Effects of Fermented Vegetable Consumption on Human Gut Microbiome Diversity—A Pilot Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analysis of Fermented Vegetables
2.2. 16 S Ribosomal RNA Gene Sequencing
2.3. Pilot Intervention Study
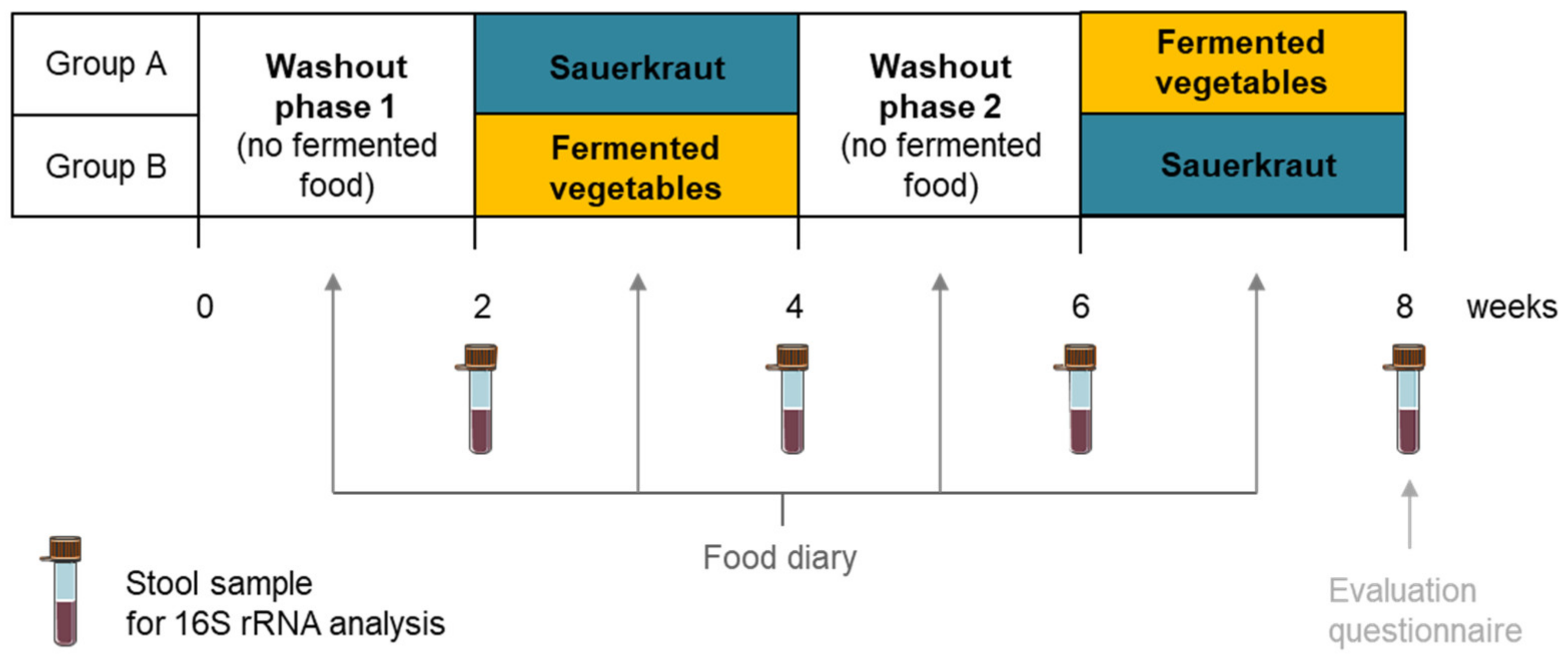
2.3.1. Study Design
2.3.2. Stool Sample Collection
2.3.3. Fecal Microbial DNA Isolation
2.3.4. 16 S Ribosomal RNA Gene Sequencing
2.4. Bioinformatics and Data Analysis
3. Results
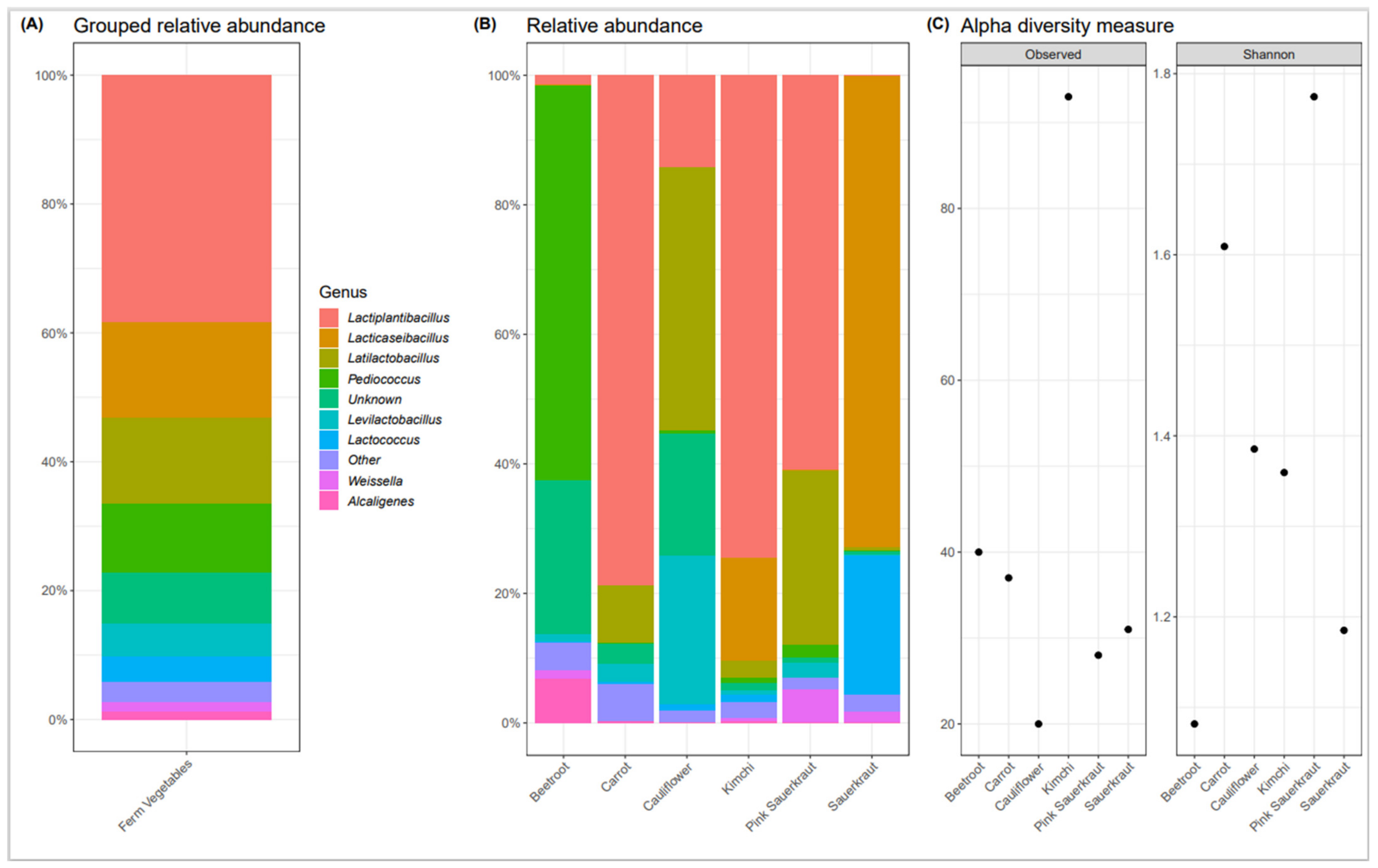
3.1. Bacterial Composition of Commercially Available Fermented Vegetables
Fermented Vegetable Microbiome
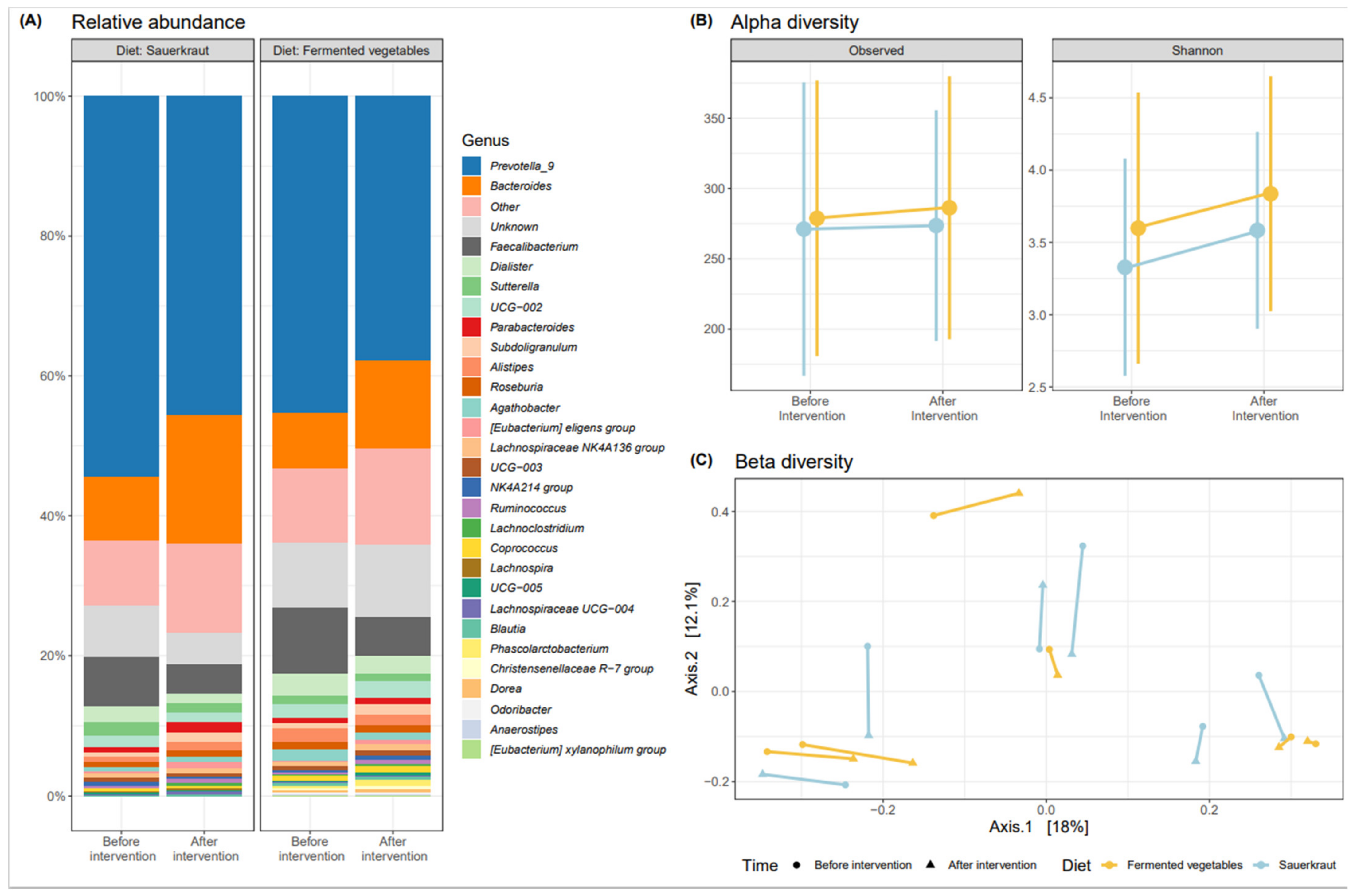
3.2. Effects of Fermented Vegetable Consumption on the Human Gut Microbiome
Gut Microbiome Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Type of Fermented Vegetable | Ingredients | Manufacturer |
|---|---|---|
| Cauliflower | Cauliflower (96%), natural sea salt, fresh turmeric, spices (turmeric, cumin) | completeorganics GmbH, Munich, Germany |
| Pink Sauerkraut | White cabbage (72%), red cabbage, natural sea salt, cranberries (dried) | completeorganics GmbH, Munich, Germany |
| Carrots | Carrots (96%), natural sea salt, ginger (fresh) | completeorganics GmbH, Munich, Germany |
| Beet Root | Beetroot (93%), horseradish, natural sea salt, goji berries (dried) | completeorganics GmbH, Munich, Germany |
| Daikon Kimchi | Radish (84%), carrots, red peppers, natural sea salt, spices (ginger, chili) | completeorganics GmbH, Munich, Germany |
| Traditional Sauerkraut | White cabbage, sea salt, lactic acid bacteria | Eden Frischkost-(L+)-Sauerkraut, Radolfszell, Germany |
References
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligne, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Dimidi, E.; Cox, S.R.; Rossi, M.; Whelan, K. Fermented Foods: Definitions and Characteristics, Impact on the Gut Microbiota and Effects on Gastrointestinal Health and Disease. Nutrients 2019, 11, 1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, E.S.; Garnås, E.; Jensen, K.J.; Hansen, L.H.; Olsen, P.S.; Ritz, C.; Krych, L.; Nielsen, D.S. Lacto-fermented sauerkraut improves symptoms in IBS patients independent of product pasteurisation—A pilot study. Food Funct. 2018, 9, 5323–5335. [Google Scholar] [CrossRef] [PubMed]
- Septembre-Malaterre, A.; Remize, F.; Poucheret, P. Fruits and vegetables, as a source of nutritional compounds and phytochem-icals: Changes in bioactive compounds during lactic fermentation. Food Res. Int. 2018, 104, 86–99. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Kim, J.M.; Park, M.S.; Bae, J.-W.; Hahn, Y.; Madsen, E.L.; Jeon, C.O. Metagenomic Analysis of Kimchi, a Traditional Korean Fermented Food. Appl. Environ. Microbiol. 2011, 77, 2264–2274. [Google Scholar] [CrossRef] [Green Version]
- Zabat, M.A.; Sano, W.H.; Wurster, J.I.; Cabral, D.J.; Belenky, P. Microbial Community Analysis of Sauerkraut Fermentation Reveals a Stable and Rapidly Established Community. Foods 2018, 7, 77. [Google Scholar] [CrossRef] [Green Version]
- Rezac, S.; Kok, C.R.; Heermann, M.; Hutkins, R. Fermented Foods as a Dietary Source of Live Organisms. Front. Microbiol. 2018, 9, 1785. [Google Scholar] [CrossRef]
- Zmora, N.; Suez, J.; Elinav, E. You are what you eat: Diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 35–56. [Google Scholar] [CrossRef]
- Müller, M.; Canfora, E.E.; Blaak, E.E. Gastrointestinal Transit Time, Glucose Homeostasis and Metabolic Health: Modulation by Dietary Fibers. Nutrients 2018, 10, 275. [Google Scholar] [CrossRef] [Green Version]
- Roager, H.M.; Hansen, L.B.S.; Bahl, M.I.; Frandsen, H.L.; Carvalho, V.; Gøbel, R.J.; Dalgaard, M.D.; Plichta, D.R.; Sparholt, M.H.; Vestergaard, H.; et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat. Microbiol. 2016, 1, 16093. [Google Scholar] [CrossRef]
- Koopman, M.; El Aidy, S. Depressed gut? The microbiota-diet-inflammation trialogue in depression. Curr. Opin. Psychiatry 2017, 30, 369–377. [Google Scholar] [CrossRef] [Green Version]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vázquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The Treatment-Naive Microbiome in New-Onset Crohn’s Disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Arrieta, M.-C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef]
- Greenblum, S.; Turnbaugh, P.J.; Borenstein, E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc. Natl. Acad. Sci. USA 2011, 109, 594–599. [Google Scholar] [CrossRef] [Green Version]
- Reitmeier, S.; Kiessling, S.; Neuhaus, K.; Haller, D. Comparing Circadian Rhythmicity in the Human Gut Microbiome. STAR Protoc. 2020, 1, 100148. [Google Scholar] [CrossRef]
- Haring, E.; Andrieux, G.; Uhl, F.M.; Krausz, M.; Proietti, M.; Sauer, B.; Esser, P.R.; Martin, S.F.; Pfeifer, D.; Schmitt-Graeff, A.; et al. Therapeutic targeting of endoplasmic reticulum stress in acute graft-versus-host disease. Haematologica 2021, 4, 2782–2858. [Google Scholar] [CrossRef]
- Norona, J.; Apostolova, P.; Schmidt, D.; Ihlemann, R.; Reischmann, N.; Taylor, G.; Köhler, N.; De Heer, J.; Heeg, S.; Andrieux, G.; et al. Glucagon-like peptide 2 for intestinal stem cell and Paneth cell repair during graft-versus-host disease in mice and humans. Blood 2020, 136, 1442–1455. [Google Scholar] [CrossRef]
- Haring, E.; Uhl, F.M.; Andrieux, G.; Proietti, M.; Bulashevska, A.; Sauer, B.; Braun, L.M.; Gomez, E.D.V.; Esser, P.R.; Martin, S.F.; et al. Bile acids regulate intestinal antigen presentation and reduce graft-versus-host disease without impairing the graft-versus-leukemia effect. Haematologica 2020, 106, 2131–2146. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Gustaw, K.; Niedźwiedź, I.; Rachwał, K.; Polak-Berecka, M. New Insight into Bacterial Interaction with the Matrix of Plant-Based Fermented Foods. Foods 2021, 10, 1603. [Google Scholar] [CrossRef]
- Porto, M.C.W.; Kuniyoshi, T.M.; Azevedo, P.O.S.; Vitolo, M.; Oliveira, R.P.S. Pediococcus spp.: An important genus of lactic acid bacteria and pediocin producers. Biotechnol. Adv. 2017, 35, 361–374. [Google Scholar] [CrossRef]
- Kavitake, D.; Devi, P.B.; Shetty, P.H. Overview of exopolysaccharides produced by Weissella genus—A review. Int. J. Biol. Macromol. 2020, 164, 2964–2973. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Volokh, O.; Klimenko, N.; Berezhnaya, Y.; Tyakht, A.; Nesterova, P.; Popenko, A.; Alexeev, D. Human Gut Microbiome Response Induced by Fermented Dairy Product Intake in Healthy Volunteers. Nutrients 2019, 11, 547. [Google Scholar] [CrossRef] [Green Version]
- Stiemsma, L.T.; Nakamura, R.E.; Nguyen, J.G.; Michels, K.B. Does Consumption of Fermented Foods Modify the Human Gut Microbiota? J. Nutr. 2020, 150, 1680–1692. [Google Scholar] [CrossRef]
- Kwak, H.; Park, S.; Kim, D. Biostabilization of Kefir with a Nonlactose-Fermenting Yeast. J. Dairy Sci. 1996, 79, 937–942. [Google Scholar] [CrossRef]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.; Cotter, P.D. Sequencing-Based Analysis of the Bacterial and Fungal Composition of Kefir Grains and Milks from Multiple Sources. PLoS ONE 2013, 8, e69371. [Google Scholar] [CrossRef] [Green Version]
- Bourrie, B.C.T.; Willing, B.P.; Cotter, P.D. The Microbiota and Health Promoting Characteristics of the Fermented Beverage Kefir. Front. Microbiol. 2016, 7, 647. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E. Prevotella in the gut: Choose carefully. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 69–70. [Google Scholar] [CrossRef] [PubMed]
- Conlon, M.A.; Bird, A.R. The Impact of Diet and Lifestyle on Gut Microbiota and Human Health. Nutrients 2014, 7, 17–44. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Bose, S.; Wang, J.-H.; Kim, B.-S.; Kim, M.J.; Kim, E.-J.; Kim, H. Contrasting effects of fresh and fermented kimchi consumption on gut microbiota composition and gene expression related to metabolic syndrome in obese Korean women. Mol. Nutr. Food Res. 2015, 59, 1004–1008. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thriene, K.; Hansen, S.S.; Binder, N.; Michels, K.B. Effects of Fermented Vegetable Consumption on Human Gut Microbiome Diversity—A Pilot Study. Fermentation 2022, 8, 118. https://doi.org/10.3390/fermentation8030118
Thriene K, Hansen SS, Binder N, Michels KB. Effects of Fermented Vegetable Consumption on Human Gut Microbiome Diversity—A Pilot Study. Fermentation. 2022; 8(3):118. https://doi.org/10.3390/fermentation8030118
Chicago/Turabian StyleThriene, Kerstin, Sina S. Hansen, Nadine Binder, and Karin B. Michels. 2022. "Effects of Fermented Vegetable Consumption on Human Gut Microbiome Diversity—A Pilot Study" Fermentation 8, no. 3: 118. https://doi.org/10.3390/fermentation8030118
APA StyleThriene, K., Hansen, S. S., Binder, N., & Michels, K. B. (2022). Effects of Fermented Vegetable Consumption on Human Gut Microbiome Diversity—A Pilot Study. Fermentation, 8(3), 118. https://doi.org/10.3390/fermentation8030118






