Growth and Metabolism of Lacticaseibacillus casei and Lactobacillus kefiri Isolated from Qymyz, a Traditional Fermented Central Asian Beverage
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria Identification
2.2. Growth Medium Composition
2.3. Growth Experiments
2.3.1. Effect of Temperature
2.3.2. Effect of Carbohydrate Source and pH
2.3.3. Plate-Counting Methods for Calibration Curve
2.4. Chemical Analyses
2.5. Mathematical Modeling
2.5.1. Microorganism Growth
2.5.2. Limiting Substrate
2.5.3. Metabolite Production Modeling
2.5.4. Model Fitting and Estimation of Model Parameters
3. Results and Discussion
3.1. Bacterial Identification and General Metabolism Features
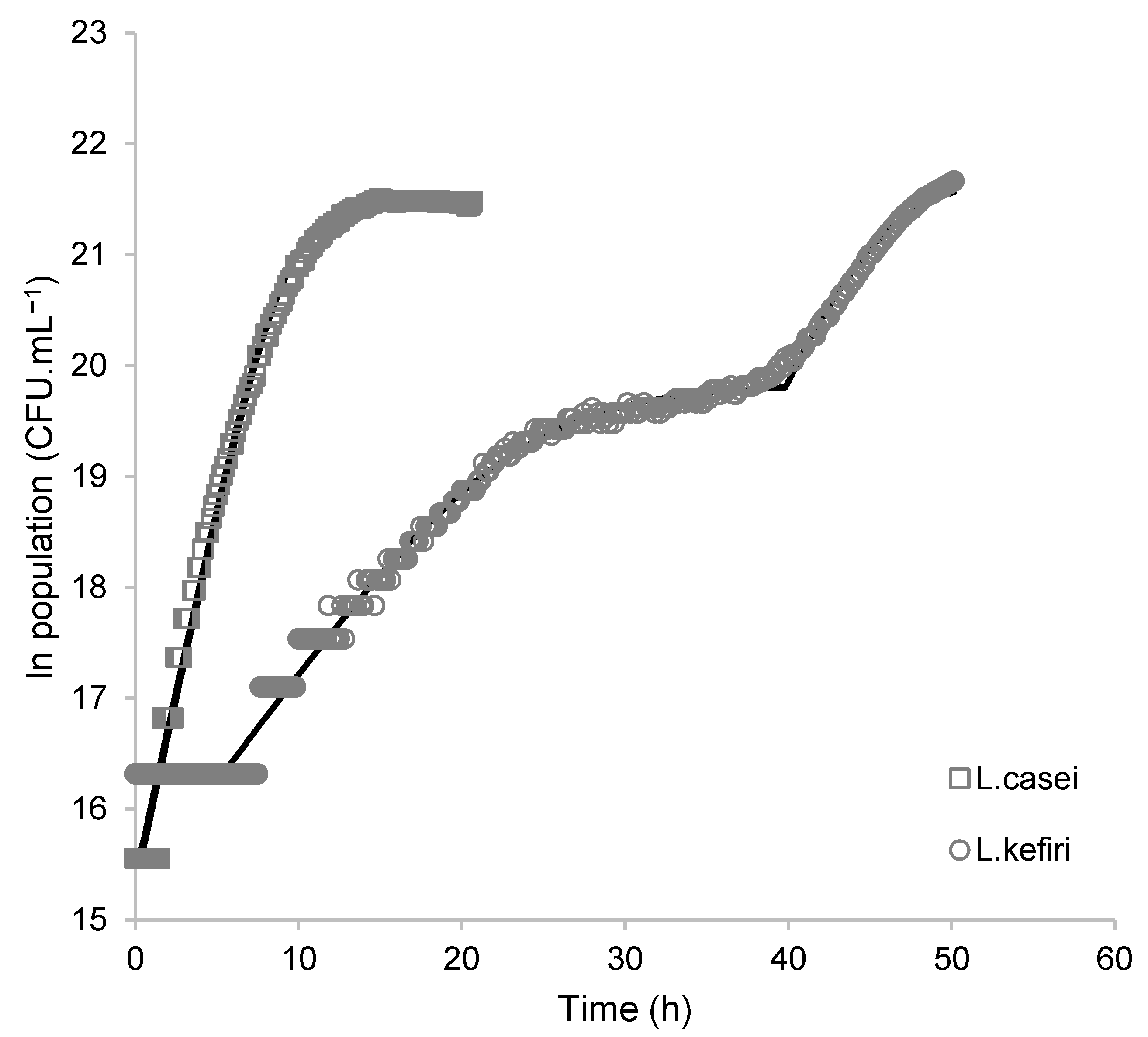
3.2. Growth Characteristic of L. casei and L. kefiri
3.2.1. Microorganism Growth Modeling
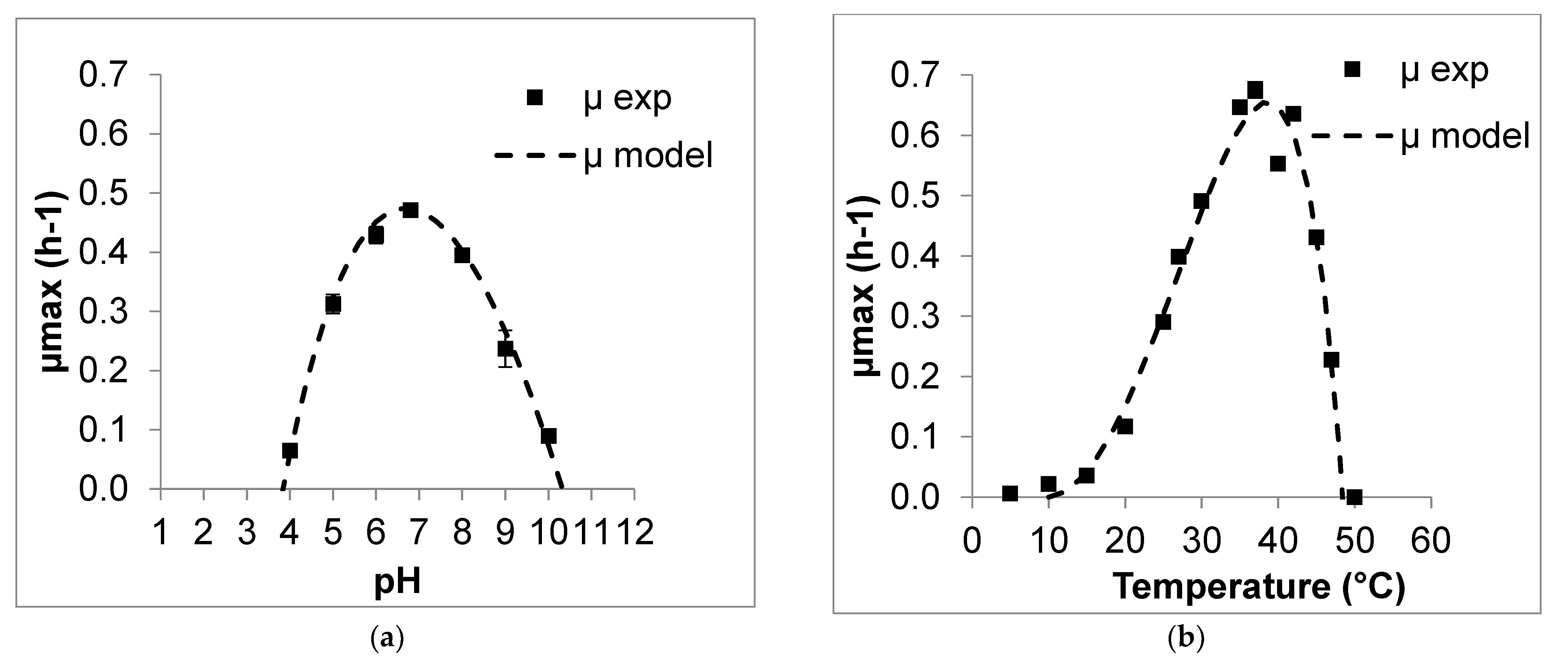
3.2.2. Modeling Effect of pH and Temperature for L. casei
3.2.3. Modeling Effect of pH and Temperature for L. kefiri
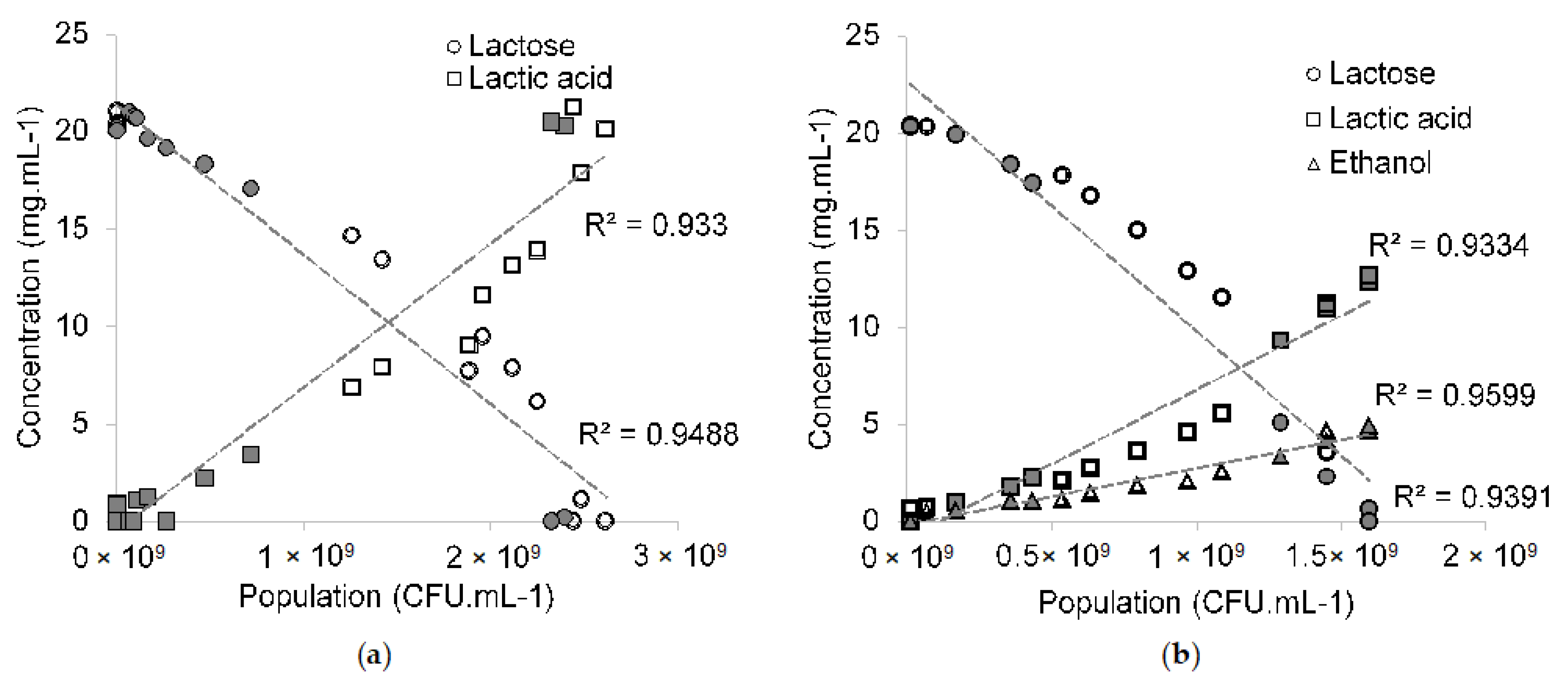
3.3. Metabolite Production
3.4. Resulting Parameters of Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rizzoli, R.; Biver, E. Effects of fermented milk products on bone. Calcif. Tissue Int. 2018, 102, 489–500. [Google Scholar] [CrossRef]
- Beena Divya, J.; Kulangara Varsha, K.; Madhavan Nampoothiri, K.; Ismail, B.; Pandey, A. Probiotic fermented foods for health benefits. Eng. Life Sci. 2012, 12, 377–390. [Google Scholar] [CrossRef]
- Rakhmanova, A.; Wang, T.; Xing, G.; Ma, L.; Hong, Y.; Lu, Y.; Xin, L.; Xin, W.; Zhu, Q.; Lü, X. Isolation and identification of microorganisms in Kazakhstan koumiss and their application in preparing cow-milk koumiss. J. Dairy Sci. 2021, 104, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Hosino, B.; Komiyama, H.; Uehara, A.; Nurtazin, S. Study on Production and Properties of Kumiss of Herders in Mongolian Dry Steppe. J. Arid Land Stud. 2014, 24, 3. [Google Scholar]
- Wu, Y.; Li, Y.; Gesudu, Q.; Zhang, J.; Sun, Z.; Halatu, H.; Menghe, B.; Liu, W. Bacterial composition and function during fermentation of Mongolia koumiss. Food Sci. Nutr. 2021, 9, 4146–4155. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Yu, J.; Hou, Q.; Hui, W.; Liu, W.; Kwok, L.-Y.; Menghe, B.; Sun, T.; Zhang, H.; Zhang, W. A Perspective Study of Koumiss Microbiome by Metagenomics Analysis Based on Single-Cell Amplification Technique. Front. Microbiol. 2017, 8, 165. [Google Scholar] [CrossRef] [PubMed]
- Dzialo, M.C.; Park, R.; Steensels, J.; Lievens, B.; Verstrepen, K.J. Physiology, ecology and industrial applications of aroma formation in yeast. FEMS Microbiol. Rev. 2017, 41, S95–S128. [Google Scholar] [CrossRef] [PubMed]
- Mu, Z.; Yang, X.; Yuan, H. Detection and identification of wild yeast in Koumiss. Food Microbiol. 2012, 31, 301–308. [Google Scholar] [CrossRef]
- Leporq, B.; Membré, J.-M.; Dervin, C.; Buche, P.; Guyonnet, J. The “Sym’Previus” software, a tool to support decisions to the foodstuff safety. Int. J. Food Microbiol. 2005, 100, 231–237. [Google Scholar] [CrossRef]
- Sumner, J.; Krist, K. The use of predictive microbiology by the Australian meat industry. Int. J. Food Microbiol. 2002, 73, 363–366. [Google Scholar] [CrossRef][Green Version]
- Leroy, F.; Degeest, B.; De Vuyst, L. A novel area of predictive modelling: Describing the functionality of beneficial microorganisms in foods. Int. J. Food Microbiol. 2002, 73, 251–259. [Google Scholar] [CrossRef]
- Kouamé, C.; Loiseau, G.; Grabulos, J.; Boulanger, R.; Mestres, C. Development of a model for the alcoholic fermentation of cocoa beans by a Saccharomyces cerevisiae strain. Int. J. Food Microbiol. 2021, 337, 108917. [Google Scholar] [CrossRef]
- Mestres, C.; Munanga, B.d.J.C.; Grabulos, J.; Loiseau, G. Modeling mixed fermentation of gowé using selected Lactobacillus plantarum and Pichia kluyveri strains. Food Microbiol. 2019, 84, 103242. [Google Scholar] [CrossRef]
- Infantes, D.; del Campo, A.G.; Villaseñor, J.; Fernández, F. Kinetic model and study of the influence of pH, temperature and undissociated acids on acidogenic fermentation. Biochem. Eng. J. 2012, 66, 66–72. [Google Scholar] [CrossRef]
- Béal, C.; Spinnler, H.E.; Corrieu, G. Comparison of growth, acidification and productivity of pure and mixed cultures of Streptococcus salivarius subsp. thermophilus 404 and Lactobacillus delbrueckii subsp. bulgaricus 398. Appl. Microbiol. Biotechnol. 1994, 41, 95–98. [Google Scholar] [CrossRef]
- Committee for Standardization, Metrology and Certification of the Ministry of Energy, Industry and Trade of the Republic of Kazakhstan. ST RK 1004-98. In Natural koumiss. Specifications; SMCC: Almaty, Kazakhstan, 1999. [Google Scholar]
- Kondybayev, A.; Loiseau, G.; Achir, N.; Mestres, C.; Konuspayeva, G. Fermented mare milk product (Qymyz, Koumiss). Int. Dairy J. 2021, 119, 105065. [Google Scholar] [CrossRef]
- Clarridge, J.E., 3rd. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef]
- Augustin, J.-C.; Rosso, L.; Carlier, V. Estimation of temperature dependent growth rate and lag time of Listeria monocytogenes by optical density measurements. J. Microbiol. Methods 1999, 38, 137–146. [Google Scholar] [CrossRef]
- Munanga, B.D.J.C.; Loiseau, G.; Grabulos, J.; Mestres, C. Modeling Lactic Fermentation of Gowé Using Lactobacillus Starter Culture. Microorganisms 2016, 4, 44. [Google Scholar] [CrossRef]
- Delhalle, L.; Daube, G.; Adolphe, Y.; Crevecoeur, S.; Clinquart, A. Les modèles de croissance en microbiologie prévisionnelle pour la maitrise de la sécurité des aliments (synthèse bibliographique). Biotechnol. Agron. Soc. Environ. 2012, 16, 13. [Google Scholar]
- Zwietering, M.H.; Wijtzes, T.; Rombouts, F.M.; Riet, K. A decision support system for prediction of microbial spoilage in foods. J. Ind. Microbiol. 1993, 12, 324–329. [Google Scholar] [CrossRef]
- Rosso, L.; Lobry, J.R.; Bajard, S.; Flandrois, J.P. Convenient Model to Describe the Combined Effects of Temperature and pH on Microbial Growth. Appl. Environ. Microbiol. 1995, 61, 610–616. [Google Scholar] [CrossRef]
- Monod, J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef]
- Willems, R.J.; Top, J.; Marga van Santen, D.; Coque, T.M.; Baquero, F.; Grundmann, H.; Bonten, M.J. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 2005, 11, 821. [Google Scholar] [CrossRef] [PubMed]
- Berditsch, M.; Afonin, S.; Ulrich, A.S. The ability of Aneurinibacillus migulanus (Bacillus brevis) to produce the antibiotic gramicidin S is correlated with phenotype variation. Appl. Environ. Microbiol. 2007, 73, 6620–6628. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, J.M.; Munekata, P.E.; Dominguez, R.; Pateiro, M.; Saraiva, J.A.; Franco, D. Main Groups of Microorganisms of Relevance for Food Safety and Stability. In Innovative Technologies for Food Preservation; Elsevier: Amsterdam, The Netherlands, 2018; pp. 53–107. [Google Scholar]
- Ibrahim, S.A. Lactic Acid Bacteria: Lactobacillus spp.: Other Species. In Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Alexandraki, V.; Kazou, M.; Angelopoulou, A.; Arena, M.P.; Capozzi, V.; Russo, P.; Fiocco, D.; Spano, G.; Papadimitriou, K.; Tsakalidou, E. Chapter 6—The Microbiota of Non-cow Milk and Products. In Non-Bovine Milk and Milk Products; Tsakalidou, E., Papadimitriou, K., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 117–159. [Google Scholar]
- Baubekova, A.; Akhmetsadykova, S.; Kondybayev, A.; Faye, B.; Konuspayeva, G. Volatile organic compounds profile of Lactobacillus casei and Streptococcus thermophilus in fermented mare milk of Kazakhstan. Int. J. Biol. Chem. 2018, 11, 28–35. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Kim, D.-H.; Jeong, D.; Kang, I.-B.; Kim, H.; Song, K.-Y.; Seo, K.-H. Dual function of Lactobacillus kefiri DH5 in preventing high-fat-diet-induced obesity: Direct reduction of cholesterol and upregulation of PPAR-α in adipose tissue. Mol. Nutr. Food Res. 2017, 61, 1700252. [Google Scholar] [CrossRef]
- Kandler, O.; Kunath, P. Lactobacillus kefir sp.nov., a component of the microflora of Kefir. Syst. Appl. Microbiol. 1983, 4, 286–294. [Google Scholar] [CrossRef]
- Alvarez, M.M.; Aguirre-Ezkauriatza, E.J.; Ramírez-Medrano, A.; Rodríguez-Sánchez, Á. Kinetic analysis and mathematical modeling of growth and lactic acid production of Lactobacillus casei var. rhamnosus in milk whey. J. Dairy Sci. 2010, 93, 5552–5560. [Google Scholar] [CrossRef]
- Møller, C.O.d.A.; Christensen, B.B.; Rattray, F.P. Modelling the biphasic growth of non-starter lactic acid bacteria on starter-lysate as a substrate. Int. J. Food Microbiol. 2021, 337, 108937. [Google Scholar] [CrossRef]
- Shen, J.-P.; Chou, C.-F. Morphological plasticity of bacteria—Open questions. Biomicrofluidics 2016, 10, 031501. [Google Scholar] [CrossRef]
- Rajoka, M.S.R.; Mehwish, H.M.; Fang, H.; Padhiar, A.A.; Zeng, X.; Khurshid, M.; He, Z.; Zhao, L. Characterization and anti-tumor activity of exopolysaccharide produced by Lactobacillus kefiri isolated from Chinese kefir grains. J. Funct. Foods 2019, 63, 103588. [Google Scholar] [CrossRef]
- Montanari, G.; Zambonelli, C.; Grazia, L.; Kamesheva, G.K.; Shigaeva, M.K. Saccharomyces unisporus as the principal alcoholic fermentation microorganism of traditional koumiss. J. Dairy Res. 1996, 63, 327. [Google Scholar] [CrossRef]

| Temperature (°C) | Phase 1 | Phase 2 | µ2/µ1 | ||||
|---|---|---|---|---|---|---|---|
| λ (h) | µmax (h−1) | Nmax (CFU·mL−1) | λ h | µmax (h−1) | Nmax (CFU·mL−1) | ||
| 20 | 26.30 ± 0.38 | 0.08 ± 0.01 | 3.83·× 108 | 55.17 ± 0.01 | 0.02 ± 0.00 | 9·× 108 | 0.28 |
| 27 | 10.55 ± 0.09 | 0.20 ± 0.01 | 6.10·× 108 | 44.33 ± 0.01 | 0.03 ± 0.00 | 1.32·× 109 | 0.15 |
| 30 | 1.91 ± 0.08 | 0.25 ± 0.01 | 7.60·× 108 | 28.66 ± 0.01 | 0.07 ± 0.01 | 1.32·× 109 | 0.28 |
| 35 | 3.69 ± 0.09 | 0.23 ± 0.01 | 7.16·× 108 | 35.83 ± 0.01 | 0.07 ± 0.01 | 1.60·× 109 | 0.30 |
| 37 | 11.57 ± 0.11 | 0.20 ± 0.01 | 4.10·× 108 | 46.09 ± 0.10 | 0.35 ± 0.01 | 2.67·× 109 | 1.75 |
| 40 | 0.00 ± 0.00 | 0.16 ± 0.01 | 3.28·× 108 | 42.49 ± 0.47 | 0.64 ± 0.20 | 1.19·× 109 | 4.00 |
| Parameters | Lacticaseibacillus casei (Value ± SD/SE) | 1st Phase of Lactobacillus kefiri (Value ± SD/SE) |
|---|---|---|
| pH min | 3.84 ± 0.12 | 3.35 ± 0.12 |
| pH max | 10.33 ± 0.18 | 7.64 ± 0.05 |
| pH opt | 6.69 ± 0.20 | 5.93 ± 0.08 |
| T min (°C) | 9.66 ± 1.18 | 8.10 ± 1.27 |
| T max (°C) | 48.48 ± 0.13 | 44.94 ± 0.31 |
| T opt (°C) | 38.63 ± 0.32 | 33.15 ± 0.53 |
| µopt (h−1) | 0.66 ± 0.01 | 0.29 ± 0.01 |
| Glucose µopt (h−1) | 0.49 ± 0.01 | 0.26 ± 0.01 |
| Glucose Ks (g L−1) | 0.43 ± 0.07 | 0.21 ± 0.03 |
| Galactose µopt (h−1) | 0.47 ± 0.01 | 0.27 ± 0.01 |
| Galactose Ks (g L−1) | 0.19 ± 0.05 | 0.18 ± 0.05 |
| Lactose µopt (h−1) | 0.43 ± 0.01 | 0.24 ± 0.01 |
| Lactose Ks (g L−1) | 0.12 ± 0.04 | 0.72 ± 0.57 |
| Lactic Acid Production * (10−9 mg·CFU−1) | 7.36 ± 0.43 | 7.64 ± 0.54 |
| Ethanol Production* (10−9 mg·CFU−1) | 0 | 2.90 ± 0.15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kondybayev, A.; Konuspayeva, G.; Strub, C.; Loiseau, G.; Mestres, C.; Grabulos, J.; Manzano, M.; Akhmetsadykova, S.; Achir, N. Growth and Metabolism of Lacticaseibacillus casei and Lactobacillus kefiri Isolated from Qymyz, a Traditional Fermented Central Asian Beverage. Fermentation 2022, 8, 367. https://doi.org/10.3390/fermentation8080367
Kondybayev A, Konuspayeva G, Strub C, Loiseau G, Mestres C, Grabulos J, Manzano M, Akhmetsadykova S, Achir N. Growth and Metabolism of Lacticaseibacillus casei and Lactobacillus kefiri Isolated from Qymyz, a Traditional Fermented Central Asian Beverage. Fermentation. 2022; 8(8):367. https://doi.org/10.3390/fermentation8080367
Chicago/Turabian StyleKondybayev, Askar, Gaukhar Konuspayeva, Caroline Strub, Gerard Loiseau, Christian Mestres, Joel Grabulos, Marie Manzano, Shynar Akhmetsadykova, and Nawel Achir. 2022. "Growth and Metabolism of Lacticaseibacillus casei and Lactobacillus kefiri Isolated from Qymyz, a Traditional Fermented Central Asian Beverage" Fermentation 8, no. 8: 367. https://doi.org/10.3390/fermentation8080367
APA StyleKondybayev, A., Konuspayeva, G., Strub, C., Loiseau, G., Mestres, C., Grabulos, J., Manzano, M., Akhmetsadykova, S., & Achir, N. (2022). Growth and Metabolism of Lacticaseibacillus casei and Lactobacillus kefiri Isolated from Qymyz, a Traditional Fermented Central Asian Beverage. Fermentation, 8(8), 367. https://doi.org/10.3390/fermentation8080367






