Lignin Biodegradation and Its Valorization
Abstract
:1. Introduction
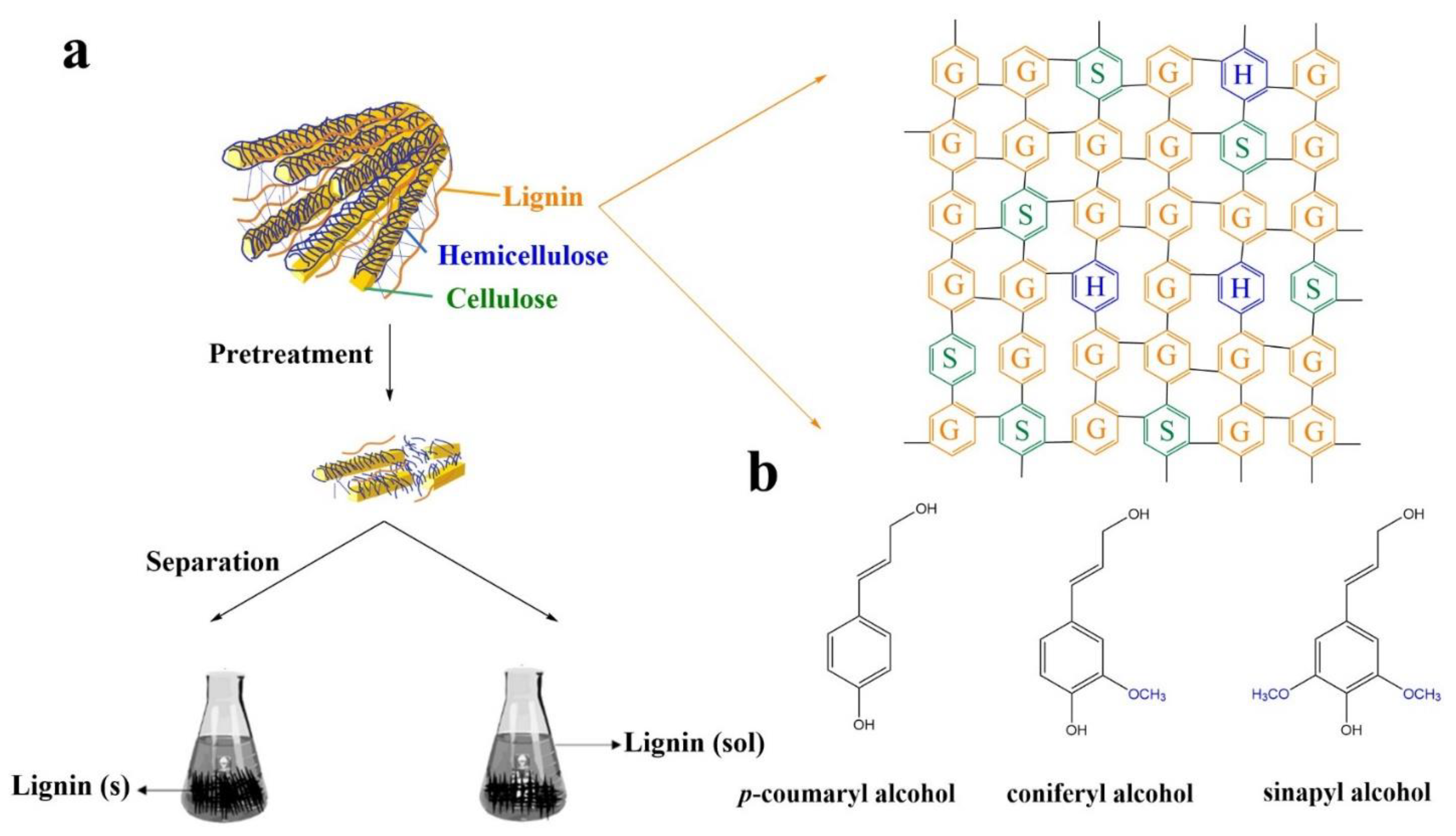
2. Lignin
3. Enzymes for Lignin Depolymerization
3.1. Laccase (EC 1.10.3.2)
3.2. Lignin Peroxidase (EC 1.11.1.14)
3.3. Manganese Peroxidase (EC 1.11.1.13)
3.4. Versatile Peroxidase (EC 1.11.1.16)
4. Degradative Pathways for Lignin-Derived Aromatic Compounds
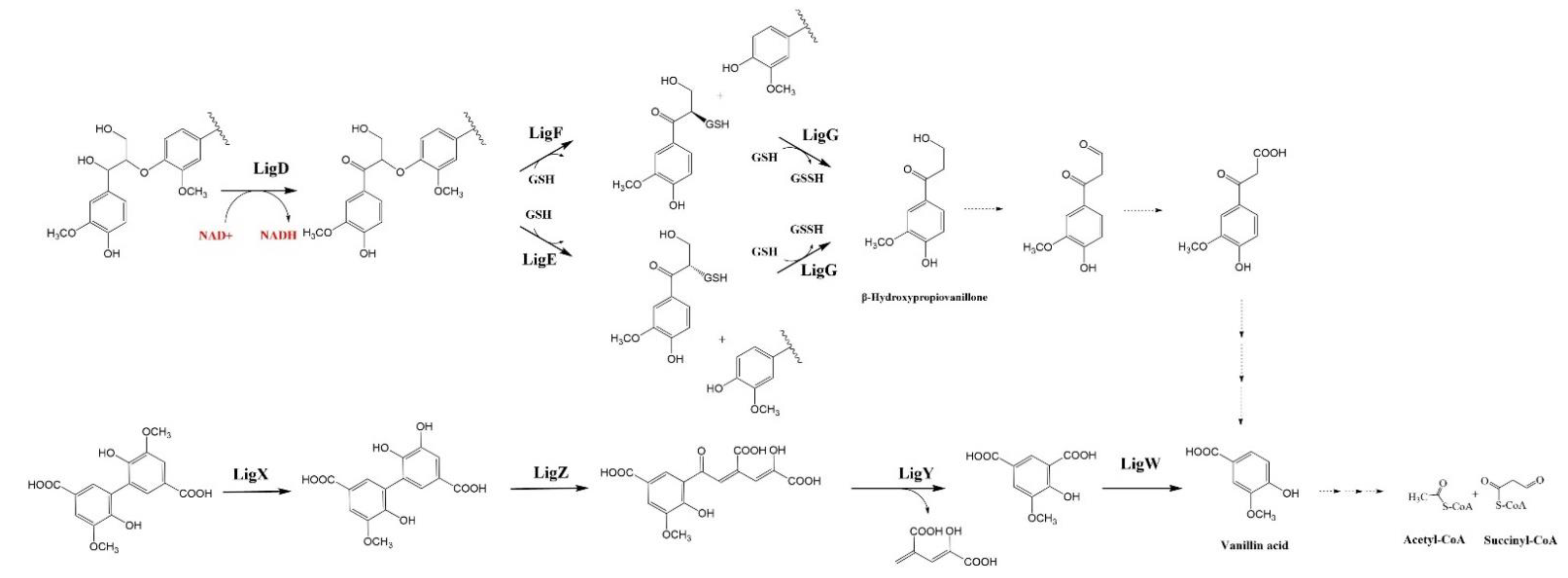
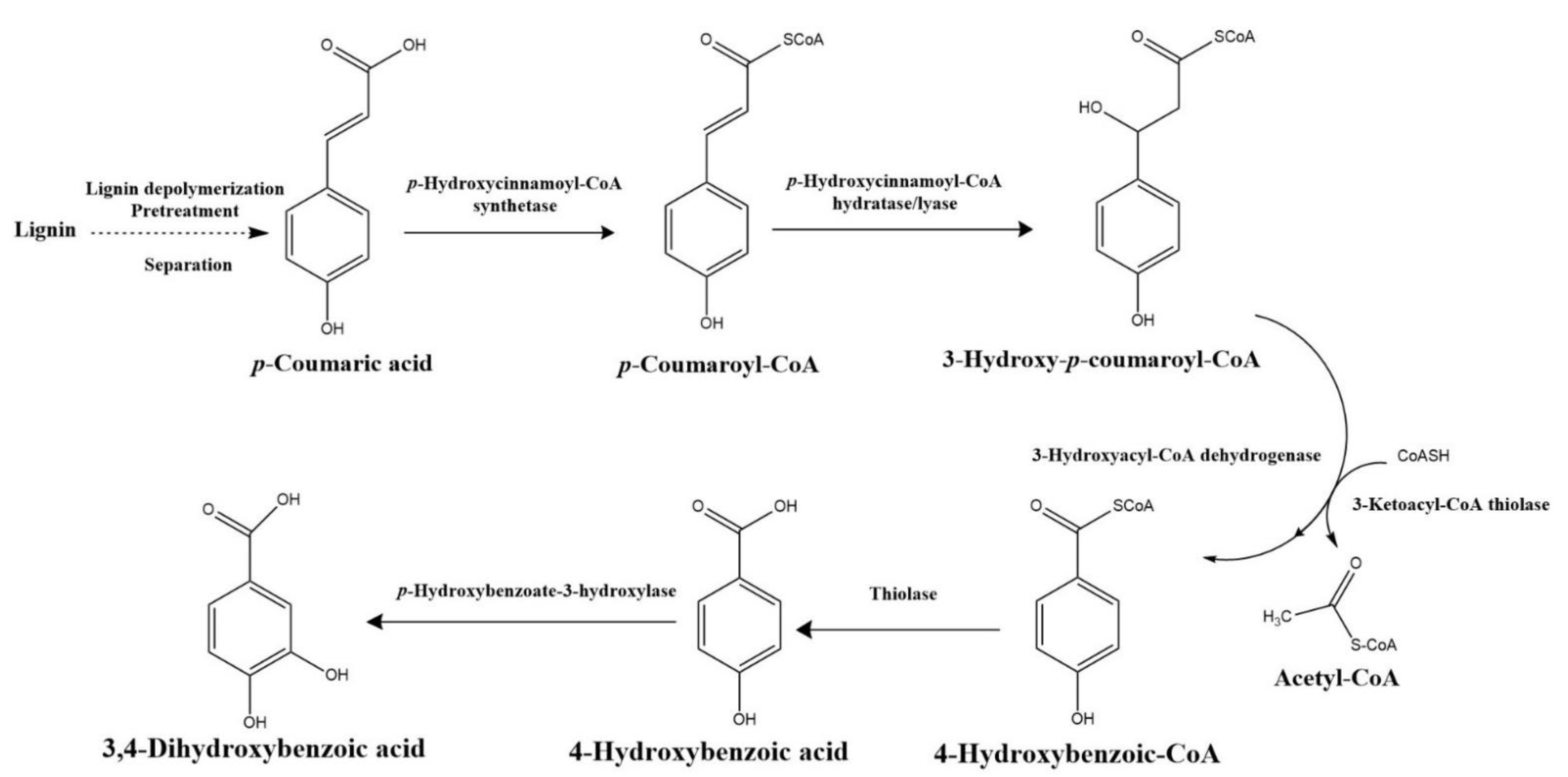
4.1. β-Aryl ether (β-O-4) Catabolic Pathway
4.2. Biphenyl Catabolic Pathway
4.3. β-ketoadipate Pathway
5. Auxiliary Pathways
5.1. β-oxidation-like Pathway
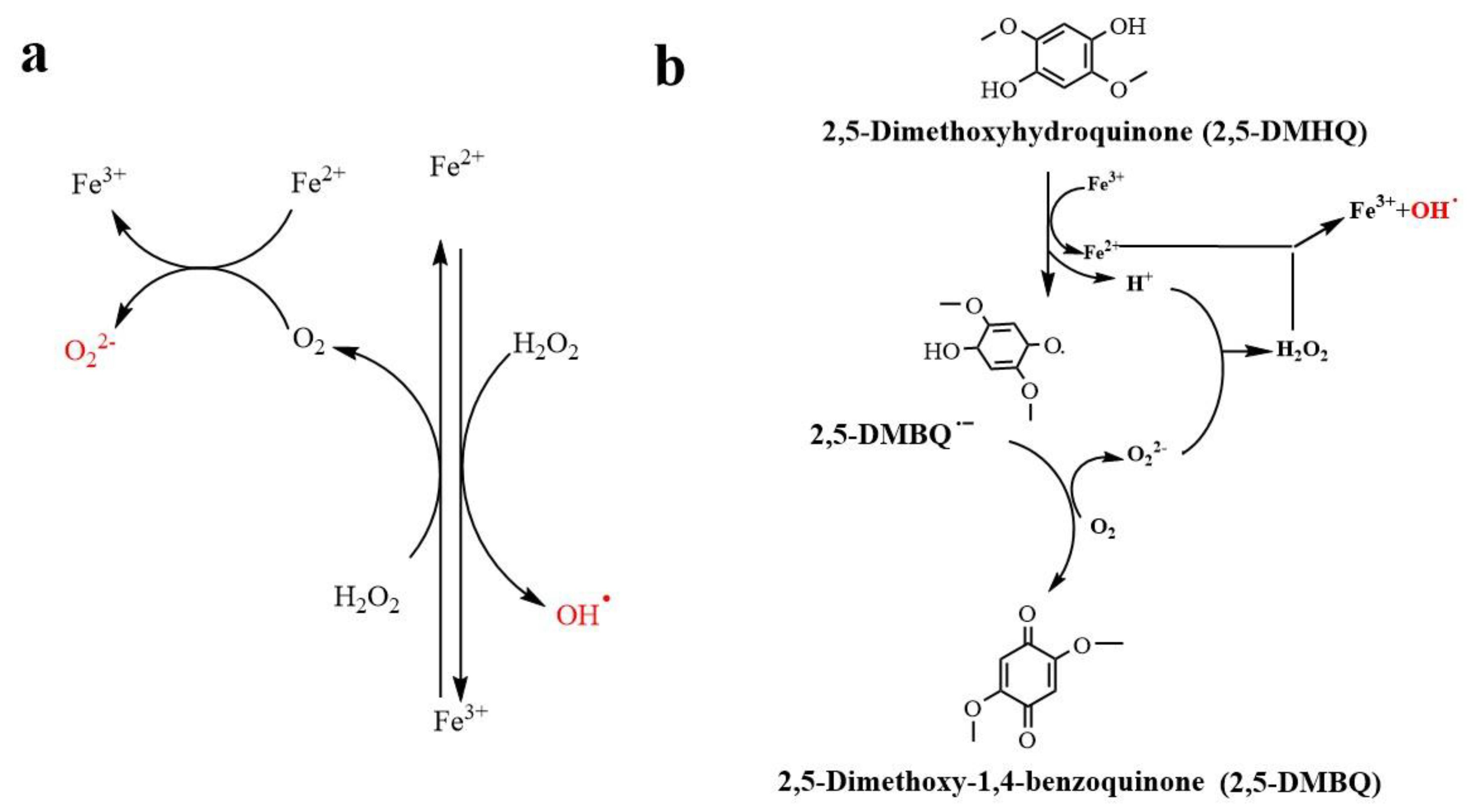
5.2. Fenton Reaction
5.3. Demethylation
6. Value-Added Chemicals Produced from Lignin
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdelaziz, O.Y.; Brink, D.P.; Prothmann, J.; Ravi, K.; Sun, M.; García-Hidalgo, J.; Sandahl, M.; Hulteberg, C.P.; Turner, C.; Lidén, G. Biological valorization of low molecular weight lignin. Biotechnol. Adv. 2016, 34, 1318–1346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, T.; Qi, W.; Su, R.; He, Z. Promising techniques for depolymerization of lignin into value-added chemicals. ChemCatChem 2019, 11, 639–654. [Google Scholar] [CrossRef]
- Marinović, M.; Nousiainen, P.; Dilokpimol, A.; Kontro, J.; Moore, R.; Sipilä, J.; De Vries, R.P.; Mäkelä, M.R.; Hildén, K. Selective cleavage of lignin β-O-4 aryl ether bond by β-etherase of the white-rot fungus Dichomitus squalens. ACS Sustain. Chem. Eng. 2018, 6, 2878–2882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dashtban, M.; Maki, M.; Leung, K.T.; Mao, C.; Qin, W. Cellulase activities in biomass conversion: Measurement methods and comparison. Crit. Rev. Biotechnol. 2010, 30, 302–309. [Google Scholar] [CrossRef]
- Wang, Z.; Li, N.; Pan, X. Transformation of ammonia fiber expansion (AFEX) corn stover lignin into microbial lipids by Rhodococcus opacus. Fuel 2019, 240, 119–125. [Google Scholar] [CrossRef]
- Wang, H.; Pu, Y.; Ragauskas, A.; Yang, B. From lignin to valuable products–strategies, challenges, and prospects. Bioresour. Technol. 2019, 271, 449–461. [Google Scholar] [CrossRef]
- Stewart, J.J.; Akiyama, T.; Chapple, C.; Ralph, J.; Mansfield, S.D. The effects on lignin structure of overexpression of ferulate 5-hydroxylase in hybrid poplar1. Plant Physiol. 2009, 150, 621–635. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhuo, C.; Xiao, X.; Wang, X.; Docampo-Palacios, M.; Chen, F.; Dixon, R.A. Substrate specificity of LACCASE8 facilitates polymerization of caffeyl alcohol for C-lignin biosynthesis in the seed coat of Cleome hassleriana. Plant Cell 2020, 32, 3825–3845. [Google Scholar] [CrossRef]
- Ayuso-Fernández, I.; Rencoret, J.; Gutiérrez, A.; Ruiz-Dueñas, F.J.; Martínez, A.T. Peroxidase evolution in white-rot fungi follows wood lignin evolution in plants. Proc. Natl. Acad. Sci. USA 2019, 116, 17900–17905. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Kelley, S.S.; Venditti, R.A. Lignin-based thermoplastic materials. ChemSusChem 2016, 9, 770–783. [Google Scholar] [CrossRef]
- Kazzaz, A.E.; Feizi, Z.H.; Fatehi, P. Grafting strategies for hydroxy groups of lignin for producing materials. Green Chem. 2019, 21, 5714–5752. [Google Scholar] [CrossRef] [Green Version]
- Meister, J.J. Modification of lignin. J. Macromol. Sci. Part C Polym. Rev. 2002, 42, 235–289. [Google Scholar] [CrossRef]
- Ruwoldt, J. A critical review of the physicochemical properties of lignosulfonates: Chemical structure and behavior in aqueous solution, at surfaces and interfaces. Surfaces 2020, 3, 622–648. [Google Scholar] [CrossRef]
- Kienberger, M.; Maitz, S.; Pichler, T.; Demmelmayer, P. Systematic review on isolation processes for technical lignin. Processes 2021, 9, 804. [Google Scholar] [CrossRef]
- Demuner, I.F.; Gomes, F.J.B.; Gomes, J.S.; Coura, M.R.; Borges, F.P.; Carvalho, A.M.M.L.; Silva, C.M. Improving kraft pulp mill sustainability by lignosulfonates production from processes residues. J. Clean. Prod. 2021, 317, 128286. [Google Scholar] [CrossRef]
- Hanhikoski, S.; Niemelä, K.; Vuorinen, T. Biorefining of Scots pine using neutral sodium sulphite pulping: Investigation of fibre and spent liquor compositions. Ind. Crops Prod. 2019, 129, 135–141. [Google Scholar] [CrossRef]
- Tribot, A.; Amer, G.; Alio, M.A.; de Baynast, H.; Delattre, C.; Pons, A.; Mathias, J.-D.; Callois, J.-M.; Vial, C.; Michaud, P. Wood-lignin: Supply, extraction processes and use as bio-based material. Eur. Polym. J. 2019, 112, 228–240. [Google Scholar] [CrossRef]
- Guadix-Montero, S.; Sankar, M. Review on catalytic cleavage of C–C inter-unit linkages in lignin model compounds: Towards lignin depolymerisation. Top. Catal. 2018, 61, 183–198. [Google Scholar] [CrossRef] [Green Version]
- Wörmeyer, K.; Ingram, T.; Saake, B.; Brunner, G.; Smirnova, I. Comparison of different pretreatment methods for lignocellulosic materials. Part II: Influence of pretreatment on the properties of rye straw lignin. Bioresour. Technol. 2011, 102, 4157–4164. [Google Scholar] [CrossRef] [PubMed]
- Vishtal, A.G.; Kraslawski, A. Challenges in industrial applications of technical lignins. BioResources 2011, 6, 3547–3568. [Google Scholar] [CrossRef]
- Chen, Z.; Wan, C. Biological valorization strategies for converting lignin into fuels and chemicals. Renew. Sustain. Energy Rev. 2017, 73, 610–621. [Google Scholar] [CrossRef]
- Pang, B.; Lam, S.S.; Shen, X.J.; Cao, X.F.; Liu, S.J.; Yuan, T.Q.; Sun, R.C. Valorization of technical lignin for the production of desirable resins with high substitution rate and controllable viscosity. ChemSusChem 2020, 13, 4446–4454. [Google Scholar] [CrossRef]
- Qian, Y.; Zuo, C.; Tan, J.; He, J. Structural analysis of bio-oils from sub-and supercritical water liquefaction of woody biomass. Energy 2007, 32, 196–202. [Google Scholar] [CrossRef]
- Kloekhorst, A.; Heeres, H.J. Catalytic hydrotreatment of alcell lignin using supported Ru, Pd, and Cu catalysts. ACS Sustain. Chem. Eng. 2015, 3, 1905–1914. [Google Scholar] [CrossRef]
- Morpurgo, L.; Graziani, M.; Finazzi-Agro, A.; Rotilio, G.; Mondovi, B. Optical properties of japanese-lacquer-tree (Rhus vernicifera) laccase depleted of type 2 copper (II). Involvement of type-2 copper (II) in the 330nm chromophore. Biochem. J. 1980, 187, 361–366. [Google Scholar] [CrossRef] [Green Version]
- Claus, H. Laccases and their occurrence in prokaryotes. Arch. Microbiol. 2003, 179, 145–150. [Google Scholar] [CrossRef]
- Mandic, M.; Djokic, L.; Nikolaivits, E.; Prodanovic, R.; O’Connor, K.; Jeremic, S.; Topakas, E.; Nikodinovic-Runic, J. Identification and characterization of new laccase biocatalysts from Pseudomonas species suitable for degradation of synthetic textile dyes. Catalysts 2019, 9, 629. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Zeng, G.; Lai, C.; Li, J.; Xu, P.; Wu, H. Molecular basis of laccase bound to lignin: Insight from comparative studies on the interaction of Trametes versicolor laccase with various lignin model compounds. RSC Adv. 2015, 5, 52307–52313. [Google Scholar] [CrossRef]
- Perna, V.; Agger, J.W.; Andersen, M.L.; Holck, J.; Meyer, A.S. Laccase induced lignin radical formation kinetics evaluated by electron paramagnetic resonance spectroscopy. ACS Sustain. Chem. Eng. 2019, 7, 10425–10434. [Google Scholar] [CrossRef]
- Gutiérrez, A.; Rencoret, J.; Cadena, E.M.; Rico, A.; Barth, D.; José, C.; Martínez, Á.T. Demonstration of laccase-based removal of lignin from wood and non-wood plant feedstocks. Bioresour. Technol. 2012, 119, 114–122. [Google Scholar] [CrossRef] [Green Version]
- Braunschmid, V.; Binder, K.; Fuerst, S.; Subagia, R.; Danner, C.; Weber, H.; Schwaiger, N.; Nyanhongo, G.S.; Ribitsch, D.; Guebitz, G.M. Comparison of a fungal and a bacterial laccase for lignosulfonate polymerization. Process Biochem. 2021, 109, 207–213. [Google Scholar] [CrossRef]
- Mattinen, M.-L.; Maijala, P.; Nousiainen, P.; Smeds, A.; Kontro, J.; Sipilä, J.; Tamminen, T.; Willför, S.; Viikari, L. Oxidation of lignans and lignin model compounds by laccase in aqueous solvent systems. J. Mol. Catal. B: Enzym. 2011, 72, 122–129. [Google Scholar] [CrossRef]
- Kawai, S.; Iwatsuki, M.; Nakagawa, M.; Inagaki, M.; Hamabe, A.; Ohashi, H. An alternative β-ether cleavage pathway for a non-phenolic β-O-4 lignin model dimer catalyzed by a laccase-mediator system. Enzym. Microb. Technol. 2004, 35, 154–160. [Google Scholar] [CrossRef]
- Hilgers, R.; Vincken, J.-P.; Gruppen, H.; Kabel, M.A. Laccase/mediator systems: Their reactivity toward phenolic lignin structures. ACS Sustain. Chem. Eng. 2018, 6, 2037–2046. [Google Scholar] [CrossRef] [Green Version]
- Bujanovic, B.; Ralph, S.; Reiner, R.; Hirth, K.; Atalla, R. Polyoxometalates in oxidative delignification of chemical pulps: Effect on lignin. Materials 2010, 3, 1888–1903. [Google Scholar] [CrossRef] [Green Version]
- Rencoret, J.; Pereira, A.; del Río, J.C.; Martínez, A.N.T.; Gutiérrez, A. Delignification and saccharification enhancement of sugarcane byproducts by a laccase-based pretreatment. ACS Sustain. Chem. Eng. 2017, 5, 7145–7154. [Google Scholar] [CrossRef]
- Rencoret, J.; Pereira, A.; Del Río, J.C.; Martínez, A.T.; Gutiérrez, A. Laccase-mediator pretreatment of wheat straw degrades lignin and improves saccharification. BioEnergy Res. 2016, 9, 917–930. [Google Scholar] [CrossRef] [Green Version]
- Xie, B.; Tobimatsu, Y.; Kamitakahara, H.; Takano, T. Reaction Selectivity in Electro-oxidation of Lignin Dimer Model Compounds and Synthetic Lignin with Different Mediators for the Laccase Mediator System (PZH, NHPI, ABTS). ACS Sustain. Chem. Eng. 2022, 10, 6633–6641. [Google Scholar] [CrossRef]
- Kumar, A.; Chandra, R. Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon 2020, 6, e03170. [Google Scholar] [CrossRef]
- Guo, M.; Lu, F.; Du, L.; Pu, J.; Bai, D. Optimization of the expression of a laccase gene from Trametes versicolor in Pichia methanolica. Appl. Microbiol. Biotechnol. 2006, 71, 848–852. [Google Scholar] [CrossRef]
- Eggert, C.; Temp, U.; Eriksson, K.-E.L. Laccase is essential for lignin degradation by the white-rot fungus Pycnoporus cinnabarinus. Febs Lett. 1997, 407, 89–92. [Google Scholar] [CrossRef] [Green Version]
- Kawai, S.; Nakagawa, M.; Ohashi, H. Degradation mechanisms of a nonphenolic β-O-4 lignin model dimer by Trametes versicolor laccase in the presence of 1-hydroxybenzotriazole. Enzym. Microb. Technol. 2002, 30, 482–489. [Google Scholar] [CrossRef]
- Arias, M.; Blánquez, A.; Hernandez, M.; Rodriguez, J.; Ball, A.; Jiménez-Morillo, N.; González-Vila, F.; González-Pérez, J. Role of a thermostable laccase produced by Streptomyces ipomoeae in the degradation of wheat straw lignin in solid state fermentation. J. Anal. Appl. Pyrolysis 2016, 122, 202–208. [Google Scholar] [CrossRef]
- Guan, Z.-B.; Shui, Y.; Song, C.-M.; Zhang, N.; Cai, Y.-J.; Liao, X.-R. Efficient secretory production of CotA-laccase and its application in the decolorization and detoxification of industrial textile wastewater. Environ. Sci. Pollut. Res. 2015, 22, 9515–9523. [Google Scholar] [CrossRef]
- Shi, X.; Liu, Q.; Ma, J.; Liao, H.; Xiong, X.; Zhang, K.; Wang, T.; Liu, X.; Xu, T.; Yuan, S. An acid-stable bacterial laccase identified from the endophyte Pantoea ananatis Sd-1 genome exhibiting lignin degradation and dye decolorization abilities. Biotechnol. Lett. 2015, 37, 2279–2288. [Google Scholar] [CrossRef]
- Asina, F.; Brzonova, I.; Voeller, K.; Kozliak, E.; Kubátová, A.; Yao, B.; Ji, Y. Biodegradation of lignin by fungi, bacteria and laccases. Bioresour. Technol. 2016, 220, 414–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colao, M.C.; Lupino, S.; Garzillo, A.M.; Buonocore, V.; Ruzzi, M. Heterologous expression of lcc1 gene from Trametes trogii in Pichia pastoris and characterization of the recombinant enzyme. Microb. Cell Factories 2006, 5, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bo, W.; Yan, Y.; Xu, J.; Fu, X.; Han, H.; Gao, J.; Li, Z.; Wang, L.; Tian, Y.; Peng, R. Heterologous expression and characterization of a laccase from Laccaria bicolor in Pichia pastoris and Arabidopsis thaliana. J. Microbiol. Biotechnol. 2018, 28, 2057–2063. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Zhang, P.; Xie, C.; Zhang, W.; Sun, J.; Qian, W.-J.; Yang, B. Biodegradation of alkaline lignin by Bacillus ligniniphilus L1. Biotechnol. Biofuels 2017, 10, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirk, T.K.; Croan, S.; Tien, M.; Murtagh, K.E.; Farrell, R.L. Production of multiple ligninases by Phanerochaete chrysosporium: Effect of selected growth conditions and use of a mutant strain. Enzym. Microb. Technol. 1986, 8, 27–32. [Google Scholar] [CrossRef]
- Leisola, M.; Kozulic, B.; Meussdoerffer, F.; Fiechter, A. Homology among multiple extracellular peroxidases from Phanerochaete chrysosporium. J. Biol. Chem. 1987, 262, 419–424. [Google Scholar] [CrossRef]
- Fujii, K.; Uemura, M.; Hayakawa, C.; Funakawa, S.; Kosaki, T. Environmental control of lignin peroxidase, manganese peroxidase, and laccase activities in forest floor layers in humid Asia. Soil Biol. Biochem. 2013, 57, 109–115. [Google Scholar] [CrossRef]
- Sayadi, S.; Ellouz, R. Roles of lignin peroxidase and manganese peroxidase from Phanerochaete chrysosporium in the decolorization of olive mill wastewaters. Appl. Environ. Microbiol. 1995, 61, 1098–1103. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yao, B.; Su, X. Linking enzymatic oxidative degradation of lignin to organics detoxification. Int. J. Mol. Sci. 2018, 19, 3373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calvo-Flores, F.G.; Dobado, J.A. Lignin as renewable raw material. ChemSusChem 2010, 3, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.J.; Khan, M.F.; Kim, Y.H. Recombinant lignin peroxidase-catalyzed decolorization of melanin using in-situ generated H2O2 for application in whitening cosmetics. Int. J. Biol. Macromol. 2019, 136, 20–26. [Google Scholar] [CrossRef]
- Majeke, B.; Collard, F.-X.; Tyhoda, L.; Görgens, J. The synergistic application of quinone reductase and lignin peroxidase for the deconstruction of industrial (technical) lignins and analysis of the degraded lignin products. Bioresour. Technol. 2021, 319, 124152. [Google Scholar] [CrossRef] [PubMed]
- Liu, E.; Segato, F.; Wilkins, M.R. Fed-batch production of Thermothelomyces thermophilus lignin peroxidase using a recombinant Aspergillus nidulans strain in stirred-tank bioreactor. Bioresour. Technol. 2021, 325, 124700. [Google Scholar] [CrossRef]
- Kuwahara, M.; Glenn, J.K.; Morgan, M.A.; Gold, M.H. Separation and characterization of two extracelluar H2O2-dependent oxidases from ligninolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 1984, 169, 247–250. [Google Scholar] [CrossRef] [Green Version]
- Wariishi, H.; Valli, K.; Gold, M.H. Oxidative cleavage of a phenolic diarylpropane lignin model dimer by manganese peroxidase from Phanerochaete chrysosporium. Biochemistry 1989, 28, 6017–6023. [Google Scholar] [CrossRef]
- Wang, Y.; Vazquez-Duhalt, R.; Pickard, M.A. Purification, characterization, and chemical modification of manganese peroxidase from Bjerkandera adusta UAMH 8258. Curr. Microbiol. 2002, 45, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Hofrichter, M. Lignin conversion by manganese peroxidase (MnP). Enzym. Microb. Technol. 2002, 30, 454–466. [Google Scholar] [CrossRef]
- Masarin, F.; Norambuena, M.; Ramires, H.O.; Demuner, B.J.; Pavan, P.C.; Ferraz, A. Manganese peroxidase and biomimetic systems applied to in vitro lignin degradation in Eucalyptus grandis milled wood and kraft pulps. J. Chem. Technol. Biotechnol. 2016, 91, 1422–1430. [Google Scholar] [CrossRef]
- Min, K.; Kim, Y.H.; Kim, J.; Kim, Y.; Gong, G.; Um, Y. Effect of manganese peroxidase on the decomposition of cellulosic components: Direct cellulolytic activity and synergistic effect with cellulase. Bioresour. Technol. 2022, 343, 126138. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.S.; Al-Tohamy, R.; Khalil, M.A.; Ho, S.-H.; Fu, Y.; Sun, J. Exploring the potential of a newly constructed manganese peroxidase-producing yeast consortium for tolerating lignin degradation inhibitors while simultaneously decolorizing and detoxifying textile azo dye wastewater. Bioresour. Technol. 2022, 351, 126861. [Google Scholar]
- Camarero, S.; Sarkar, S.; Ruiz-Dueñas, F.J.; Martínez, M.J.; Martínez, A.T. Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese peroxidase and lignin peroxidase substrate interaction sites. J. Biol. Chem. 1999, 274, 10324–10330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sáez-Jiménez, V.; Acebes, S.; Guallar, V.; Martínez, A.T.; Ruiz-Dueñas, F.J. Improving the oxidative stability of a high redox potential fungal peroxidase by rational design. PLoS ONE 2015, 10, e0124750. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.C.; Paice, M.; Zhang, X. Enzymatic oxidation of lignin: Challenges and barriers toward practical applications. ChemCatChem 2020, 12, 401–425. [Google Scholar] [CrossRef]
- Bugg, T.D.; Rahmanpour, R. Enzymatic conversion of lignin into renewable chemicals. Curr. Opin. Chem. Biol. 2015, 29, 10–17. [Google Scholar] [CrossRef]
- Beckham, G.T.; Johnson, C.W.; Karp, E.M.; Salvachúa, D.; Vardon, D.R. Opportunities and challenges in biological lignin valorization. Curr. Opin. Biotechnol. 2016, 42, 40–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, H.; Li, T.; Wu, S.; Zhang, X. Effect of the interaction of phenolic hydroxyl with the benzene rings on lignin pyrolysis. Bioresour. Technol. 2020, 309, 123351. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Mimura, N.; Shirai, M.; Sato, O. Effect of metal catalysts on bond cleavage reactions of lignin model compounds in supercritical water. Waste Biomass Valorization 2020, 11, 669–674. [Google Scholar] [CrossRef]
- Reiter, J.; Strittmatter, H.; Wiemann, L.O.; Schieder, D.; Sieber, V. Enzymatic cleavage of lignin β-O-4 aryl ether bonds via net internal hydrogen transfer. Green Chem. 2013, 15, 1373–1381. [Google Scholar] [CrossRef]
- Gall, D.L.; Kim, H.; Lu, F.; Donohue, T.J.; Noguera, D.R.; Ralph, J. Stereochemical features of glutathione-dependent enzymes in the Sphingobium sp. strain SYK-6 β-aryl etherase pathway. J. Biol. Chem. 2014, 289, 8656–8667. [Google Scholar] [CrossRef] [Green Version]
- Hilgers, R.; Van Dam, A.; Zuilhof, H.; Vincken, J.-P.; Kabel, M.A. Controlling the competition: Boosting laccase/HBT-catalyzed cleavage of a β-O-4′ linked lignin model. ACS Catal. 2020, 10, 8650–8659. [Google Scholar] [CrossRef]
- Liu, Z.-H.; Olson, M.L.; Shinde, S.; Wang, X.; Hao, N.; Yoo, C.G.; Bhagia, S.; Dunlap, J.R.; Pu, Y.; Kao, K.C. Synergistic maximization of the carbohydrate output and lignin processability by combinatorial pretreatment. Green Chem. 2017, 19, 4939–4955. [Google Scholar] [CrossRef]
- Peng, X.; Egashira, T.; Hanashiro, K.; Masai, E.; Nishikawa, S.; Katayama, Y.; Kimbara, K.; Fukuda, M. Cloning of a Sphingomonas paucimobilis SYK-6 gene encoding a novel oxygenase that cleaves lignin-related biphenyl and characterization of the enzyme. Appl. Environ. Microbiol. 1998, 64, 2520–2527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Higuchi, Y.; Aoki, S.; Takenami, H.; Kamimura, N.; Takahashi, K.; Hishiyama, S.; Lancefield, C.S.; Ojo, O.S.; Katayama, Y.; Westwood, N.J. Bacterial catabolism of β-hydroxypropiovanillone and β-hydroxypropiosyringone produced in the reductive cleavage of arylglycerol-β-aryl ether in lignin. Appl. Environ. Microbiol. 2018, 84, e02670-17. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Liu, Z.-H.; Zhang, R.-K.; Yuan, J.S.; Li, B.-Z.; Yuan, Y.-J. Bacterial conversion routes for lignin valorization. Biotechnol. Adv. 2022, 60, 108000. [Google Scholar] [CrossRef]
- Peng, X.; Masai, E.; Katayama, Y.; Fukuda, M. Characterization of the meta-cleavage compound hydrolase gene involved in degradation of the lignin-related biphenyl structure by Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 1999, 65, 2789–2793. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Masai, E.; Kasai, D.; Miyauchi, K.; Katayama, Y.; Fukuda, M. A second 5-carboxyvanillate decarboxylase gene, ligW2, is important for lignin-related biphenyl catabolism in Sphingomonas paucimobilis SYK-6. Appl. Environ. Microbiol. 2005, 71, 5014–5021. [Google Scholar] [CrossRef] [Green Version]
- Mohn, W.W.; Westerberg, K.; Cullen, W.R.; Reimer, K.J. Aerobic biodegradation of biphenyl and polychlorinated biphenyls by Arctic soil microorganisms. Appl. Environ. Microbiol. 1997, 63, 3378–3384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shuai, J.; Yu, X.; Zhang, J.; Xiong, A.-s.; Xiong, F. Regional analysis of potential polychlorinated biphenyl degrading bacterial strains from China. Br. J. Microbiol. 2016, 47, 536–541. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Chen, C.; Wu, X.; Tsang, C.-W.; Mou, J.; Yan, J.; Liu, Y.; Lin, C.S.K. Recent advancement in lignin biorefinery: With special focus on enzymatic degradation and valorization. Bioresour. Technol. 2019, 291, 121898. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, Y.; Zhang, L.; Xu, Z.; Ben, H.; Gaffrey, M.J.; Yang, Y.; Yang, S.; Yuan, J.S.; Qian, W.-J. Discovery of potential pathways for biological conversion of poplar wood into lipids by co-fermentation of Rhodococci strains. Biotechnol. Biofuels 2019, 12, 60. [Google Scholar] [CrossRef] [Green Version]
- Niu, J.; Li, X.; Qi, X.; Ren, Y. Pathway analysis of the biodegradation of lignin by Brevibacillus thermoruber. Bioresour. Technol. 2021, 341, 125875. [Google Scholar] [CrossRef] [PubMed]
- Venkatesagowda, B.; Dekker, R.F. Enzymatic demethylation of Kraft lignin for lignin-based phenol-formaldehyde resin applications. Biomass Convers. Biorefinery 2020, 10, 203–225. [Google Scholar] [CrossRef]
- Cao, L.; Iris, K.; Liu, Y.; Ruan, X.; Tsang, D.C.; Hunt, A.J.; Ok, Y.S.; Song, H.; Zhang, S. Lignin valorization for the production of renewable chemicals: State-of-the-art review and future prospects. Bioresour. Technol. 2018, 269, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Cheng, Y.; Zang, H.; Chen, X.; Wang, Y.; Zhang, Y.; Wang, J.; Shen, X.; Li, C. Biodegradation of lignin and the associated degradation pathway by psychrotrophic Arthrobacter sp. C2 from the cold region of China. Cellulose 2020, 27, 1423–1440. [Google Scholar] [CrossRef]
- Cafaro, V.; Izzo, V.; Scognamiglio, R.; Notomista, E.; Capasso, P.; Casbarra, A.; Pucci, P.; Di Donato, A. Phenol hydroxylase and toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1: Interplay between two enzymes. Appl. Environ. Microbiol. 2004, 70, 2211–2219. [Google Scholar] [CrossRef] [Green Version]
- Wells, T., Jr.; Ragauskas, A.J. Biotechnological opportunities with the β-ketoadipate pathway. Trends Biotechnol. 2012, 30, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Dikshit, P.K.; Jun, H.-B.; Kim, B.S. Biological conversion of lignin and its derivatives to fuels and chemicals. Korean J. Chem. Eng. 2020, 37, 387–401. [Google Scholar] [CrossRef]
- Prousek, J. Fenton chemistry in biology and medicine. Pure Appl. Chem. 2007, 79, 2325–2338. [Google Scholar] [CrossRef]
- Jensen, K.A., Jr.; Houtman, C.J.; Ryan, Z.C.; Hammel, K.E. Pathways for extracellular Fenton chemistry in the brown rot basidiomycete Gloeophyllum trabeum. Appl. Environ. Microbiol. 2001, 67, 2705–2711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, R.; Jensen, K.A.; Houtman, C.J.; Hammel, K.E. Significant levels of extracellular reactive oxygen species produced by brown rot basidiomycetes on cellulose. FEBS Lett. 2002, 531, 483–488. [Google Scholar] [CrossRef] [Green Version]
- Sahni, M.; Finney, W.C.; Locke, B.R. Degradation of aqueous phase polychlorinated biphenyls (PCB) using pulsed corona discharges. J. Adv. Oxid. Technol. 2005, 8, 105–111. [Google Scholar] [CrossRef]
- Zhou, C.; Dong, A.; Wang, Q.; Yu, Y.; Fan, X.; Cao, Y.; Li, T. Effect of common metal ions and anions on laccase catalysis of guaiacol and lignocellulosic fiber. BioResources 2017, 12, 5102–5117. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Xiang, W.; Ma, M.; Zhang, X.; Bao, Z.; Xie, S.; Yan, S. The role of laccase in stabilization of soil organic matter by iron in various plant-dominated peatlands: Degradation or sequestration? Plant Soil 2019, 443, 575–590. [Google Scholar] [CrossRef]
- Niladevi, K.N.; Jacob, N.; Prema, P. Evidence for a halotolerant-alkaline laccase in Streptomyces psammoticus: Purification and characterization. Process Biochem. 2008, 43, 654–660. [Google Scholar] [CrossRef]
- Kim, S.; Oh, S.; Lee, J.; Roh, H.-g.; Park, J. Changes of lignin molecular structures in a modification of kraft lignin using acid catalyst. Materials 2016, 9, 657. [Google Scholar] [CrossRef]
- Lai, C.; Jia, Y.; Zhou, C.; Yang, C.; Shen, B.; Zhang, D.; Yong, Q. Facilitating enzymatic digestibility of larch by in-situ lignin modification during combined acid and alkali pretreatment. Bioresour. Technol. 2020, 311, 123517. [Google Scholar] [CrossRef] [PubMed]
- Ferhan, M.; Yan, N.; Sain, M. Bark depolymerization during submerged fermentation using monofloral honey, a natural mediator substitute, and integration between laccases vs. bark biopolymers, characterized by Py-GC-MS. RSC Adv. 2015, 5, 14937–14952. [Google Scholar] [CrossRef]
- Zou, L.; Ross, B.M.; Hutchison, L.J.; Christopher, L.P.; Dekker, R.F.; Malek, L. Fungal demethylation of Kraft lignin. Enzym. Microb. Technol. 2015, 73, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ma, Y.; Wang, C.; Li, S.; Zhang, M.; Chu, F. Preparation and properties of lignin–phenol–formaldehyde resins based on different biorefinery residues of agricultural biomass. Ind. Crops Prod. 2013, 43, 326–333. [Google Scholar] [CrossRef]
- Zhang, W.; Ma, Y.; Xu, Y.; Wang, C.; Chu, F. Lignocellulosic ethanol residue-based lignin–phenol–formaldehyde resin adhesive. Int. J. Adhes. Adhes. 2013, 40, 11–18. [Google Scholar] [CrossRef]
- Zhao, M.; Jing, J.; Zhu, Y.; Yang, X.; Wang, X.; Wang, Z. Preparation and performance of lignin–phenol–formaldehyde adhesives. Int. J. Adhes. Adhes. 2016, 64, 163–167. [Google Scholar] [CrossRef]
- Yang, Z.; Peng, H.; Wang, W.; Liu, T. Crystallization behavior of poly (ε-caprolactone)/layered double hydroxide nanocomposites. J. Appl. Polym. Sci. 2010, 116, 2658–2667. [Google Scholar] [CrossRef]
- Zhang, Y.; Yuan, Z.; Mahmood, N.; Huang, S.; Xu, C.C. Sustainable bio-phenol-hydroxymethylfurfural resins using phenolated de-polymerized hydrolysis lignin and their application in bio-composites. Ind. Crops Prod. 2016, 79, 84–90. [Google Scholar] [CrossRef]
- Cheng, S.; Wilks, C.; Yuan, Z.; Leitch, M.; Xu, C.C. Hydrothermal degradation of alkali lignin to bio-phenolic compounds in sub/supercritical ethanol and water–ethanol co-solvent. Polym. Degrad. Stab. 2012, 97, 839–848. [Google Scholar] [CrossRef]
- Sawamura, K.; Tobimatsu, Y.; Kamitakahara, H.; Takano, T. Lignin functionalization through chemical demethylation: Preparation and tannin-like properties of demethylated guaiacyl-type synthetic lignins. ACS Sustain. Chem. Eng. 2017, 5, 5424–5431. [Google Scholar] [CrossRef]
- Kim, K.H.; Jeong, K.; Zhuang, J.; Jeong, H.J.; Kim, C.S.; Koo, B.; Yoo, C.G. Tandem conversion of lignin to catechols via demethylation and catalytic hydrogenolysis. Ind. Crops Prod. 2021, 159, 113095. [Google Scholar] [CrossRef]
- Venkatesagowda, B. Enzymatic Kraft lignin demethylation and fungal O-demethylases like vanillate-O-demethylase and syringate O-demethylase catalyzed catechol-Fe3+ complexation method. J. Microbiol. Methods 2018, 152, 126–134. [Google Scholar] [CrossRef]
- Studenik, S.; Vogel, M.; Diekert, G. Characterization of an O-demethylase of Desulfitobacterium hafniense DCB-2. J. Bacteriol. 2012, 194, 3317–3326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashtan-Kandybovich, I.; Venkatesagowda, B.; Barbosa, A.M.; Malek, L.; Dekker, R.F. Modification of Kraft lignin by biological demethylation. J-FOR 2012, 2, 16–27. [Google Scholar]
- Venkatesagowda, B. Enzymatic demethylation of lignin for potential biobased polymer applications. Fungal Biol. Rev. 2019, 33, 190–224. [Google Scholar] [CrossRef]
- Ibrahim, V.; Mendoza, L.; Mamo, G.; Hatti-Kaul, R. Blue laccase from Galerina sp.: Properties and potential for Kraft lignin demethylation. Process Biochem. 2011, 46, 379–384. [Google Scholar] [CrossRef]
- Ma, H.; Li, T.; Wu, S.; Zhang, X. Demethylation of a methoxy group to inhibit repolymerization during alkaline lignin pyrolysis. Fuel 2021, 286, 119394. [Google Scholar] [CrossRef]
- Song, Y.; Wang, Z.; Yan, N.; Zhang, R.; Li, J. Demethylation of wheat straw alkali lignin for application in phenol formaldehyde adhesives. Polymers 2016, 8, 209. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Eberhardt, T.L.; Wang, C.; Gao, S.; Pan, H. Demethylation of alkali lignin with halogen acids and its application to phenolic resins. Polymers 2019, 11, 1771. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Zeng, G.; Huang, F.; Kosa, M.; Huang, D.; Ragauskas, A.J. Bioconversion of oxygen-pretreated Kraft lignin to microbial lipid with oleaginous Rhodococcus opacus DSM 1069. Green Chem. 2015, 17, 2784–2789. [Google Scholar] [CrossRef]
- Kosa, M.; Ragauskas, A.J. Lignin to lipid bioconversion by oleaginous Rhodococci. Green Chem. 2013, 15, 2070–2074. [Google Scholar] [CrossRef]
- Liu, Z.-H.; Xie, S.; Lin, F.; Jin, M.; Yuan, J.S. Combinatorial pretreatment and fermentation optimization enabled a record yield on lignin bioconversion. Biotechnol. Biofuels 2018, 11, 21. [Google Scholar] [CrossRef]
- Spence, E.M.; Calvo-Bado, L.; Mines, P.; Bugg, T.D. Metabolic engineering of Rhodococcus jostii RHA1 for production of pyridine-dicarboxylic acids from lignin. Microb. Cell Factories 2021, 20, 15. [Google Scholar] [CrossRef] [PubMed]
- Mallinson, S.J.; Machovina, M.M.; Silveira, R.L.; Garcia-Borràs, M.; Gallup, N.; Johnson, C.W.; Allen, M.D.; Skaf, M.S.; Crowley, M.F.; Neidle, E.L. A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion. Nat. Commun. 2018, 9, 2487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.-H.; Hao, N.; Wang, Y.-Y.; Dou, C.; Lin, F.; Shen, R.; Bura, R.; Hodge, D.B.; Dale, B.E.; Ragauskas, A.J. Transforming biorefinery designs with ‘Plug-In Processes of Lignin’to enable economic waste valorization. Nat. Commun. 2021, 12, 3912. [Google Scholar] [CrossRef]
- Clarkson, S.M.; Giannone, R.J.; Kridelbaugh, D.M.; Elkins, J.G.; Guss, A.M.; Michener, J.K. Construction and optimization of a heterologous pathway for protocatechuate catabolism in Escherichia coli enables bioconversion of model aromatic compounds. Appl. Environ. Microbiol. 2017, 83, e01313-17. [Google Scholar] [CrossRef] [Green Version]
- Contreras-Jácquez, V.; Rodríguez-González, J.; Mateos-Díaz, J.C.; Valenzuela-Soto, E.M.; Asaff-Torres, A. Differential activation of ferulic acid catabolic pathways of Amycolatopsis sp. ATCC 39116 in submerged and surface cultures. Appl. Biochem. Biotechnol. 2020, 192, 494–516. [Google Scholar] [CrossRef]
- Wu, W.; Liu, F.; Singh, S. Toward engineering E. coli with an autoregulatory system for lignin valorization. Proc. Natl. Acad. Sci. USA 2018, 115, 2970–2975. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Lin, L.; Dong, J.; Ling, J.; Wang, W.; Wang, H.; Zhang, Z.; Yu, X. Simultaneous improvements of Pseudomonas cell growth and polyhydroxyalkanoate production from a lignin derivative for lignin-consolidated bioprocessing. Appl. Environ. Microbiol. 2018, 84, e01469-18. [Google Scholar] [CrossRef] [Green Version]
- Becker, J.; Kuhl, M.; Kohlstedt, M.; Starck, S.; Wittmann, C. Metabolic engineering of Corynebacterium glutamicum for the production of cis, cis-muconic acid from lignin. Microb. Cell Factories 2018, 17, 115. [Google Scholar] [CrossRef]
- Borrero-López, A.M.; Blánquez, A.; Valencia, C.; Hernández, M.; Arias, M.a.E.; Eugenio, M.E.; Fillat, Ú.; Franco, J.M. Valorization of soda lignin from wheat straw solid-state fermentation: Production of oleogels. ACS Sustain. Chem. Eng. 2018, 6, 5198–5205. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, L.; Wang, Z.; Zeng, Y.; Yang, N.; Liu, M.; Zhao, Y.; Zheng, Y. Lignin Biodegradation and Its Valorization. Fermentation 2022, 8, 366. https://doi.org/10.3390/fermentation8080366
Cui L, Wang Z, Zeng Y, Yang N, Liu M, Zhao Y, Zheng Y. Lignin Biodegradation and Its Valorization. Fermentation. 2022; 8(8):366. https://doi.org/10.3390/fermentation8080366
Chicago/Turabian StyleCui, Lingwei, Zheyi Wang, Yan Zeng, Niping Yang, Mengshuang Liu, Youxi Zhao, and Yanning Zheng. 2022. "Lignin Biodegradation and Its Valorization" Fermentation 8, no. 8: 366. https://doi.org/10.3390/fermentation8080366
APA StyleCui, L., Wang, Z., Zeng, Y., Yang, N., Liu, M., Zhao, Y., & Zheng, Y. (2022). Lignin Biodegradation and Its Valorization. Fermentation, 8(8), 366. https://doi.org/10.3390/fermentation8080366





