Górska, M.; Płaza, G.; Ratman-Kłosińska, I. Effect of the Freeze-Drying Process on the Phenotypic Diversity of Pseudomonas Putida Strains Isolated from the Interior of Healthy Roots of Sida Hermaphrodita: Phenotype Microarrays (PMs). Cryobiology 2020, 96, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Courvalin, P.; Davies, J. Antimicrobials. Curr. Opin. Microbiol. 2003, 6, 425–426. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramoni, G.; Capozzoli, C.; Bava, A.; Foschi, F.; Broggini, G.; Beltrametti, F. New Avoparcin-like Molecules from the Avoparcin Producer Amycolatopsis coloradensis ATCC 53629. Fermentation 2022, 8, 44. https://doi.org/10.3390/fermentation8020044
Ramoni G, Capozzoli C, Bava A, Foschi F, Broggini G, Beltrametti F. New Avoparcin-like Molecules from the Avoparcin Producer Amycolatopsis coloradensis ATCC 53629. Fermentation. 2022; 8(2):44. https://doi.org/10.3390/fermentation8020044
Chicago/Turabian StyleRamoni, Gina, Carmine Capozzoli, Adriana Bava, Francesca Foschi, Gianluigi Broggini, and Fabrizio Beltrametti. 2022. "New Avoparcin-like Molecules from the Avoparcin Producer Amycolatopsis coloradensis ATCC 53629" Fermentation 8, no. 2: 44. https://doi.org/10.3390/fermentation8020044
APA StyleRamoni, G., Capozzoli, C., Bava, A., Foschi, F., Broggini, G., & Beltrametti, F. (2022). New Avoparcin-like Molecules from the Avoparcin Producer Amycolatopsis coloradensis ATCC 53629. Fermentation, 8(2), 44. https://doi.org/10.3390/fermentation8020044






