Recycling Food Waste and Saving Water: Optimization of the Fermentation Processes from Cheese Whey Permeate to Yeast Oil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Fed-Batch Cultivation
2.3. Sugars and Nitrogen Determination
2.4. Dry Weight Determination
2.5. β-Galactosidase Activity
2.6. Lipid Quantification
2.7. Lipid Profile Analysis
2.8. Confocal Laser Scanning Microscopy Evaluation
2.9. Flow Cytometry for Lipid Quantification
3. Results
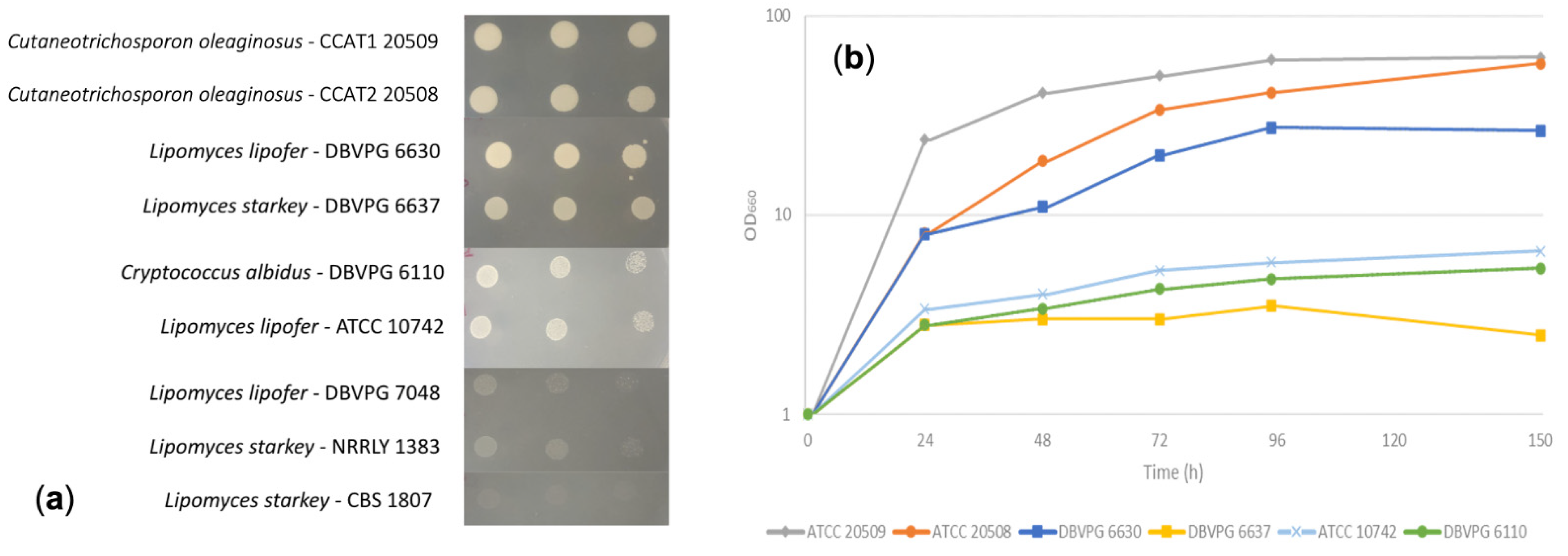
3.1. Screening of Oleaginous Species for Biomass and Lipid Biosynthesis on WP
3.2. Confocal Laser Scanning Microscopy of C. oleaginosus ATCC 20509
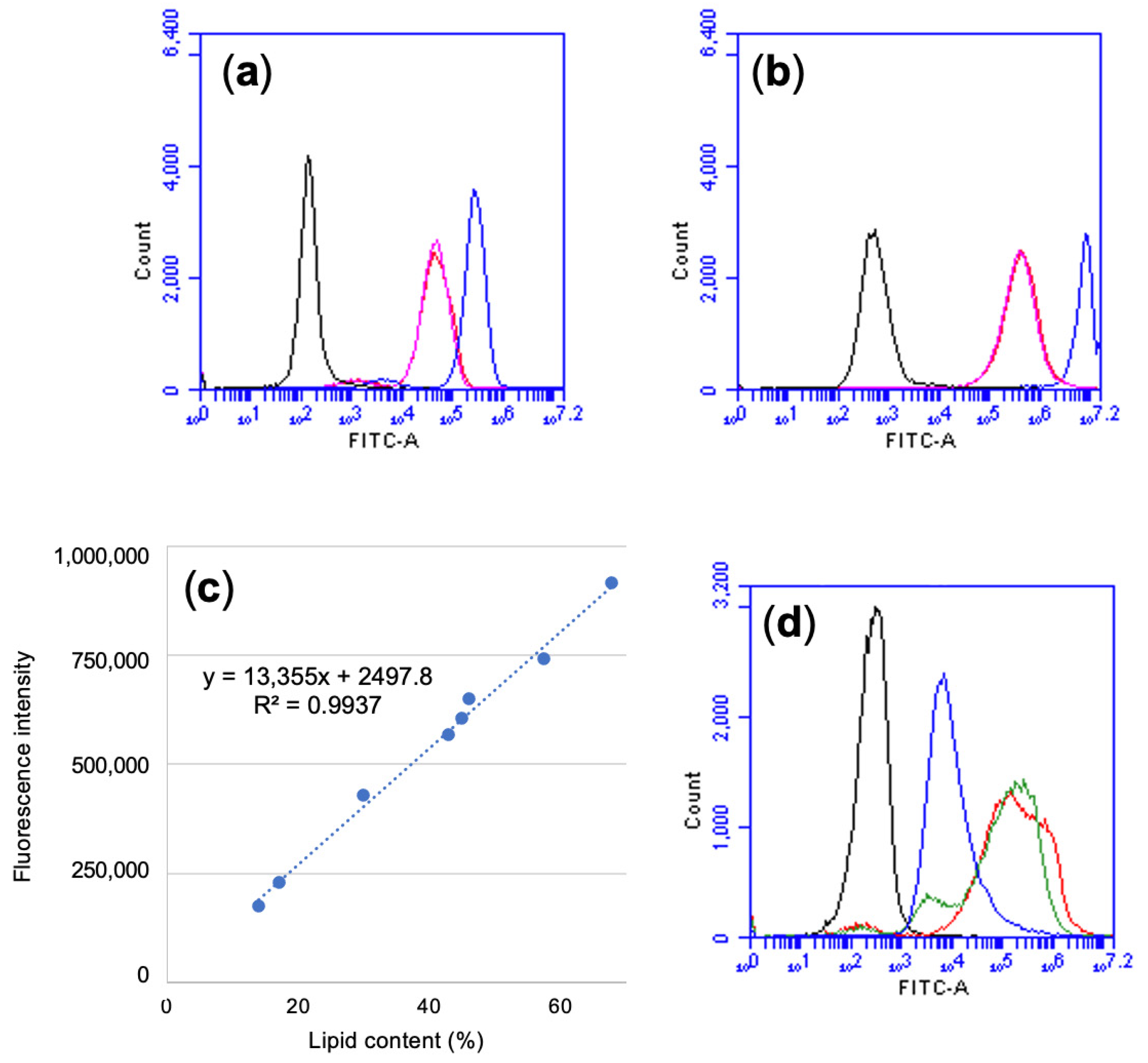
3.3. Set-up of a Lipid Quantification Protocol by Flow Cytometry
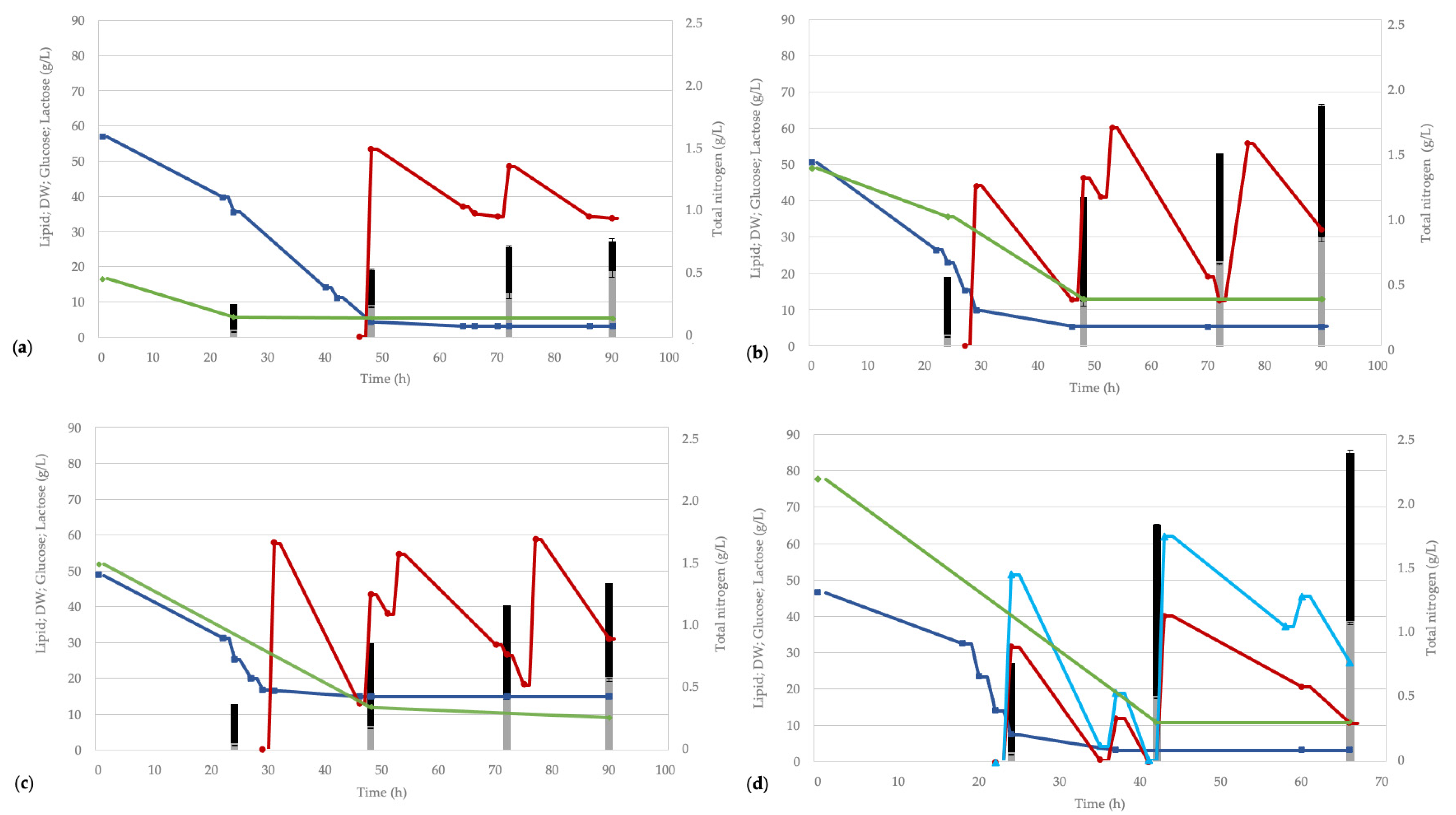
3.4. Processes in a Bioreactor on Whey Permeate-Based Media
3.5. Monitoring the Process with Flow Cytometry
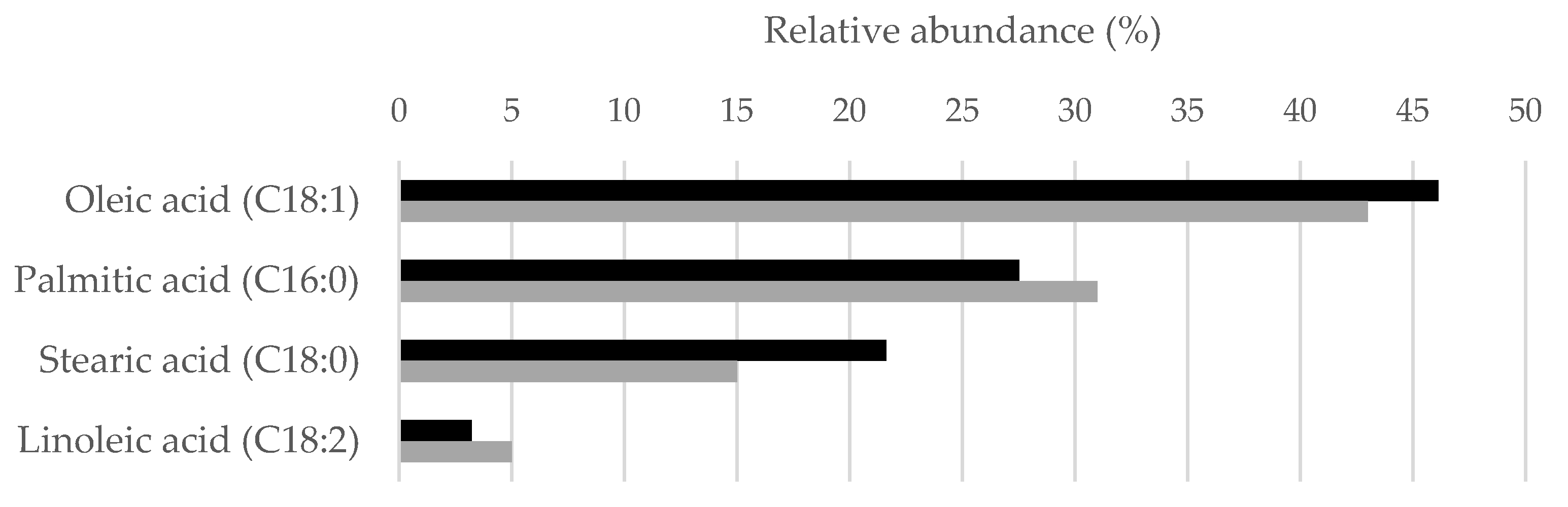
3.6. Lipid Composition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- European Commission: DG Environment. A New Circular Economy Action Plan for a Cleaner and More Competitive Europe; European Commission: DG Environment: Brussels, Belgium, 2020; Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?qid=1583933814386&uri=COM:2020:98:FIN (accessed on 27 June 2022).
- Girotto, F.; Alibardi, L.; Cossu, R. Food waste generation and industrial uses: A review. Waste Manag. 2015, 45, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Homrich, A.; Galvão, G.; Gamboa Abadia, L.; Carvalho, M.M. The circular economy umbrella: Trends and gaps on integrating pathways. J. Clean. Prod. 2018, 175, 525–543. [Google Scholar] [CrossRef]
- Fernández-Gutiérrez, D.; Veillette, M.; Giroir-Fendler, A.; Ramirez, A.A.; Faucheux, N.; Heitz, M. Biovalorization of saccharides derived from industrial wastes such as whey: A review. Rev. Environ. Sci. Bio/Technol. 2017, 16, 147–174. [Google Scholar] [CrossRef]
- Koushki, M.; Jafari, M.; Azizi, M. Comparison of ethanol production from cheese whey permeate by two yeast strains. J. Food Sci. Technol. 2012, 49, 614–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lappa, I.K.; Papadaki, A.; Kachrimanidou, V.; Terpou, A.; Koulougliotis, D.; Eriotou, E.; Kopsahelis, N. Cheese Whey Processing: Integrated Biorefinery Concepts and Emerging Food Applications. Foods 2019, 8, 347. [Google Scholar] [CrossRef] [Green Version]
- Musatti, A.; Cavicchioli, D.; Mapelli, C.; Bertoni, D.; Hogenboom, J.A.; Pellegrino, L.; Rollini, M. From Cheese Whey Permeate to Sakacin A: A circular economy approach for the food-grade biotechnological production of an anti-Listeria bacteriocin. Biomolecules 2020, 10, 597. [Google Scholar] [CrossRef] [Green Version]
- Zotta, T.; Solieri, L.; Iacumin, L.; Picozzi, C.; Gullo, M. Valorization of Cheese Whey Using Microbial Fermentations. Appl. Microbiol. Biotechnol. 2020, 104, 2749–2764. [Google Scholar] [CrossRef]
- Ratledge, C. Yeasts, molds, algae and bacteria as sources of lipids. In Technological Advances in Improved and Alternative Sources of Lipids; Springer: Boston, MA, USA, 1994; pp. 235–291. [Google Scholar] [CrossRef]
- Abeln, F.; Chuck, C.J. The history, state of the art and future prospects for oleaginous yeast research. Microb. Cell Fact. 2021, 20, 221. [Google Scholar] [CrossRef]
- Caporusso, A.; Capece, A.; De Bari, I. Oleaginous Yeasts as Cell Factories for the Sustainable Production of Microbial Lipids by the Valorization of Agri-Food wastes. Fermentation 2021, 7, 50. [Google Scholar] [CrossRef]
- Christophe, G.; Kumar, V.; Nouaille, R.; Gaudet, G.; Fontanille, P.; Pandey, A.; Soccol, C.R.; Larroche, C. Recent developments in microbial oils production: A possible alternative to vegetable oils for biodiesel without competition with human food? Brazilian Arch. Biol. Technol. 2012, 55, 29–46. [Google Scholar] [CrossRef] [Green Version]
- Atabani, A.E.; Silitonga, A.S.; Badruddin, I.A.; Mahlia, T.M.I.; Masjuki, H.H.; Mekhilef, S. A comprehensive review on biodiesel as an alternative energy resource and its characteristics. Renew. Sust. Energ. Rev. 2012, 16, 2070–2093. [Google Scholar] [CrossRef]
- Béligon, V.; Christophe, G.; Fontanille, P.; Larroche, C. Microbial lipids as potential source to food supplements. Curr. Opin. Food Sci. 2016, 7, 35–42. [Google Scholar] [CrossRef]
- Li, Q.; Du, W.; Liu, D. Perspectives of microbial oils for biodiesel production. Appl. Microbiol. Biotechnol. 2008, 80, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Moradi, H.; Shi, S.; Darvishi, F. Yeasts as microbial cell factories for sustainable production of biofuels. Renew. Sustain. Energy Rev. 2021, 143, 110907. [Google Scholar] [CrossRef]
- Zhang, Y.; Nielsen, J.; Liu, Z. Yeast based biorefineries for oleochemical production. Curr. Opin. Biotechnol. 2021, 67, 26–34. [Google Scholar] [CrossRef]
- Koutinas, A.A.; Chatzifragkou, A.; Kopsahelis, N.; Papanikolaou, S.; Kookos, I.K. Design and techno-economic evaluation of microbial oil production as a renewable resource for biodiesel and oleochemical production. Fuel 2014, 116, 566–577. [Google Scholar] [CrossRef]
- Amoah, J.; Kahar, P.; Ogino, C.; Kondo, A. Bioenergy and Biorefinery: Feedstock, Biotechnological Conversion, and Products. Biotechnol. J. 2019, 14, 1800494. [Google Scholar] [CrossRef]
- Gerbens-Leenes, W.; Hoekstra, A.Y.; van der Meer, T.H. The water footprint of bioenergy. Proc. Natl. Acad. Sci. USA 2009, 106, 10219–10223. [Google Scholar] [CrossRef] [Green Version]
- Standard 182:1999; Milk Fat. Preparation of Fatty Acid Methyl Esters. FIL-IDF International Dairy Federation: Brussels, Belgium, 1999.
- D’Incecco, P.; Ong, L.; Gras, S.; Pellegrino, L. A fluorescence in situ staining method for investigating spores and vegetative cells of Clostridia by confocal laser scanning microscopy and structured illuminated microscopy. Micron 2018, 110, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Capusoni, C.; Rodighiero, V.; Cucchetti, D.; Galafassi, S.; Bianchi, D.; Franzosi, G.; Compagno, C. Characterization of lipid accumulation and lipidome analysis in the oleaginous yeasts Rhodosporidium azoricum and Trichosporon oleaginosus. Bioresour. Technol. 2017, 238, 281–289. [Google Scholar] [CrossRef]
- Barbesti, S.; Citterio, S.; Labra, M.; Baroni, M.D.; Neri, M.G.; Sgorbati, S. Two and three-color fluorescence flow cytometric analysis of immunoidentified viable bacteria. Cytometry 2000, 40, 214–218. [Google Scholar] [CrossRef]
- Daniel, H.J.; Otto, R.T.; Binder, M.; Reuss, M.; Syldatk, C. Production of sophorolipids from whey: Development of a two-stage process with Cryptococcus curvatus ATCC 20509 and Candida bombicola ATCC 22214 using deproteinized whey concentrates as substrates. Appl. Microbiol. Biotechnol. 1999, 51, 40–45. [Google Scholar] [CrossRef]
- Meo, A.; Priebe, X.L.; Weuster-Botz, D. Lipid production with Trichosporon oleaginosus in a membrane bioreactor using microalgae hydrolysate. J. Biotechnol. 2017, 241, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kopsahelis, N.; Dimou, C.; Papadaki, A.; Xenopoulos, E.; Kyraleou, M.; Kallithraka, S.; Kotseridis, Y.; Papanikolaou, S.; Koutinas, A.A. Refining of wine lees and cheese whey for the production of microbial oil, polyphenol-rich extracts and value-added co-products. J. Chem. Technol. Biotechnol. 2018, 93, 257–268. [Google Scholar] [CrossRef]
- Brabender, M.; Hussain, M.S.; Rodriguez, G.; Blenner, M.A. Urea and urine are a viable and cost-effective nitrogen source for Yarrowia lipolytica biomass and lipid accumulation. Appl. Microbiol. Biotechnol. 2018, 102, 2313–2322. [Google Scholar] [CrossRef] [PubMed]
- Kourist, R.; Bracharz, F.; Lorenzen, J.; Kracht, O.N.; Chovatia, M.; Daum, C.; Deshpande, S.; Lipzen, A.; Nolan, M.; Ohm, R.A.; et al. Genomics and Transcriptomics Analyses of the Oil-Accumulating Basidiomycete Yeast Trichosporon oleaginosus: Insights into Substrate Utilization and Alternative Evolutionary Trajectories of Fungal Mating Systems. Mbio 2015, 6, e00918-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awad, D.; Bohnen, F.; Mehlmer, N.; Brueck, T. Multi-Factorial-Guided Media Optimization for Enhanced Biomass and Lipid Formation by the Oleaginous Yeast Cutaneotrichosporon oleaginosus. Front. Bioeng. Biotechnol. 2019, 7, 54. [Google Scholar] [CrossRef] [Green Version]
- Milne, N.; Luttik, M.A.H.; Cueto Rojas, H.F.; Wahl, A.; van Maris, A.J.A.; Pronk, J.T.; Daran, J.M. Functional expression of a heterologous nickel-dependent, ATP-independent urease in Saccharomyces cerevisiae. Metab. Eng. 2015, 130, 140. [Google Scholar] [CrossRef]
- Taiwo, A.E.; Madzimbamuto, T.N.; Ojumu, T.V. Optimization of Corn Steep Liquor Dosage and Other Fermentation Parameters for Ethanol Production by Saccharomyces cerevisiae Type 1 and Anchor Instant Yeast. Energies 2018, 11, 1740. [Google Scholar] [CrossRef] [Green Version]
- Donzella, S.; Serra, I.; Fumagalli, A.; Pellegrino, L.; Mosconi, G.; Lo Scalzo, R.; Compagno, C. Recycling industrial food wastes for lipid production by oleaginous yeasts Rhodosporidiobolus azoricus and Cutaneotrichosporon oleaginosum. Biotechnol. Biofuels Bioprod. 2022, 15, 51. [Google Scholar] [CrossRef]
- Parsons, S.; Allen, M.J.; Chuck, C.J. Coproducts of algae and yeast-derived single cell oils: A critical review of their role in improving biorefinery sustainability. Bioresour. Technol. 2020, 303, 122862. [Google Scholar] [CrossRef] [PubMed]
- Moon, N.J.; Hammond, E.G.; Glatz, B.A. Conversion of Cheese Whey and Whey Permeate to Oil and Single-Cell Protein. J. Dairy Sci. 1978, 61, 1537–1547. [Google Scholar] [CrossRef]
- Ykema, A.; Verbree, E.C.; Kater, M.M.; Smit, H. Optimization of lipid production in the oleaginous yeast Apiotrichum curvatum in whey permeate. Appl. Microbiol. Biotechnol. 1988, 29, 211–218. [Google Scholar] [CrossRef]
- Vamvakaki, A.-N.; Kandarakis, I.; Kaminarides, S.; Komaitis, M.; Papanikolaou, S. Cheese whey as a renewable substrate for microbial lipid and biomass production by Zygomycetes. Eng. Life Sci. 2010, 10, 348–360. [Google Scholar] [CrossRef]
- Demir, M.; Turhan, I.; Kucukcetin, A.; Alpkent, Z. Oil production by Mortierella isabellina from whey treated with lactase. Bioresour. Technol. 2013, 128, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.H.; Lee, I.; Jeon, S.H.; Han, J.I. Efficient Conversion from Cheese Whey to Lipid Using Cryptococcus curvatus. Biochem. Eng. J. 2014, 90, 149–153. [Google Scholar] [CrossRef]
- Carota, E.; Crognale, S.; D’Annibale, A.; Gallo, A.M.; Stazi, S.R.; Petruccioli, M. A sustainable use of Ricotta Cheese Whey for microbial biodiesel production. Sci. Total Environ. 2017, 584–585, 554–560. [Google Scholar] [CrossRef]
- Bracharz, F.; Beukhout, T.; Mehlmer, N.; Brück, T. Opportunities and challenges in the development of Cutaneotrichosporon oleaginosus ATCC 20509 as a new cell factory for custom tailored microbial oils. Microb. Cell Factor. 2017, 16, 178. [Google Scholar] [CrossRef] [Green Version]
- Di Fidio, N.; Minonne, F.; Antonetti, C.; Raspolli Galletti, A.M. Cutaneotrichosporon oleaginosus: A Versatile Whole-Cell Biocatalyst for the Production of Single-Cell Oil from Agro-Industrial Wastes. Catalysts 2021, 11, 1291. [Google Scholar] [CrossRef]
- Pham, N.; Reijnders, M.; Suarez-Diez, M.; Nijsse, B.; Springer, J.; Eggink, G.; Schaap, P.J. Genome-scale metabolic modeling underscores the potential of Cutaneotrichosporon oleaginosus ATCC 20509 as a cell factory for biofuel production. Biotechnol. Biofuels 2021, 14, 2. [Google Scholar] [CrossRef]
- Neta, N.S.; Teixeira, J.A.; Rodrigues, L.R. Sugar ester surfactants: Enzymatic synthesis and applications in food industry. Crit. Rev. Food Sc. Nutr. 2015, 55, 595–610. [Google Scholar] [CrossRef] [PubMed]
- Perfetto, S.; Chattopadhyay, P.; Roederer, M. Seventeen-colour flow cytometry: Unravelling the immune system. Nat. Rev. Immunol. 2004, 4, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Capusoni, C.; Arioli, S.; Donzella, S.; Guidi, B.; Serra, I.; Compagno, C. Hyper-Osmotic Stress Elicits Membrane Depolarization and Decreased Permeability in Halotolerant Marine Debaryomyces hansenii Strains and in Saccharomyces cerevisiae. Front. Microbiol. 2019, 10, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, J.A.; Lopes da Silva, T.; Marques, S.; Carvalheiro, F.; Roseiro, L.B.; Duarte, L.C.; Gírio, F. The use of flow cytometry to assess Rhodosporidium toruloides NCYC 921 performance for lipid production using Miscanthus sp. hydrolysates. Biotechnol. Rep. 2021, 30, e00639. [Google Scholar] [CrossRef] [PubMed]
- Rumin, J.; Bonnefond, H.; Saint-Jean, B.; Rouxel, C.; Sciandra, A.; Bernard, O.; Cadoret, J.P.; Bougaran, G. The use of fluorescent Nile red and BODIPY for lipid measurement in microalgae. Biotechnol. Biofuels 2015, 8, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raschke, D.; Knorr, D. Rapid monitoring of cell size, vitality and lipid droplet development in the oleaginous yeast Waltomyces lipofer. J. Microbiol. Methods 2009, 79, 178–183. [Google Scholar] [CrossRef]
- Bouchedja, D.N.; Danthine, S.; Kar, T.; Fickers, P.; Boudjellal, A.; Delvigne, F. Online flow cytometry, an interesting investigation process for monitoring lipid accumulation, dimorphism, and cells’ growth in the oleaginous yeast Yarrowia lipolytica JMY 775. Bioresour. Bioprocess. 2017, 4, 3. [Google Scholar] [CrossRef] [Green Version]



| Medium | Location | U/mg |
|---|---|---|
| YPD | Intracellular | 0 |
| YNB-Lactose | Supernatant | 0 |
| YNB-Lactose | Intracellular | 152 |
| WPYE | WPYEU | WPUC | WPYEU+M | |
|---|---|---|---|---|
| Initial C/N | 60.59 | 18.33 | 16.54 | 10.5 |
| Growth rate (μ) | 0.26 | 0.28 | 0.26 | 0.32 |
| Final biomass (g/L) | 26.5 ± 0.27 a | 65.5 ± 1.21 b | 46.0 ± 0.51 c | 84.0 ± 1.27 d |
| Lipids (g/L) | 18.0 ± 1.30 a | 29.4 ± 0.60 b | 19.0 ± 0.40 a | 38.1 ± 1.00 c |
| Lipid content (%) | 68% | 45% | 43% | 45% |
| Lipid yield | 0.21 | 0.16 | 0.12 | 0.10 |
| Productivity (g/L/h) | 0.20 | 0.31 | 0.20 | 0.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donzella, S.; Fumagalli, A.; Arioli, S.; Pellegrino, L.; D’Incecco, P.; Molinari, F.; Speranza, G.; Ubiali, D.; Robescu, M.S.; Compagno, C. Recycling Food Waste and Saving Water: Optimization of the Fermentation Processes from Cheese Whey Permeate to Yeast Oil. Fermentation 2022, 8, 341. https://doi.org/10.3390/fermentation8070341
Donzella S, Fumagalli A, Arioli S, Pellegrino L, D’Incecco P, Molinari F, Speranza G, Ubiali D, Robescu MS, Compagno C. Recycling Food Waste and Saving Water: Optimization of the Fermentation Processes from Cheese Whey Permeate to Yeast Oil. Fermentation. 2022; 8(7):341. https://doi.org/10.3390/fermentation8070341
Chicago/Turabian StyleDonzella, Silvia, Andrea Fumagalli, Stefania Arioli, Luisa Pellegrino, Paolo D’Incecco, Francesco Molinari, Giovanna Speranza, Daniela Ubiali, Marina S. Robescu, and Concetta Compagno. 2022. "Recycling Food Waste and Saving Water: Optimization of the Fermentation Processes from Cheese Whey Permeate to Yeast Oil" Fermentation 8, no. 7: 341. https://doi.org/10.3390/fermentation8070341
APA StyleDonzella, S., Fumagalli, A., Arioli, S., Pellegrino, L., D’Incecco, P., Molinari, F., Speranza, G., Ubiali, D., Robescu, M. S., & Compagno, C. (2022). Recycling Food Waste and Saving Water: Optimization of the Fermentation Processes from Cheese Whey Permeate to Yeast Oil. Fermentation, 8(7), 341. https://doi.org/10.3390/fermentation8070341










