Bacterial Composition and Interactions in Raw Milk and Teat Skin of Dairy Cows
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and High-Throughput Sequencing of 16S rRNA Amplicon
2.3. Sequencing Data Processing
2.4. Core Microbiota and Network Analysis
2.5. Bacterial Transfer Analysis
2.6. Statistical Methods
3. Results
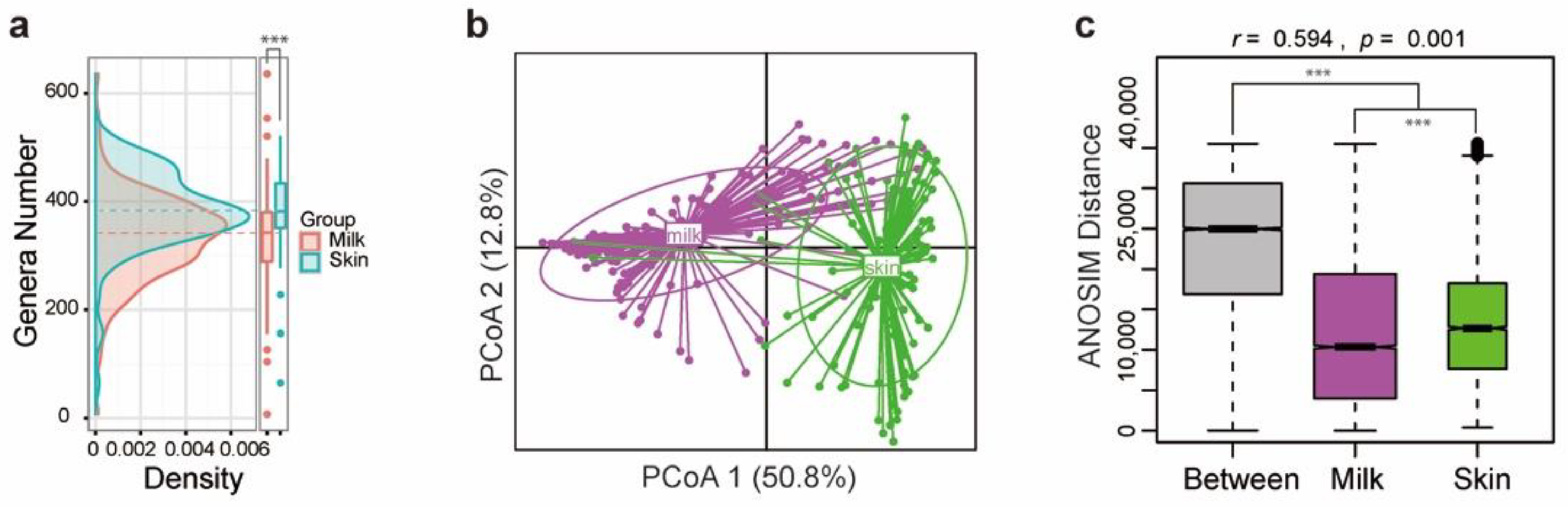
3.1. The Bacterial Communities in Milk and on Teat Skin
3.2. The Core Microbiota in Raw Milk and on Teat Skin
3.3. Co-Occurrence Network of the Bacterial Community in Milk and on Teat Skin
3.4. Bacterial Transfer between the Raw Milk and Teat Skin
3.5. Bacterial Taxa Shift on Teat Skin during Milking
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Suntinger, M.; Fuerst-Waltl, B.; Obritzhauser, W.; Firth, C.L.; Köck, A.; Egger-Danner, C. Usability of bacteriological milk analyses for genetic improvement of udder health in Austrian Fleckvieh cows. J. Dairy Sci. 2022, 105, S0022–S0302. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.H.; Cho, Y.S.; Rackerby, B.; Goddik, L.; Park, S.H. Shifts of microbiota during cheese production: Impact on production and quality. Appl. Microbiol. Biotechnol. 2021, 105, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Kable, M.E.; Srisengfa, Y.; Laird, M.; Zaragoza, J.; McLeod, J.; Heidenreich, J.; Marco, M.L. The Core and Seasonal Microbiota of Raw Bovine Milk in Tanker Trucks and the Impact of Transfer to a Milk Processing Facility. Mbio 2016, 7, e00836-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quigley, L.; O’Sullivan, O.; Stanton, C.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. The complex microbiota of raw milk. FEMS Microbiol. Rev. 2013, 37, 664–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornik, B.; Czarny, J.; Staninska-Pięta, J.; Wolko, Ł.; Cyplik, P.; Piotrowska-Cyplik, A. The Raw Milk Microbiota from Semi-Subsistence Farms Characteristics by NGS Analysis Method. Molecules 2021, 26, 5029. [Google Scholar] [CrossRef]
- Fernandez, L.; Langa, S.; Martin, V.; Maldonado, A.; Jimenez, E.; Martin, R.; Rodriguez, J.M. The human milk microbiota: Origin and potential roles in health and disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef]
- Verdiermetz, I.; Gagne, G.; Bornes, S.; Monsallier, F.; Veisseire, P.; Delbèspaus, C.; Montel, M.C. Cow teat skin, a potential source of diverse microbial populations for cheese production. Appl. Environ. Microbiol. 2012, 78, 326. [Google Scholar] [CrossRef] [Green Version]
- Doyle, C.J.; Gleeson, D.; O’Toole, P.W.; Cotter, P.D. Impacts of Seasonal Housing and Teat Preparation on Raw Milk Microbiota: A High-Throughput Sequencing Study. Appl. Environ. Microbiol. 2017, 83, e02694-16. [Google Scholar] [CrossRef] [Green Version]
- Fretin, M.; Martin, B.; Rifa, E.; Isabelle, V.M.; Pomies, D.; Ferlay, A.; Montel, M.C.; Delbes, C. Bacterial community assembly from cow teat skin to ripened cheeses is influenced by grazing systems. Sci. Rep. 2018, 8, 200. [Google Scholar] [CrossRef] [Green Version]
- Gleeson, D.; O’Brien, B.; Flynn, J.; O’Callaghan, E.; Galli, F. Effect of pre-milking teat preparation procedures on the microbial count on teats prior to cluster application. Ir. Vet. J. 2009, 62, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Martins, C.; Pinheiro, E.S.C.; Gentilini, M.; Benavides, M.L.; Santos, M.V. Efficacy of a high free iodine barrier teat disinfectant for the prevention of naturally occurring new intramammary infections and clinical mastitis in dairy cows. J. Dairy Sci. 2017, 100, 3930–3939. [Google Scholar] [CrossRef] [PubMed]
- Miciński, J.; Miciński, J.; Miseikiene, R.; Tusas, S.; Biziene, R.; Kerziene, S.; Matusevičius, P. Influence of teat disinfection with iodine preparation on bacterial contamination of teats, hygienic quality and content of iodine in milk. J. Elementol. 2019, 25, 225–236. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; De Santis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Blanton, L.V.; Charbonneau, M.R.; Salih, T.; Barratt, M.J.; Venkatesh, S.; Ilkaveya, O.; Subramanian, S.; Manary, M.J.; Trehan, I.; Jorgensen, J.M.; et al. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 2016, 351, 3311. [Google Scholar] [CrossRef] [Green Version]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef] [Green Version]
- Quigley, L.; Mccarthy, R.; O’Sullivan, O.; Beresford, T.P.; Fitzgerald, G.F.; Ross, R.P.; Stanton, C.; Cotter, P.D. The microbial content of raw and pasteurized cow milk as determined by molecular approaches. J. Dairy Sci. 2013, 96, 4928. [Google Scholar] [CrossRef]
- Aldretetapia, A.; Escobarramirez, M.C.; Tamplin, M.L.; Hernandeziturriaga, M. High-throughput sequencing of microbial communities in Poro cheese, an artisanal Mexican cheese. Food Microbiol. 2014, 44, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Falentin, H.; Rault, L.; Nicolas, A.; Bouchard, D.; Lassalas, J.; Lamberton, P.; Aubry, J.; Marnet, P.; Loir, Y.L.; Even, S. Bovine Teat Microbiome Analysis Revealed Reduced Alpha Diversity and Significant Changes in Taxonomic Profiles in Quarters with a History of Mastitis. Front. Microbiol. 2016, 7, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davis, M.Y.; Zhang, H.; Brannan, L.E.; Carman, R.J.; Boone, J.H. Rapid change of fecal microbiome and disappearance of Clostridium difficile in a colonized infant after transition from breast milk to cow milk. Mbio 2016, 4, 53. [Google Scholar] [CrossRef] [Green Version]
- van de Pol, J.A.A.; van Best, N.; Mbakwa, C.A.; Thijs, C.; Savelkoul, P.H.; Arts, I.C.W.; Hornef, M.W.; Mommers, M.; Penders, J. Gut Colonization by Methanogenic Archaea Is Associated with Organic Dairy Consumption in Children. Front. Microbiol. 2017, 8, 355. [Google Scholar] [CrossRef] [PubMed]
- Masoud, W.; Vogensen, F.K.; Lillevang, S.; Al-Soud, W.A.; Sørensen, S.J.; Jakobsen, M. The fate of indigenous microbiota, starter cultures, Escherichia coli, Listeria innocua and Staphylococcus aureus in Danish raw milk and cheeses determined by pyrosequencing and quantitative real time (qRT)-PCR. Int. J. Food Microbiol. 2012, 153, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Hantsiszacharov, E.; Halpern, M. Culturable Psychrotrophic Bacterial Communities in Raw Milk and Their Proteolytic and Lipolytic Traits. Appl. Environ. Microbiol. 2007, 73, 7162–7168. [Google Scholar] [CrossRef] [Green Version]
- Raats, D.; Offek, M.; Minz, D.; Halpern, M. Molecular analysis of bacterial communities in raw cow milk and the impact of refrigeration on its structure and dynamics. Food Microbiol. 2011, 28, 465–471. [Google Scholar] [CrossRef]
- Oikonomou, G.; Bicalho, M.L.S.; Meira, E.B.S.; Rossi, R.; Foditsch, C.; Machado, V.S.; Teixeira, A.G.V.; Santisteban, C.; Schukken, Y.H.; Bicalho, R.C. Microbiota of Cow’s Milk; Distinguishing Healthy, Sub-Clinically and Clinically Diseased Quarters. PLoS ONE 2014, 9, e85904. [Google Scholar] [CrossRef] [Green Version]
- Vacheyrou, M.; Normand, A.; Guyot, P.; Cassagne, C.; Piarroux, R.; Bouton, Y. Cultivable microbial communities in raw cow milk and potential transfers from stables of sixteen French farms. Int. J. Food Microbiol. 2011, 146, 253–262. [Google Scholar] [CrossRef]
- Braem, G.; Vliegher, S.D.; Verbist, B. Culture-independent exploration of the teat apex microbiota of dairy cows reveals a wide bacterial species diversity. Vet. Microbiol. 2012, 157, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Addis, M.F.; Tanca, A.; Uzzau, S.; Oikonomou, G.; Bicalho, R.C.; Moroni, P. The bovine milk microbiota: Insights and perspectives from -omics studies. Mol. Biosyst. 2016, 12, 2359–2372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Von Neubeck, M.; Baur, C.; Krewinkel, M.; Stoeckel, M.; Kranz, B.; Stressler, T.; Fischer, L.; Hinrichs, J.; Scherer, S.; Wenning, M. Biodiversity of refrigerated raw milk microbiota and their enzymatic spoilage potential. Int. J. Food Microbiol. 2015, 211, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, T.D.; Krause, L.; Bridge, T.C.L.; Torda, G.; Raina, J.; Zakrzewski, M.; Gates, R.D.; Padillagamino, J.L.; Spalding, H.L.; Smith, C.M. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015, 9, 2261–2274. [Google Scholar] [CrossRef]
- Hooper, S.D.; Mavromatis, K.; Kyrpides, N.C. Microbial co-habitation and lateral gene transfer: What transposases can tell us. Genome Biol. 2009, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.; Tombor, B.; Albert, R.; Oltvai, Z.N.; Barabasi, A. The large-scale organization of metabolic networks. Nature 2000, 407, 651–654. [Google Scholar] [CrossRef] [Green Version]
- Han, J.J.; Dupuy, D.; Bertin, N.; Cusick, M.E.; Vidal, M. Effect of sampling on topology predictions of protein-protein interaction networks. Nat. Biotechnol. 2005, 23, 839–844. [Google Scholar] [CrossRef]
- Cohen, R.; Erez, K.; Benavraham, D.; Havlin, S. Resilience of the internet to random breakdowns. Phys. Rev. Lett. 2000, 85, 4626. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Pan, F.; Soininen, J.; Heino, J.; Shen, J. Nutrient enrichment modifies temperature-biodiversity relationships in large-scale field experiments. Nat. Commun. 2016, 7, 13960. [Google Scholar] [CrossRef] [Green Version]
- Phillips, M.L. Gut Reaction: Environmental Effects on the Human Microbiota. Environ. Health Perspect. 2009, 117, A198–A205. [Google Scholar] [CrossRef]
- Wattiaux, M.A. Technical Dairy Guide; Babcock Institute: Wilmington, MA, USA, 2005; pp. 77–80. [Google Scholar]
- Mallon, C.A.; Van Elsas, J.D.; Salles, J.F. Microbial Invasions: The Process, Patterns, and Mechanisms. Trends Microbiol. 2015, 23, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Bashan, A.; Gibson, T.E.; Friedman, J.R.; Carey, V.J.; Weiss, S.T.; Hohmann, E.L.; Liu, Y. Universality of human microbial dynamics. Nature 2016, 534, 259–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freilich, S.; Zarecki, R.; Eilam, O.; Segal, E.S.; Henry, C.S.; Kupiec, M.; Gophna, U.; Sharan, R.; Ruppin, E. Competitive and cooperative metabolic interactions in bacterial communities. Nat. Commun. 2011, 2, 589. [Google Scholar] [CrossRef] [Green Version]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Mougi, A.; Kondoh, M. Diversity of Interaction Types and Ecological Community Stability. Science 2012, 337, 349–351. [Google Scholar] [CrossRef] [PubMed]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Rochat, F.; Chassard, C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 2014, 16, 2891–2904. [Google Scholar] [CrossRef] [PubMed]
- Galton, D.M.; Petersson, L.G.; Merrill, W.G. Effects of Premilking Udder Preparation Practices on Bacterial Counts in Milk and on Teats. J. Dairy Sci. 1986, 69, 260–266. [Google Scholar] [CrossRef]
- Baumberger, C.; Guarín, J.F.; Ruegg, P.L. Effect of 2 different premilking teat sanitation routines on reduction of bacterial counts on teat skin of cows on commercial dairy farms. J. Dairy Sci. 2016, 99, 2915. [Google Scholar] [CrossRef] [Green Version]
- Wattenburger, K.; Schmidt, R.; Placheta, L.; Middleton, J.R.; Adkins, P.R.F. Evaluation of 4 different teat disinfection methods prior to collection of milk samples for bacterial culture in dairy cattle. J. Dairy Sci. 2020, 103, 4579–4587. [Google Scholar] [CrossRef]
- Baur, C.; Krewinkel, M.; Kranz, B.; Von Neubeck, M.; Wenning, M.; Scherer, S.; Stoeckel, M.; Hinrichs, J.; Stressler, T.; Fischer, L. Quantification of the proteolytic and lipolytic activity of microorganisms isolated from raw milk. Int. Dairy J. 2015, 49, 23–29. [Google Scholar] [CrossRef]
- Machado, S.G.; Baglinière, F.; Marchand, S.; Van, C.E.; Vanetti, M.C.; De, B.J.; Heyndrickx, M. The Biodiversity of the Microbiota Producing Heat-Resistant Enzymes Responsible for Spoilage in Processed Bovine Milk and Dairy Products. Front. Microbiol. 2017, 8, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baur, C.; Krewinkel, M.; Kutzli, I.; Kranz, B.; Von Neubeck, M.; Huptas, C.; Wenning, M.; Scherer, S.; Stoeckel, M.; Hinrichs, J. Isolation and characterisation of a heat-resistant peptidase from Pseudomonas panacis withstanding general UHT processes. Int. Dairy J. 2015, 49, 46–55. [Google Scholar] [CrossRef]
- Maier, C.; Hofmann, K.; Huptas, C.; Scherer, S.; Wenning, M.; Lücking, G. Simultaneous quantification of the most common and proteolytic Pseudomonas species in raw milk by multiplex qPCR. Appl. Microbiol. Biotechnol. 2021, 105, 1693–1708. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, H.; Du, W.; Ji, S.; Guo, C.; Zhang, Y.; Wang, Y.; Cao, Z.; Li, S. Bacterial Composition and Interactions in Raw Milk and Teat Skin of Dairy Cows. Fermentation 2022, 8, 235. https://doi.org/10.3390/fermentation8050235
Yan H, Du W, Ji S, Guo C, Zhang Y, Wang Y, Cao Z, Li S. Bacterial Composition and Interactions in Raw Milk and Teat Skin of Dairy Cows. Fermentation. 2022; 8(5):235. https://doi.org/10.3390/fermentation8050235
Chicago/Turabian StyleYan, Hui, Wen Du, Shoukun Ji, Chunyan Guo, Yujing Zhang, Yajing Wang, Zhijun Cao, and Shengli Li. 2022. "Bacterial Composition and Interactions in Raw Milk and Teat Skin of Dairy Cows" Fermentation 8, no. 5: 235. https://doi.org/10.3390/fermentation8050235
APA StyleYan, H., Du, W., Ji, S., Guo, C., Zhang, Y., Wang, Y., Cao, Z., & Li, S. (2022). Bacterial Composition and Interactions in Raw Milk and Teat Skin of Dairy Cows. Fermentation, 8(5), 235. https://doi.org/10.3390/fermentation8050235





