Abstract
Microbiology has long been a keystone in fermentation, and innovative yeast molecular biotechnology continues to represent a fruitful frontier in brewing science. Consequently, modern understanding of brewer’s yeast has undergone significant refinement over the last few decades. This publication presents a condensed summation of Saccharomyces species dynamics with an emphasis on the relationship between; traditional Saccharomyces cerevisiae ale yeast, S. pastorianus interspecific hybrids used in lager production, and novel hybrid yeast progress. Moreover, introgression from other Saccharomyces species is briefly addressed. The unique history of Saccharomyces cerevisiae and Saccharomyces hybrids is exemplified by recent genomic sequencing studies aimed at categorizing brewing strains through phylogeny and redefining Saccharomyces species boundaries. Phylogenetic investigations highlight the genomic diversity of Saccharomyces cerevisiae ale strains long known to brewers for their fermentation characteristics and phenotypes. The discovery of genomic contributions from interspecific Saccharomyces species into the genome of S. cerevisiae strains is ever more apparent with increasing research investigating the hybrid nature of modern industrial and historical fermentation yeast.
Keywords:
hybrid; lager; yeast; introgression; interspecific; domestication; phylogeny; brewing; molecular; genomics 1. Species of Saccharomyces
Saccharomyces cerevisiae may be one of the oldest domesticated organisms known to humans. Domestication events imposed on brewing strains of the budding yeast species S. cerevisiae resulted in unique strains similar to the divergence seen in animal lineages of Canis familiaris breeds or the plant lineage of Brassica oleracea foods. It has been suggested that S. cerevisiae behaved as a synanthropic species, following human settlements as a commensal organism residing in gardens and vineyards, although the time period and location of the yeast’s origins has been the subject of much debate throughout history. Domesticated Saccharomyces brewing strains feature flocculation capabilities, fast fermentation rates, malt sugar utilization, pleasant aromas, and are largely negative for production of phenolic off flavors (POF) [1,2].
Nearly two centuries have passed since the first accessible description was produced regarding brewer’s yeast and its recognition in fermentation [3,4]. Recently, phylogenic research utilizing genomics and modern molecular biology techniques has shed some light on the historically convoluted nomenclature surrounding this budding yeast. Genomic analysis of the Saccharomyces genus has consolidated many variations into eight individual species: S. cerevisiae, S. paradoxus (syn. S. cariocanus, S. cerevisiae var. tetraspora, S. cerevisiae var. terrestris, S. douglasii), S. uvarum (syn. S. bayanus var. uvarum), S. mikatae, S. kudriavzevii, S. arboricola (syn. S. arboricolus), S. eubayanus, and S. jurei [3,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19] (Table 1). Moreover, two natural hybrids are recognized in the Saccharomyces clade: S. pastorianus (syn. S. carlsbergensis, S. monacensis) and S. bayanus [20,21,22]. Most modern lager fermentations utilize S. patorianus yeasts.

Table 1.
Current Saccharomyces Yeast Species History.
2. Hybrid Nature of Yeast
Interspecific hybrids are not unique to lager brewing. For example, the livestock and agricultural industries commonly employ selective breeding to alter species’ properties or increase yields [23,24,25]. A time-honored showpiece of hybrid vigor is the mule, a great pack animal known for its hardiness and longevity. For over 4000 years, the mule has been bred as the hybrid progeny of a male donkey and a female horse. Since the early 1900s, maize has been hybridized to increase yields and introduce biodiversity [26]. Similarly, hybrid yeasts have been isolated from fermentation processes on numerous occasions [27] (Figure 1). A hybrid between S. cerevisiae and S. kudriavzevii was isolated from Belgian Trappist beers [28]. Popular in wine production, strain VIN7 is a hybrid of S. cerevisiae and S. kudriavzevii [29]. Other interspecific S. cerevisiae and S. uvarum hybrids are also regularly used for production of wines [29,30]. Spontaneous fermentations have yielded Pichia apotheca, a hybrid of P. membranifaciens and an unknown species [31]. Hybrid vigor, or heterosis, confers a competitive advantage by facilitating transgressive phenotypes in changing environments, and is known to be a driver of fungal evolution and adaptation [32]. This is especially important for yeast as the many stages of fermentation and maturation create a microbially competitive environment in which rapid adaptation may be advantageous.
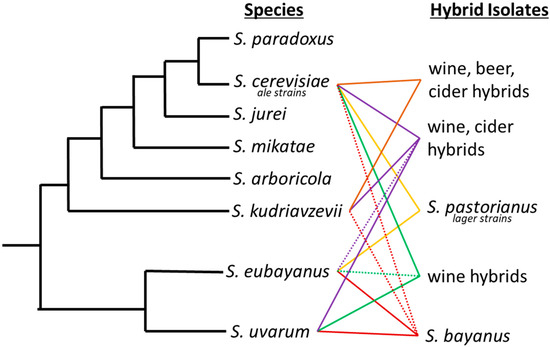
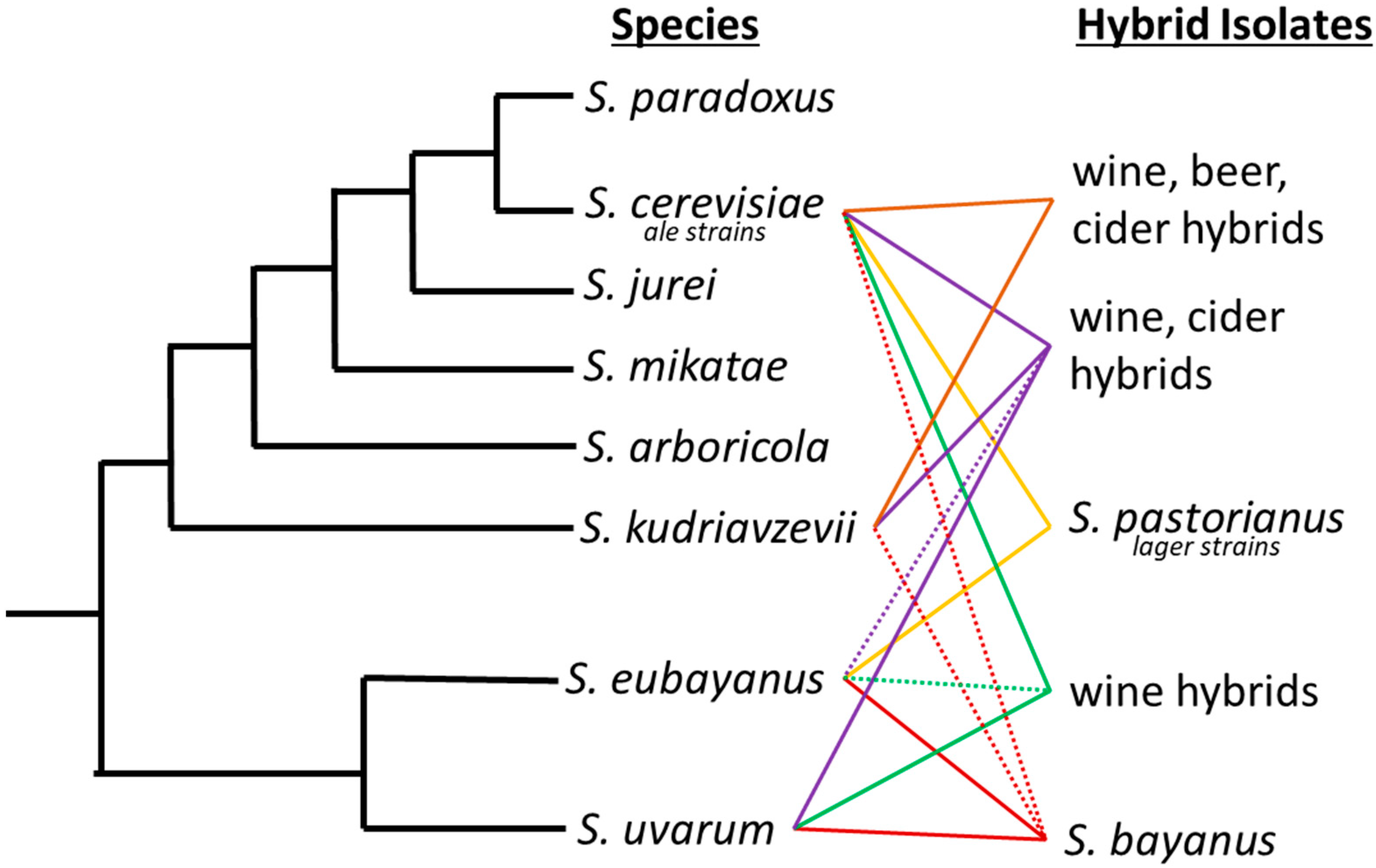
Figure 1.
Saccharomyces phylogenetic tree with industrially important hybrids. Industrial hybrids are listed to the right by the fermentation they have predominately been associated with. Solid lines between two species signifies the interspecific hybrids and dashed lines denote introgression from a third or fourth species that may not always be present in each hybrid strain.
The mule of the brewing industry is the lager yeast S. pastorianus, an interspecific hybrid that produces the lion’s share, in volume, of the global beer production. Although its use is widespread, the biodiversity is limited to two main lineages, Saaz/group I (syn. S. carlsbergensis, (L12: Noble, Imperial Yeast Culture Collection, type strain CBS1513) and Frohberg/group II (L13: Global, Imperial Yeast Culture Collection, type strain-Weihenstephan 34/70). Saaz and Frohberg lineages vary in their genomic composition from each parent species, S. eubayanus and S. cerevisiae, which influences important fermentation characteristics. Genomic analysis demonstrated a genomic composition of 1:2 S. cerevisiae to S. eubayanus sub genome in the Saaz lineage and 2:2 S. cerevisiae to S. eubayanus sub genome in the Frohberg lineage supporting the traditional designations used by brewers [33,34,35]. Saaz lineage hybrids are very well adapted to cold fermentations and many of these strains lack maltotriose utilization [27]. The Frohberg hybrids contain more S. cerevisiae genomic content conferring greater attenuation, higher ethanol production, differing ester profiles, and higher typical viabilities [36]. The composition of genetic material transferred and retained in these hybrids impart important fermentation characteristics and phenotypes such as the POF (phenolic off flavor) trait, efficient fermentation of maltose and maltotriose, reduction of diacetyl, flocculation, and production of unique volatile metabolite profiles that are low in off-aroma/flavors. Investigation into these hybrid lineages supports bolstering the fermentation capacity of S. cerevisiae with hybrid vigor from S. eubayanus incorporation and conveyance of a positive phenotype.
3. Novel Hybrid Development
The first yeast breeding experiments aimed at combining desirable traits of brewing strains were conducted by Ojvind Winge during his tenure at the Carlsberg Laboratory in the 1930s [37]. Hybrid yeast development has been carried out for over half a century since then, aimed predominantly at increasing attenuation and fermentation rates via intraspecific crosses with ale and lab strains [38,39,40]. Modern fermentations benefit from many innate and acquired hybrids that have been isolated or developed [11,27,28,41,42,43,44,45,46,47,48,49,50,51,52]. Early efforts in brewing science established the fundamentals necessary to explore the phylogeny, genomics, and strain development for Saccharomyces fermentation. During typical rich nutrient propagations of yeast in a brewing environment, mother cells reproduce asexually to bud off small daughter clones (Figure 2). Under poor nitrogen conditions, such as proline, yeast growth changes to a pseudohyphal form [53,54]. The complete absence of a nitrogen source and the presence of a non-fermentable carbon source, such as acetate, will sporulate yeast cells [55]. Sporulation transforms the cell wall into the ascus, or sack, that holds four spores termed a tetrad. Analogous to the human egg and sperm, these spores divide equally into mating types as either a or α [56]. When conditions improve for yeast growth, new haploid (1n) yeast can conjugate with the opposite mating type yeast as they form a shmoo. Depending on the genomic make up of each parental strain or species, there is some genomic instability or rewiring that occurs during the following mitotic budding growth.
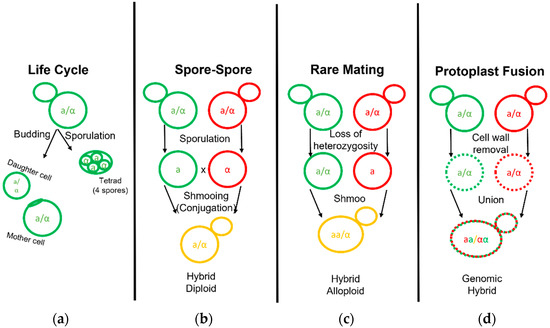
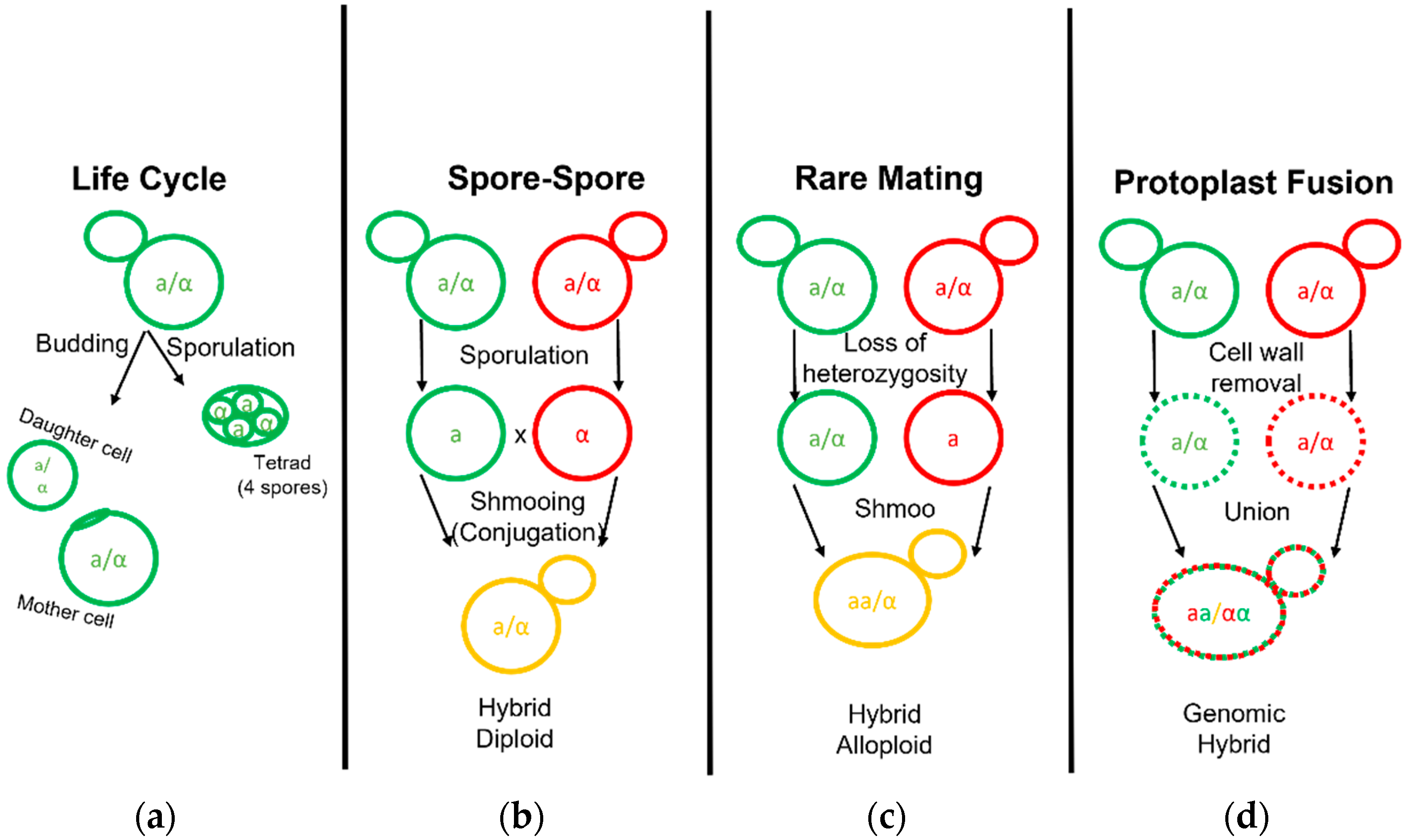
Figure 2.
Life and Mating of Saccharomyces Yeast. Diagram pertaining to the clonal growth typical of yeast fermentation cultures and the various known techniques employed to generate yeast hybrids. (a) Diploid yeast cells may bud and grow clonally to form a mother and daughter cell or undergo sporulation to form a tetrad. (b) Yeast hybridization may form by direct spore to spore mating. (c) Yeast hybridization may form by rare mating events in which one or both diploid parent cells gain competency by becoming hemizygous or MATa/MATa and MATα/MATα diploids. (d) Yeast hybridization may also form by fusion of two separate yeast cell protoplasts with their cell wall removed.
Interspecific hybridization is seen as a valuable tool for yeast strain development, enabling the combination and enhancement of characteristics from both parental strains or species [57]. The development of hybrids is executed via three primary methodologies: spore–spore mating, rare mating, and protoplast fusion (Figure 2). Spore to spore mating is most similar to what would be considered natural mating, as outlined in Figure 2b. This approach bears a high success rate, high genomic stability, and can avoid the aid of selection markers such as drug resistance or autotrophies. Rare mating utilizes a described spontaneous loss of heterozygosity at the mating type locus. Normal diploid cells carry two sets of chromosomes with both the MATa and MATα genetic alleles and do not respond to sex pheromones for mating purposes. The spontaneous loss of either sex allele tolerates yeast mating to a yeast cell of complimentary sex. This results in yeast with high chromosome counts, influencing gene dosage during cellular processes and partially explains the outperformance over a diploid yeast of the same background [57]. Rare mating, as the name implies, is uncommon and selection markers are needed to perform this technique. The frequency of rare mating is estimated to occur in 1 out of 10 million cells [58]. This procedure is beneficial in overcoming poor sporulation but produces hybrids prone to high genomic instability. Lastly, protoplast fusion is performed by removing the cell wall and fusing the protoplasts of two cells together before the cell wall is repaired. This technique generates cells with a high chromosome copy number and higher genomic instability but overcomes low sporulation. This technique can be used in the laboratory to combine yeast from different genera such as the brewer’s yeast S. cerevisiae and other yeast outside of the Saccharomyces genera which are otherwise incompatible [59]. Protoplast fusion is considered genetic engineering in many parts of the world. Recent select investigations into Saccharomyces hybrid application in beverage fermentations are listed in table format (Table 2).
Utilizing advanced molecular biology techniques for the modification of yeast strains presents an ongoing endeavor by many academic labs and some commercial yeast laboratories. Several research groups have developed protocols that use plasmids carrying genetic markers for drug resistance or functional enzymes that target the mating system in yeast. Industrial strains are well-known for their poor ability to adhere to laboratory techniques including sporulation and transformation [60]. One strategy developed at the University of Wisconsin generates allotetraploid strains of prototrophic yeast without the need for sporulation or modification to the nuclear genome of parental yeast strains [61]. This method leverages a series of inducible plasmids coined HyPr (Hybrid Production) containing compilatory drug markers and an inducible HO cassette. HO encodes an endonuclease that performs a double stranded break in the DNA that determines the Saccharomyces sex type. To repair this damage, the yeast cell will copy the silenced sex type already present in the genome, effectively performing a sex change and allowing the cell to mate with other cells of the opposite mating type. Using drug resistance markers and the HO cassette, hybrids are produced and serial growth in the absence of drug selection purges cells of exogenous DNA [61]. Similarly at the Advanced Industrial Science and Technology Center of the Biomedical Research Institute of Japan, another plasmid utilizing system aimed at exploiting the loss of heterozygosity of yeast cells was developed. A series of three to four plasmids allows for isolating a- and α- type cells from mixed cell populations and subsequent continual cross breeding [62]. CRISPR/Cas9 has also been utilized to force double stranded breaks in the MAT locus in order to increase the diversity of industrial yeast strains and their hybrids [63]. In the coming years, industrial yeasts will be designed by utilizing these protocols and molecular toolkits.
Currently, the options for strain selection in S. cerevisiae yeast are plentiful, but the criteria for fermentation performance in the brewing environment remains selective. Strains are employed largely by beer style, equipment availability, and supporting knowledge base. Certain beer styles also contain defining features from specific yeast flavor active molecules [64]. Banana and clove flavors are derived from isoamyl acetate and 4-vinyl guaiacol (4VG) in Weissbier [64,65,66]. Hybridization of yeast bears several advantages in brewing to include transgressive phenotypes such as increased ethanolic fermentation performance or stress tolerance, shifting fermentation temperatures beyond traditional inhibitory conditions, and creating a mosaic blend of parental fermentation profiles [24,51,57,63,67]. Targets of yeast hybridization may include increased formation of glycerol for an enhanced mouthfeel, alternative carbon source metabolic ability, reduced off-flavor production, increased formation of antioxidants that increase beer flavor stability, or increase production of yeast longevity molecules such as trehalose. The methodologies to create yeast hybrids vary in their specificity to target genetic or phenotypic results, but efforts to harness yeast hybrids in brewing broadly increase the biodiversity of fermentation yeast, add depth to the complexity of fermentation profiles, and advance brewing science knowledge.

Table 2.
Interspecific Yeast Hybrids in Fermentation.
Table 2.
Interspecific Yeast Hybrids in Fermentation.
| History | Parents | Reference |
|---|---|---|
| Isolated | S. cerevisiae × S. eubayanus | [11] |
| Isolated | S. cerevisiae × S. eubayanus × S. uvarum | [41] |
| Isolated | S. cerevisiae × S. uvarum | [68] |
| Isolated | S. cerevisiae × S. kudriavzevii | [27,28,42] |
| Isolated | S. uvarum × S. eubayanus | [27,42] |
| Developed | S. cerevisiae × S. eubayanus | [43,45,46,47,69] |
| Developed | S. cerevisiae × S. mikatae | [48] |
| Developed | S. cerevisiae × S. kudriavzevii × S. paradoxus | [49] |
| Developed | S. cerevisiae × S. kudriavzevii | [52] |
| Developed | S. cerevisiae × S. arboricola | [48,51] |
| Developed | S. cerevisiae × S. jurei | [70] |
4. Natural Considerations and State of the Field
Outside of industrial fermentations and laboratory settings, Saccharomyces yeast is readily obtained from tree sap exuded seasonally from oaks and beech tree bark, part of the Fagaceae family. The natural fluctuation of available carbon, nutrition, and climate influences the state cells between active replication, a non-dividing state termed quiescence, and sporulation [71]. During industrial use, yeast spend their time in active growth and metabolism.
The fermentation of sugars into ethanol, carbon dioxide, and flavor molecules is utilized to prepare food or drink for human consumption. The brewer’s and baker’s yeast have secured a niche ecological space in industry via an advantage in high sugar environments. The Crabtree effect is a critical microbial factor for beer production and occurs when fermentation of sugars is the preferred metabolic route instead of aerobic respiration in the presence of oxygen. This metabolic capability is estimated to have evolved during the Cretaceous period, ca. 125 million years ago, when modern fruiting plants also appeared and humans were absent [72,73]. Bacteria present in these environments competed for resources, but the make–accumulate–consume provided a winning strategy for yeast. By producing inhibitory concentrations of alcohol for later consumption via a diauxic shift the yeast continued to occupy these environments over time [71,72,74].
The production of fruity esters may be an advantageous evolutionary trait developed to allow the passage of non-mobile unicellular fungi yeast from one rich environment to the next either via insect or brewer [75,76,77,78]. One theory suggests insect vectors’ intestines act as a vessel for facilitating natural yeast hybridization events [76,77]. Unfortunately, the microscopic nature of yeast impose a limit for scientists to infer retrospectively on yeast mating modes and frequency by analyzing genomic data sets [79,80].
Interspecific hybridizations facilitate exchange of DNA which intertwine lineages and blur traditional species boundaries [32]. In yeast, the newly developed hybrids can rapidly adapt by filtering their diverse chromosomes and retaining advantageous portions via the loss of heterozygosity [81,82,83]. Cells experiencing a stressor, such as a drug or environmental condition, appear to prioritize genomic filtering early in growth, demonstrated by Saccharomyces interspecific hybrids subjected to various temperatures. S. cerevisiae and S. uvarum hybrids retained the cryotolerant subgenome of S. uvarum during cold adaptation, but retained the S. cerevisiae subgenome during warm environmental growth [83,84]. The dynamics between interspecific hybridization, adaptation, and evolution in Saccharomyces is not fully understood, but recent evidence suggests that this exchange of DNA components has and will continue to play a pivotal role as new yeast hybrids are described.
5. Conclusions
The quest to gain diverse and novel fermentation characteristics from a pure culture remains an overarching goal for brewing molecular biologists. Flavor attributes and temperature phenotypes were investigated by many research groups; however, gaps in knowledge remain concerning yeast fermentation and the evolution field. Efforts large and small are being exerted worldwide to explore the awesome power of yeast genetics in brewing sciences, yet many questions continue to surround the budding fungus akin to the brewers and bakers of the globe. Are interspecific hybrids naturally abundant under the correct environmental conditions or are they rare success stories? To what extent are yeast present in various natural climates and substrates, what vectors are involved in mobility, what interactions occur with other microbes, what is the typical lifecycle timeline of wild yeast, do Saccharomyces yeast originate out of Asia, or will researchers ever obtain adequate and representative environmental samples? Yeast phenotypes that, to the authors knowledge, remain unexplored by targeted hybridization include formation of antioxidants that increase flavor stability, formation of glycerol for an enhanced mouthfeel in low alcohol beverages, and reduced off-flavor production.
Many recent interspecific hybrids developed have largely focused on reinventing the lager yeast, S. pastorianus, by crossing S. cerevisiae with S. eubayanus. As research continues, understudied Saccharomyces species may serve as a reservoir for diverse genomic contributions. Natural isolates of Saccharomyces hybrids suggest that interspecific mating is not as uncommon as previously thought and recent investigations continue to redefine species whilst uncovering genetic exchange events. The ability of yeast to participate in interspecific hybridization and avoid hinderance by large genetic distances between parent species is promising and illustrative of the introgression likely yet to be discovered in many preserved isolates. While there are many yeast strains favored for production of quality fermented foods and beverages, their status as species or hybrids may be adjusted over time owing to the continual refinement of phylogenetics, the advancement in genome sequencing technology, and increased accessibility to genomic sequencing capabilities. By increasing yeast sampling, focusing on metabolic specifics, and facilitating collaboration, the yeast of tomorrow will be driven by scientific innovation in the laboratory today.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
A special acknowledgement is deserved to every team member of Imperial Yeast for supporting fundamental yeast research in and outside of the facility. Additional acknowledgment is needed to recognize the time needed to pursue and refine this educational outreach from Jess Caudil, Owen Lingley, and Jason Stepper. Thank you to Scott Forbes for translation of old German manuscripts.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Nomenclature
| Terminology | Description |
| a/α | The two mating types of S. cerevisiae that enable cellular fusion when the complimentary pheromone is detected. The mating type and sexual state is largely determined by the MAT locus on chromosome III. |
| Allele | A variant of any particular gene found at the same genomic location. The mating type of yeast is determined by which allele, MATa or MATα, is present at the MAT locus. |
| Alloploids | A hybrid organism or cell composed of two or more sets of chromosomes obtained from two separate species. |
| Auxotrophy | The loss of a functional gene needed for growth. This is particularly relevant for amino acid synthesis and metabolism during yeast genetics experiments where controlling growth is needed. |
| Ascus | The sack like structure enclosing the four spores of Saccharomyces, this characteristic is important in classification of the Ascomycota (sac fungi) yeast. |
| Brassica oleracea | Important food crop plant species. It is wild cabbage in the uncultivated form, but cultivation has yielded varieties over time to include broccoli, brussels sprouts, kale, cauliflower, cabbage, and collards. |
| Crabtree Positive | Microorganism that preferentially metabolizes sugar into ethanol in the presence of oxygen instead of cellular respiration. |
| Diauxic Shift | The shift from fermentation to utilizing ethanol for cellular respiration and cell growth. |
| Diploid | An organism with a paired set of chromosomes creating a (2n) genome, such as a human with a set of genomes from each parent. |
| Fagaceae spp. | Family of Angiosperms (flowering plants) that include the beech and oak trees. |
| Frohberg | One of two primary lineages of modern lager yeast known to brewers originally used in Germany. |
| Gene | The basic unit of heredity which is composed of a sequence of DNA. It is characterized by a start sequence, genomic segment that often is translatable into a protein of function, and a stop sequence. |
| Genomics | The study of all of an organism’s complete sequences of genetic materials composed of DNA (deoxyribonucleic acid) including structure, function, evolution, mapping, editing, and environmental interactions. |
| Haploid | An organism with one set of chromosomes composing their genome (1n). |
| Hemizygous | A condition of diploid cells where only one copy of a gene or other genetic component is present. |
| Hybrid Vigor (syn. Heterosis) | The tendency for the hybrid progeny between two species to outperform their parents in traits such as strength, size, or yield. |
| Interspecific | Arising or existing between separate species. |
| Intraspecific | Arising or existing within a species or individuals from the same species. |
| Locus | A specific fixed position on an individual chromosome where particular genetic content is present. |
| Make Accumulate Consume | S. cerevisiae yeast survival strategy that invokes the ability to make ethanol from saccharides, the ability to survive accumulation of the toxin, and the ability to consume ethanol for energy post fermentation. |
| Maltotriose | Prominent trisaccharide typical of beer wort and is enzymatically derived from starch. It is composed of three glucose molecules linked together by α-1,4 glycosidic bonds. |
| Nomenclature | The terminology and language used to categorize and communicate in various disciplines. |
| Out of Asia/Silk Road Hypothesis | The idea that Saccharomyces yeast originate from an Asian geographical region because of the abundance natural biodiversity found in that region. |
| Phenotype | A characterized trait, best exemplified by measurable qualities such as color or yield. |
| Phylogeny | The lineage and evolution of relative organisms. |
| Progeny | the offspring or descendants of an organism. |
| Quiescence | A state of inactivity, dormancy, or a period of idleness in yeast ecology. |
| Saaz | one of two primary lineages of modern lager yeast known to brewers originally used in Bohemia region of the Czech Republic where the town Žatec/Saaz is located. |
| Shmoo | The distinct physical form of two Saccharomyces yeast cells mating. This terminology originated from the morphological similarity to an Al Capp cartoon popular in America during the early 1950s. |
| Synanthropic Species | An undomesticated organism that habitually exists with human populations and benefit from non-natural environments. Their life cycles are adapted fully or in part to conditions created by human activity. |
| Tetrad | Four spores produced via meisis of Ascomycota yeast, specifically focused on yeast S. cerevisiae in this article. |
| Transgressive Phenotype | Formation of extreme phenotypes that surpass the ability of parental lineages, often found in hybrids and can be positive or negative for fitness of the individual. |
References
- Gallone, B.; Steensels, J.; Baele, G.; Maere, S.; Verstrepen, K.J.; Prahl, T.; Soriaga, L.; Saels, V.; Herrera-Malaver, B.; Merlevede, A.; et al. Domestication and Divergence of Saccharomyces cerevisiae Beer Yeasts. Cell 2016, 166, 1397–1410.e16. [Google Scholar] [CrossRef] [PubMed]
- Preiss, R.; Tyrawa, C.; Krogerus, K.; Garshol, L.M.; Van Der Merwe, G. Traditional Norwegian Kveik are a genetically distinct group of domesticated Saccharomyces cerevisiae brewing yeasts. Front. Microbiol. 2018, 9, 2137. [Google Scholar] [CrossRef] [PubMed]
- Pasteur, L. Nouveaux faits concernant l’histoire de la fermentation alcoolique. Comptes Rendus Chim. 1858, 47, 1011–1013. [Google Scholar]
- Meyen, F.J.F. Jahresbericht über die Resultate der Arbeiten im Felde der physiologischen Botanik von dem Jahre 1837. In Berlin: Nicolai’sche Buchhandlung; Nicolai: Berlin, Germany, 1838; pp. 1–186. [Google Scholar]
- Martini, A.V.; Martini, A. Three newly delimited species of Saccharomyces sensu stricto. Antonie Van Leeuwenhoek 1987, 53, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.A.; Bai, F.Y. Saccharomyces arboricolus sp. nov., a yeast species from tree bark. Int. J. Syst. Evol. Microbiol. 2008, 58, 510–514. [Google Scholar] [CrossRef]
- Naumova, E.S.; Roberts, I.N.; James, S.A.; Naumov, G.I.; Louis, E.J. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Syst. Evol. Microbiol. 2015, 50, 1931–1942. [Google Scholar] [CrossRef]
- Naumov, G.I.; Naumova, E.S.; Hagler, A.N.; Mendonça-Hagler, L.C.; Louis, E.J. A new genetically isolated population of the Saccharomyces sensu stricto complex from Brazil. Antonie Van Leeuwenhoek 1995, 67, 351–355. [Google Scholar] [CrossRef]
- Naumov, G.I.; Lee, C.F.; Naumova, E.S. Molecular genetic diversity of the Saccharomyces yeasts in Taiwan: Saccharomyces arboricola, Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2013, 103, 217–228. [Google Scholar] [CrossRef]
- Naumov, G.I.; Naumova, E.S.; Louis, E.J. Two New Genetically Isolated Popoulations of the Saccharomyces Sensu Stricto Complex from Japan. J. Gen. Appl. Microbiol. 1995, 41, 499–505. [Google Scholar] [CrossRef]
- Libkind, D.; Hittinger, C.T.; Valério, E.; Gonçalves, C.; Dover, J.; Johnston, M.; Gonçalves, P.; Sampaio, J.P. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 14539–14544. [Google Scholar] [CrossRef]
- Boynton, P.J.; Greig, D. The ecology and evolution of non-domesticated Saccharomyces species Primrose. Yeast 2014, 31, 449–462. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M. Interspecies hybridisation and genome chimerisation in Saccharomyces: Combining of gene pools of species and its biotechnological perspectives. Front. Microbiol. 2018, 9, 3071. [Google Scholar] [CrossRef] [PubMed]
- Naseeb, S.; James, S.A.; Alsammar, H.; Michaels, C.J.; Gini, B.; Nueno-Palop, C.; Bond, C.J.; McGhie, H.; Roberts, I.N.; Delneri, D. Saccharomyces jurei sp. Nov., isolation and genetic identification of a novel yeast species from Quercus robur. Int. J. Syst. Evol. Microbiol. 2017, 67, 2046–2052. [Google Scholar] [CrossRef] [PubMed]
- Batschinskaya, A. Entwicklungsgeschichte und Kultur des neuen Hefepilzes Saccharomyces paradoxus. J. Microbiol. Epidemiol. Immunobiol. 1914, 1, 231–247. [Google Scholar]
- Beijerinck, M. Uber Regeneration der Sporenbildung bei Alkohol Hefen, wo diese Funktion im Verchwinden begriffen ist. Cent. Bakt 1898, 4, 657–731. [Google Scholar]
- Beijerinck, M. Sur la generation de la faculte de produire des spores chez des levures en voie de la perdre. Extr. Des Arch. Neelandaises Des Sci. Exactes Nat. Ser. II 1898, 2, 1–81. [Google Scholar]
- Kaneko, Y.; Banno, I. Re-examination of Saccharomyces bayanus strains by DNA-DNA hybridization and electrophoretic karyotyping. IFO Res. Commun. 1991, 15, 30–41. [Google Scholar]
- Yamada, Y.; Mikata, K.; Banno, I. Reidentification of 121 strains of the genus Saccharomyces. Bull JFCC 1993, 9, 95–119. [Google Scholar]
- Masneuf, I.; Hansen, J.; Groth, C.; Piskur, J.; Dubourdieu, D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 1998, 64, 3887–3892. [Google Scholar] [CrossRef]
- Nguyen, H.V.; Legras, J.L.; Neuvéglise, C.; Gaillardin, C. Deciphering the hybridisation history leading to the lager lineage based on the mosaic genomes of Saccharomyces bayanus strains NBRC1948 and CBS380 T. PLoS One 2011, 6, e25821. [Google Scholar] [CrossRef]
- Querol, A.; Bond, U. The complex and dynamic genomes of industrial yeasts: MINIREVIEW. FEMS Microbiol. Lett. 2009, 293, 1–10. [Google Scholar] [CrossRef]
- Chen, Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013, 14, 471–482. [Google Scholar] [CrossRef]
- Fu, D.; Xiao, M.; Hayward, A.; Jiang, G.; Zhu, L.; Zhou, Q.; Li, J.; Zhang, M. What is crop heterosis: New insights into an old topic. J. Appl. Genet. 2015, 56, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Springer, N.M. Progress toward understanding heterosis in crop plants. Annu. Rev. Plant Biol. 2013, 64, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Crow, J.F. 90 Years Ago: The Beginning of Hybrid Maize. Genetics 1998, 148, 923–928. [Google Scholar] [CrossRef]
- Gallone, B.; Steensels, J.; Mertens, S.; Dzialo, M.C.; Gordon, J.L.; Wauters, R.; Theßeling, F.A.; Bellinazzo, F.; Saels, V.; Herrera-Malaver, B.; et al. Interspecific hybridization facilitates niche adaptation in beer yeast. Nat. Ecol. Evol. 2019, 3, 1562–1575. [Google Scholar] [CrossRef] [PubMed]
- González, S.S.; Barrio, E.; Querol, A. Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Appl. Environ. Microbiol. 2008, 74, 2314–2320. [Google Scholar] [CrossRef]
- Borneman, A.R.; Desany, B.A.; Riches, D.; Affourtit, J.P.; Forgan, A.H.; Pretorius, I.S.; Egholm, M.; Chambers, P.J. The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS Yeast Res. 2012, 12, 88–96. [Google Scholar] [CrossRef]
- Le Jeune, C.; Lollier, M.; Demuyter, C.; Erny, C.; Legras, J.-L.; Aigle, M.; Masneuf Pomarède, I. Characterization of natural hybrids of Saccharomyces cerevisiae and Saccharomyces bayanus var. uvarum. FEMS Yeast Res. 2007, 7, 540–549. [Google Scholar] [CrossRef][Green Version]
- Heil, C.S.; Burton, J.N.; Liachko, I.; Friedrich, A.; Hanson, N.A.; Morrise, C.L.; Schacherer, J.; Shendurer, J.; Thomas, J.H.; Maitreya, J. Dunham Identification of a novel interspecific hybrid yeast from a metagenomic spontaneously inoculated beer sample using Hi-C. Yeast 2018, 35, 71–84. [Google Scholar] [CrossRef]
- Steensels, J.; Gallone, B.; Verstrepen, K.J. Interspecific hybridization as a driver of fungal evolution and adaptation. Nat. Rev. Microbiol. 2021, 19, 485–500. [Google Scholar] [CrossRef] [PubMed]
- Liti, G.; Peruffo, A.; James, S.A.; Roberts, I.N.; Louis, E.J. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast 2005, 22, 177–192. [Google Scholar] [CrossRef] [PubMed]
- Dunn, B.; Sherlock, G. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 2008, 18, 1610–1623. [Google Scholar] [CrossRef] [PubMed]
- Glendinning, T. Some Practical Aspects of the Fermentable Matter. J. Fed. Inst. Brew. 1898, 5, 20–34. [Google Scholar] [CrossRef]
- Walther, A.; Hesselbart, A.; Wendland, J. Genome Sequence of Saccharomyces carlsbergensis, the World’s First Pure Culture Lager Yeast. G3: Genes|Genomes|Genet. 2014, 4, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J.A. A history of research on yeasts 10: Foundations of yeast genetics. Yeast 2007, 24, 799–845. [Google Scholar] [CrossRef]
- Hammond, B.J.R.M.; Eckersley, K.W. Fermentation Properties of Brewing Yeast with Killer Character. J. Insta. Brew. 1984, 90, 167–177. [Google Scholar] [CrossRef]
- Johnston, J.R. Breeding yeasts for brewing. J. Inst. Brew. 1965, 71, 135–137. [Google Scholar] [CrossRef]
- Spencer, J.F.T.; Spencer, D.M. Hybridization of non-sporulating and weakly sporulating strains of brewer’s and distiller’s yeasts. J. Inst. Brew. 1977, 83, 287–289. [Google Scholar] [CrossRef]
- Peŕez-Través, L.; Lopes, C.A.; Querol, A.; Barrio, E. On the complexity of the Saccharomyces bayanus taxon: Hybridization and potential hybrid speciation. PLoS ONE 2014, 9, e93729. [Google Scholar] [CrossRef]
- Langdon, Q.K.; Peris, D.; Baker, E.C.P.; Opulente, D.A.; Nguyen, H.V.; Bond, U.; Gonçalves, P.; Sampaio, J.P.; Libkind, D.; Hittinger, C.T. Fermentation innovation through complex hybridization of wild and domesticated yeasts. Nat. Ecol. Evol. 2019, 3, 1576–1586. [Google Scholar] [CrossRef] [PubMed]
- Mertens, S.; Steensels, J.; Saels, V.; De Rouck, G.; Aerts, G.; Verstrepen, K.J. A large set of newly created interspecific Saccharomyces hybrids increases aromatic diversity in lager beers. Appl. Environ. Microbiol. 2015, 81, 8202–8214. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, F.; Krogerus, K.; Vidgren, V.; Sandell, M.; Gibson, B. Improved cider fermentation performance and quality with newly generated Saccharomyces cerevisiae × Saccharomyces eubayanus hybrids. J. Ind. Microbiol. Biotechnol. 2017, 44, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
- Krogerus, K.; Magalhães, F.; Vidgren, V.; Gibson, B. New lager yeast strains generated by interspecific hybridization. J. Ind. Microbiol. Biotechnol. 2015, 42, 769–778. [Google Scholar] [CrossRef]
- Krogerus, K.; Arvas, M.; De Chiara, M.; Magalhães, F.; Mattinen, L.; Oja, M.; Vidgren, V.; Yue, J.X.; Liti, G.; Gibson, B. Ploidy influences the functional attributes of de novo lager yeast hybrids. Appl. Microbiol. Biotechnol. 2016, 100, 7203–7222. [Google Scholar] [CrossRef]
- Hebly, M.; Brickwedde, A.; Bolat, I.; Driessen, M.R.M.; de Hulster, E.A.F.; van den Broek, M.; Pronk, J.T.; Geertman, J.M.; Daran, J.M.; Daran-Lapujade, P. S. cerevisiae × S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 2015, 15, 1–14. [Google Scholar] [CrossRef]
- Nikulin, J.; Krogerus, K.; Gibson, B. Alternative Saccharomyces interspecies hybrid combinations and their potential for low-temperature wort fermentation. Yeast 2018, 35, 113–127. [Google Scholar] [CrossRef]
- Bellon, J.R.; Eglinton, J.M.; Siebert, T.E.; Pollnitz, A.P.; Rose, L.; De Barros Lopes, M.; Chambers, P.J. Newly generated interspecific wine yeast hybrids introduce flavour and aroma diversity to wines. Appl. Microbiol. Biotechnol. 2011, 91, 603–612. [Google Scholar] [CrossRef]
- Bellon, J.R.; Schmid, F.; Capone, D.L.; Dunn, B.L.; Chambers, P.J. Introducing a New Breed of Wine Yeast: Interspecific Hybridisation between a Commercial Saccharomyces cerevisiae Wine Yeast and Saccharomyces mikatae. PLoS ONE 2013, 8, e62053. [Google Scholar] [CrossRef]
- Winans, M.J.; Yamamoto, Y.; Fujimaru, Y.; Kusaba, Y.; Gallagher, J.E.G.; Kitagaki, H. Saccharomyces arboricola and Its Hybrids’ Propensity for Sake Production: Interspecific hybrids reveal increased fermentation abilities and a mosaic metabolic profile. Fermentation 2020, 6, 14. [Google Scholar] [CrossRef]
- Bizaj, E.; Cordente, A.G.; Bellon, J.R.; Raspor, P.; Curtin, C.D.; Pretorius, I.S. A breeding strategy to harness flavor diversity of Saccharomyces interspecific hybrids and minimize hydrogen sulfide production. FEMS Yeast Res. 2012, 12, 456–465. [Google Scholar] [CrossRef] [PubMed]
- Gancedo, J.M. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2001, 25, 107–123. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.; Pierce, J. Absorption of Amino Acids from Wort by Yeast. J. Inst. Brew. 1964, 70, 307–315. [Google Scholar] [CrossRef]
- Esposito, R.E.; Klapholz, S. Meiosis and Ascospore Development. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance; Strathern, J.N., Jones, E.W., Broach, E.R., Eds.; Cold Springs Harbor Laboratory Press: Cold Springs Harbor, NY, USA, 1981; pp. 211–287. [Google Scholar]
- Liti, G. The fascinating and secret wild life of the budding yeast S. cerevisiae. Elife 2015, 4, e05835. [Google Scholar] [CrossRef]
- Krogerus, K.; Magalhães, F.; Vidgren, V.; Gibson, B. Novel brewing yeast hybrids: Creation and application. Appl. Microbiol. Biotechnol. 2017, 101, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Gunge, N.; Nakatomi, Y. Genetic mechanisms of rare matings of the yeast saccharomyces cerevisiae heterozygous for mating type. Genetics 1972, 70, 41–58. [Google Scholar] [CrossRef]
- Ferenczy, L.; Maráz, A. Transfer of mitochondria by protoplast fusion in Saccharomyces cerevisiae. Nature 1977, 268, 524–525. [Google Scholar] [CrossRef]
- Ogata, T.; Okumura, Y.; Tadenuma, M.; Tamura, G. Improving Transformation Method for Industrial Yeasts: Construction of ADH1-APT2 Gene and Using Electroporation. J. Gen. Appl. Microbiol. 1993, 294, 285–294. [Google Scholar] [CrossRef][Green Version]
- Alexander, W.G.; Peris, D.; Pfannenstiel, B.T.; Opulente, D.A.; Kuang, M.; Hittinger, C.T. Efficient engineering of marker-free synthetic allotetraploids of Saccharomyces. Fungal Genet. Biol. 2016, 89, 10–17. [Google Scholar] [CrossRef]
- Fukuda, N.; Kaishima, M.; Ishii, J.; Kondo, A.; Honda, S. Continuous crossbreeding of sake yeasts using growth selection systems for a-type and α-type cells. AMB Express 2016, 6, 45. [Google Scholar] [CrossRef]
- Krogerus, K.; Fletcher, E.; Rettberg, N.; Gibson, B.; Preiss, R. Efficient breeding of industrial brewing yeast strains using CRISPR/Cas9-aided mating-type switching. Appl. Microbiol. Biotechnol. 2021, 105, 8359–8376. [Google Scholar] [CrossRef]
- Strong, G.; England, K. Beer Style Guidelines—BJCP; Brewers Association: Boulder, CO, USA, 2021. [Google Scholar]
- Verstrepen, K.J.; Derdelinckx, G.; Dufour, J.P.; Winderickx, J.; Thevelein, J.M.; Pretorius, I.S.; Delvaux, F.R. Flavor-active esters: Adding fruitiness to beer. J. Biosci. Bioeng. 2003, 96, 110–118. [Google Scholar] [CrossRef]
- Mcmurrough, I.; Madigan, D.; Donnelly, D.; Hurley, J.; Doyle, A.; Hennigan, G.; McNulty, N. Control of Ferulic Acid and 4-Vinyl Guaiacol in Brewing. J. Inst. Brew. 1996, 102, 327–332. [Google Scholar] [CrossRef]
- Gibson, B.; Geertman, J.M.A.; Hittinger, C.T.; Krogerus, K.; Libkind, D.; Louis, E.J.; Magalhães, F.; Sampaio, J.P. New yeasts-new brews: Modern approaches to brewing yeast design and development. FEMS Yeast Res. 2017, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Krogerus, K.; Preiss, R.; Gibson, B. A unique saccharomyces cerevisiae saccharomyces uvarum hybrid isolated from norwegian farmhouse beer: Characterization and reconstruction. Front. Microbiol. 2018, 9, 2253. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, F.; Krogerus, K.; Castillo, S.; Ortiz-Julien, A.; Dequin, S.; Gibson, B. Exploring the potential of Saccharomyces eubayanus as a parent for new interspecies hybrid strains in winemaking. FEMS Yeast Res. 2017, 17, 1–10. [Google Scholar] [CrossRef]
- Giannakou, K.; Visinoni, F.; Zhang, P.; Nathoo, N.; Jones, P.; Cotterrell, M.; Vrhovsek, U.; Delneri, D. Biotechnological exploitation of Saccharomyces jurei and its hybrids in craft beer fermentation uncovers new aroma combinations. Food Microbiol. 2021, 100, 103838. [Google Scholar] [CrossRef]
- Gray, J.V.; Petsko, G.A.; Johnston, G.C.; Ringe, D.; Singer, R.A.; Werner-washburne, M. “Sleeping Beauty”: Quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004, 68, 187–206. [Google Scholar] [CrossRef]
- Dashko, S.; Zhou, N.; Compagno, C.; Piškur, J. Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res. 2014, 14, 826–832. [Google Scholar] [CrossRef]
- Fleming, T.H.; John Kress, W. A brief history of fruits and frugivores. Acta Oecologica 2011, 37, 521–530. [Google Scholar] [CrossRef]
- Hagman, A.; Sall, T.; Compagno, C.; Piskur, J. Yeast “Make-Accumulate-Consume ” Life Strategy Evolved as a Multi-Step Process That Predates the Whole Genome Duplication. PLoS ONE 2013, 8, e68734. [Google Scholar] [CrossRef]
- Christiaens, J.F.; Franco, L.M.; Cools, T.L.; de Meester, L.; Michiels, J.; Wenseleers, T.; Hassan, B.A.; Yaksi, E.; Verstrepen, K.J. The fungal aroma gene ATF1 promotes dispersal of yeast cells through insect vectors. Cell Rep. 2014, 9, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, I.; Dapporto, L.; Berń, L.; Polsinelli, M.; Turillazzi, S.; Cavalieri, D. Social wasps are a saccharomyces mating nest. Proc. Natl. Acad. Sci. USA 2016, 113, 2247–2251. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, I.; Dapporto, L.; Legras, J.-L.; Calabretta, A.; Di Paola, M.; De Filippo, C.; Viola, R.; Capretti, P.; Polsinelli, M.; Turillazzi, S.; et al. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. USA 2012, 109, 13398–13403. [Google Scholar] [CrossRef] [PubMed]
- Reuter, M.; Bell, G.; Greig, D. Increased outbreeding in yeast in response to dispersal by an insect vector. Curr. Biol. 2007, 17, R81–R83. [Google Scholar] [CrossRef]
- Tsai, I.J.; Bensasson, D.; Burt, A.; Koufopanou, V. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc. Natl. Acad. Sci. USA 2008, 105, 4957–4962. [Google Scholar] [CrossRef]
- Ruderfer, D.M.; Pratt, S.C.; Seidel, H.S.; Kruglyak, L. Population genomic analysis of outcrossing and recombination in yeast. Nat. Genet. 2006, 38, 1077–1081. [Google Scholar] [CrossRef]
- James, T.Y.; Michelotti, L.A.; Glasco, A.D.; Clemons, R.A.; Powers, R.A.; James, E.S.; Simmons, D.R.; Bai, F.; Ge, S. Adaptation by Loss of Heterozygosity in Saccharomyces cerevisae Clones Under Divergent Selection. Genetics 2019, 213, 665–683. [Google Scholar] [CrossRef]
- Lancaster, S.M.; Payen, C.; Heil, C.S.; Dunham, M.J. Fitness benefits of loss of heterozygosity in Saccharomyces hybrids. Genome Res. 2019, 29, 1685–1692. [Google Scholar] [CrossRef]
- Smukowski Heil, C.S.; DeSevo, C.G.; Pai, D.A.; Tucker, C.M.; Hoang, M.L.; Dunham, M.J. Loss of Heterozygosity Drives Adaptation in Hybrid Yeast. Mol. Biol. Evol. 2017, 34, 1596–1612. [Google Scholar] [CrossRef]
- Smukowski Heil, C.S.; Large, C.R.L.; Patterson, K.; Hickey, A.S.M.; Yeh, C.L.C.; Dunham, M.J. Temperature preference can bias parental genome retention during hybrid evolution. PLoS Genet. 2019, 15, e1008383. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).