Application of Milk Permeate as an Inducer for the Production of Microbial Recombinant Lipolytic Enzymes
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Bacterial Strains and Plasmids
2.3. Culture Conditions
2.4. Induction of Recombinant Enzyme Synthesis
2.5. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
2.6. Zymography and Lipolytic Activity Assay
3. Results
3.1. Optimization of Milk Permeate (MP) Concentration
3.2. Influence of Lower Antibiotic Concentrations on Recombinant Enzyme Production
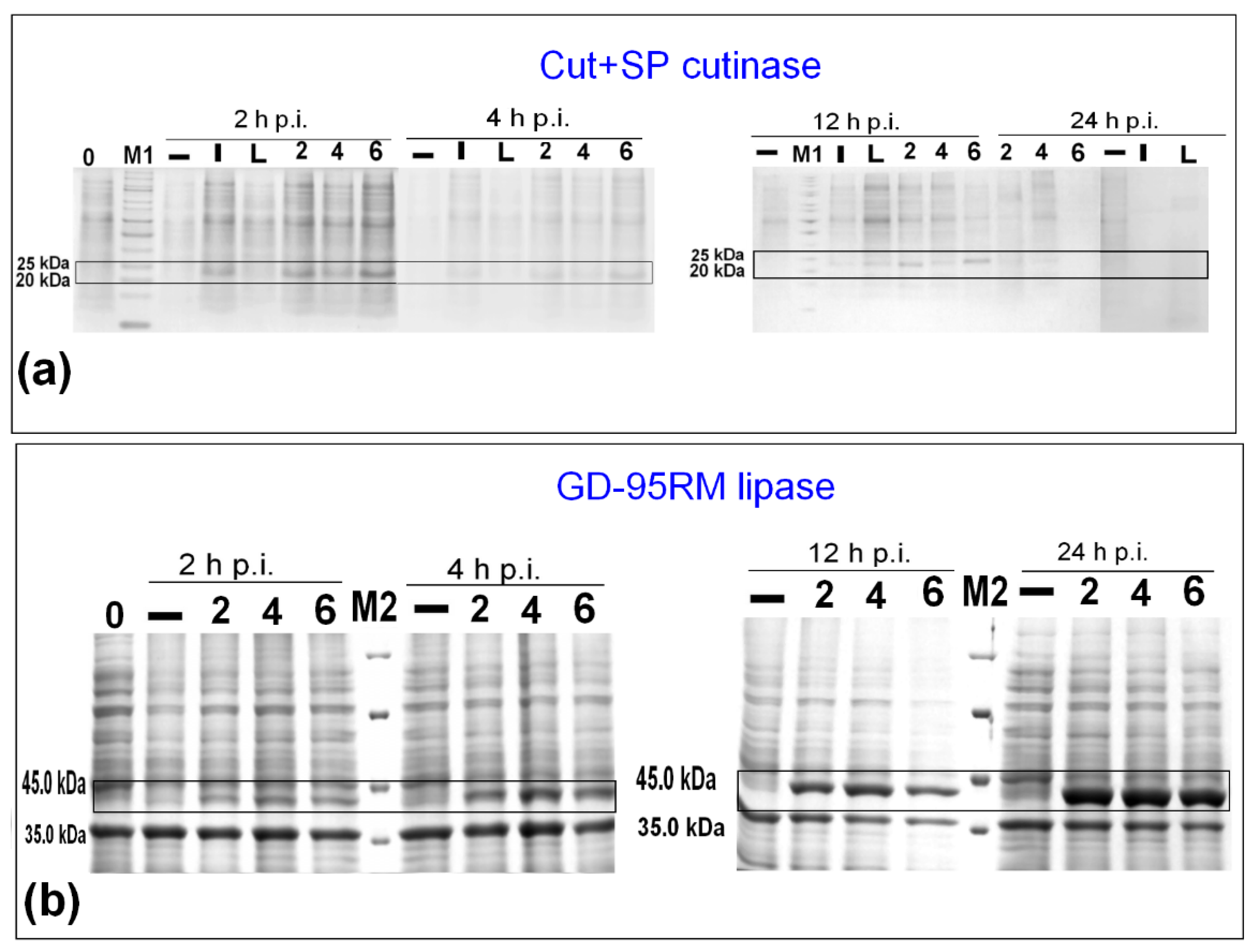
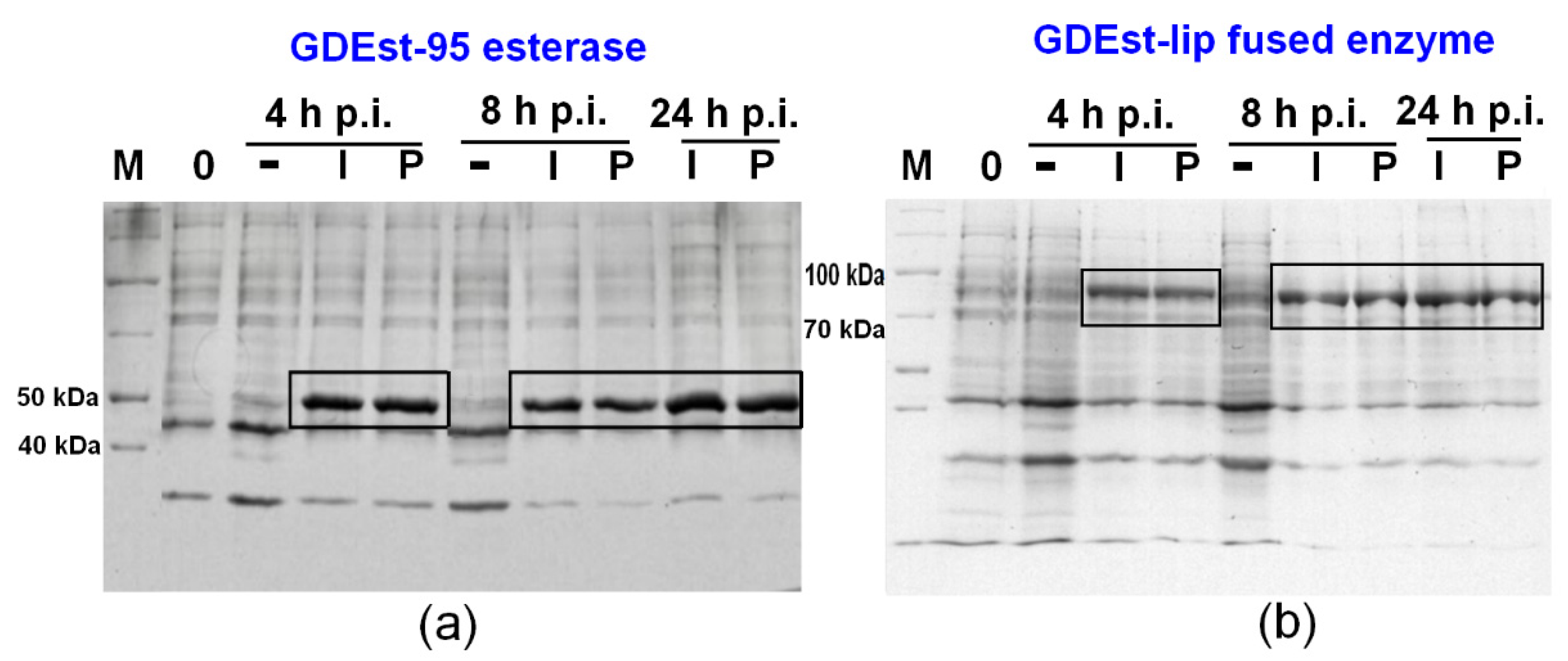
3.3. Synthesis of GDEst-lip and GDEst-95 in Low Scale
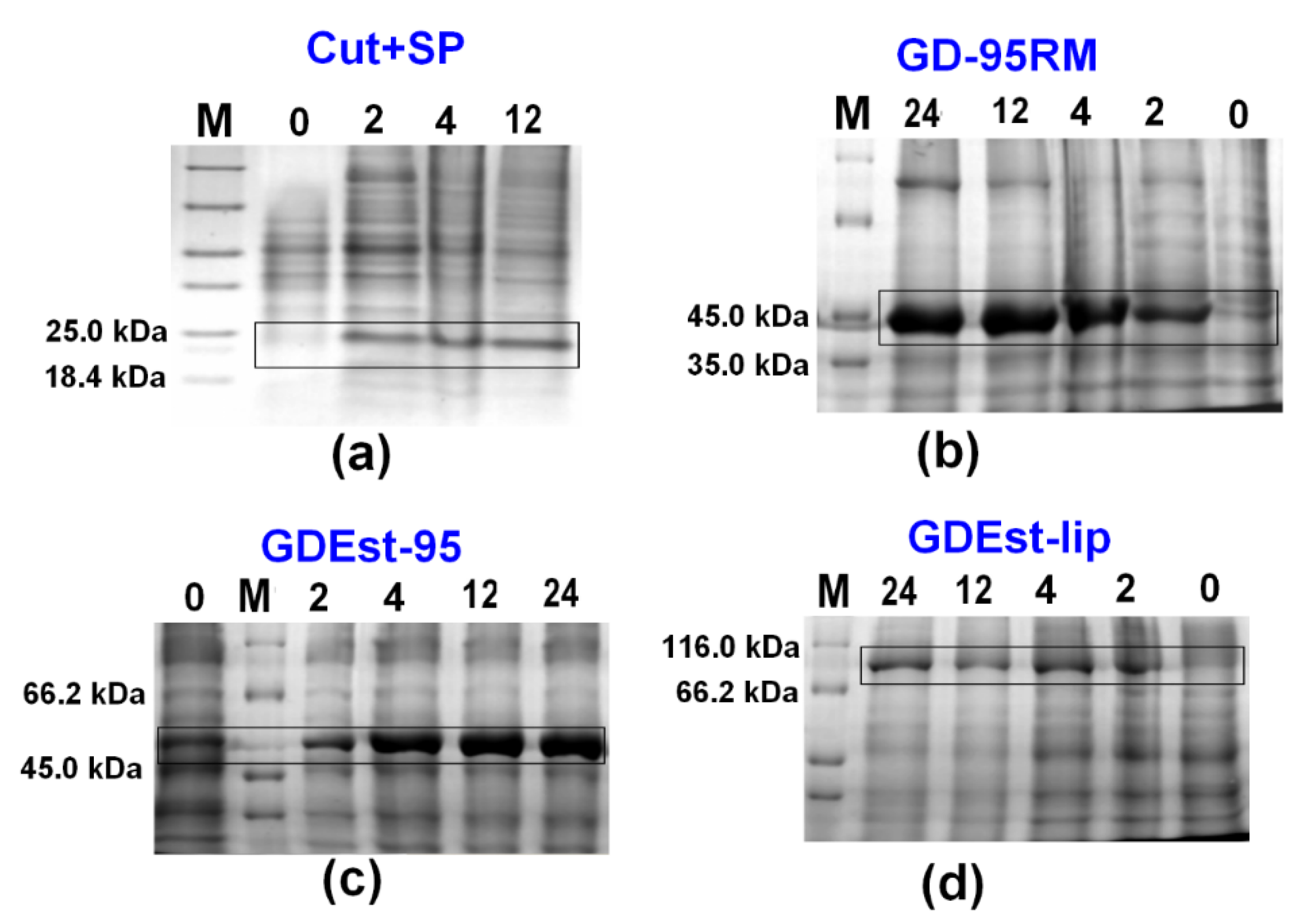
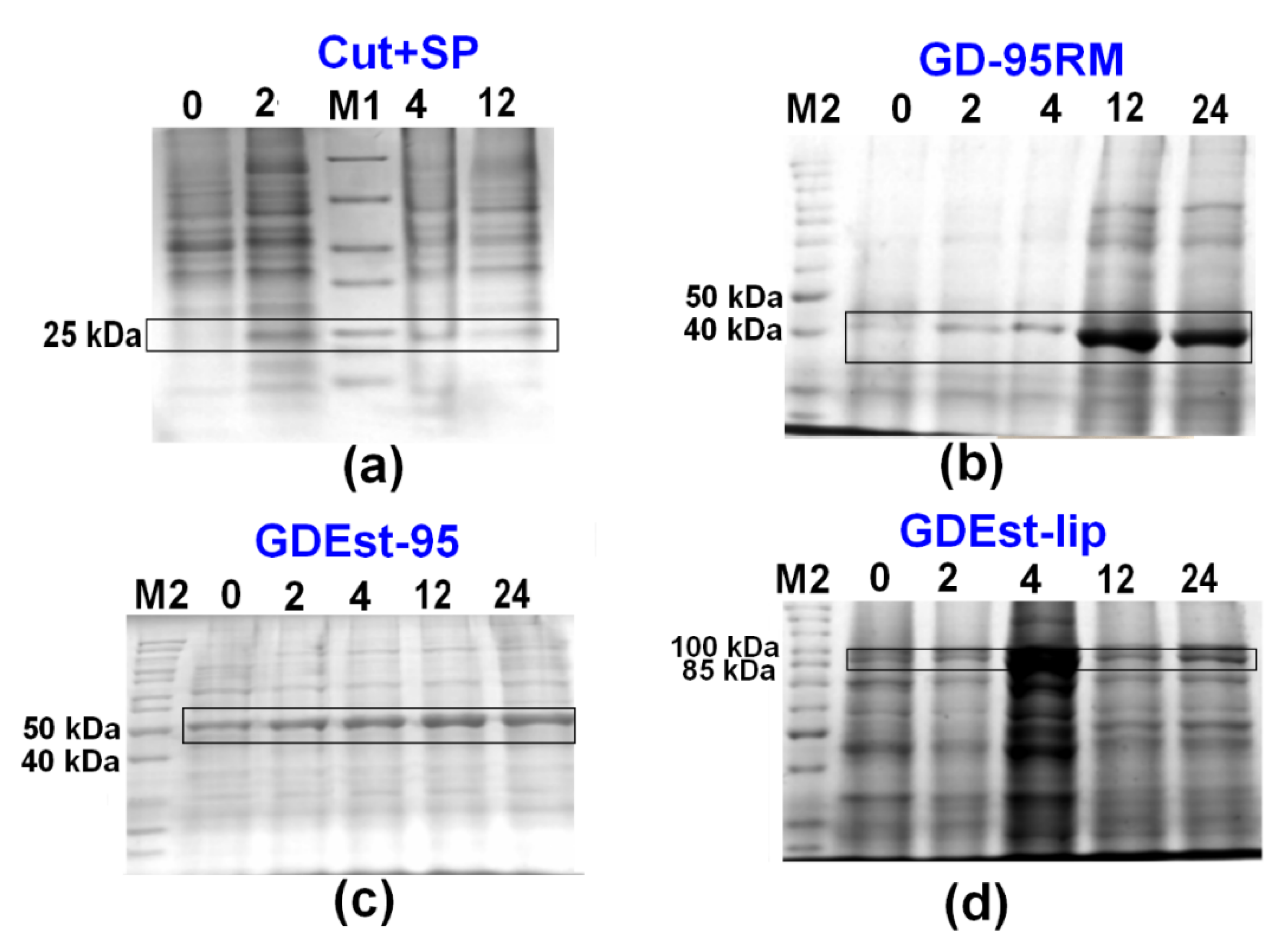
3.4. Up-Scale Synthesis of Cut+SP, GD-95RM, GDEst-lip and GDEst-95 Enzymes
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Demain, A.L.; Vaishnav, P. Production of recombinant enzymes. In Reference Module in Food Science; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar] [CrossRef]
- Liu, L.; Yang, H.; Shin, H.-D.; Chen, R.R.; Li, J.; Du, G.; Chen, J. How to achieve high-level expression of microbial enzymes. Bioengineered 2013, 4, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Hausjell, J.; Weissensteiner, J.; Molitor, C.; Halbwirth, H.; Spadiut, O. E. coli HMS174(DE3) is a sustainable alternative to BL21(DE3). Microb. Cell Factories 2018, 17, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Makino, T.; Skretas, G.; Georgiou, G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb. Cell Factories 2011, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Fakruddin, M.; Mazumdar, R.M.; Mannan, K.S.; Chowdhury, A.; Hossain, M.N. Critical Factors Affecting the Success of Cloning, Expression, and Mass Production of Enzymes by Recombinant E. coli. ISRN Biotechnol. 2013, 2013, 590587. [Google Scholar] [CrossRef] [PubMed]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed]
- Idalia, V.-M.N.; Bernardo, F. Escherichia coli as a model organism and its application in biotechnology. In Escherichia coli-Recent Advances on Physiology, Pathogenesis and Biotechnological Applications; Amidou, S., Ed.; IntchOpen: London, UK, 2017; pp. 253–274. [Google Scholar] [CrossRef]
- Castiñeiras, T.S.; Williams, S.G.; Hitchcock, A.G.; Smith, D.C. E. coli strain engineering for the production of advanced biopharmaceutical products. FEMS Microbiol. Lett. 2018, 365, fny162. [Google Scholar] [CrossRef]
- Terol, G.L.; Gallego-Jara, J.; Martínez, R.A.S.; Vivancos, A.M.; Díaz, M.C.; Puente, T.D.D. Impact of the Expression System on Recombinant Protein Production in Escherichia coli BL21. Front. Microbiol. 2021, 12, 682001. [Google Scholar] [CrossRef]
- Huang, C.-J.; Lin, H.; Yang, X. Industrial production of recombinant therapeutics in Escherichia coli and its recent advancements. J. Ind. Microbiol. Biotechnol. 2012, 39, 383–399. [Google Scholar] [CrossRef]
- Theisen, M.; Liao, J.C. Industrial biotechnology: Escherichia coli as a host. In Industrial Biotechnology: Microorganisms, 1st ed.; Wittmann, C., Liao, J.C., Eds.; Wiley-VCH Verlag GmbH & Co. KgaA: Weinheim, Germany, 2017; Volume 1, pp. 149–181. [Google Scholar] [CrossRef]
- Rosano, G.L.; Morales, E.S.; Ceccarelli, E.A. New tools for recombinant protein production in Escherichia coli: A 5-year update. Protein Sci. 2019, 28, 1412–1422. [Google Scholar] [CrossRef]
- Baeshen, M.N.; Al-Hejin, A.M.; Bora, R.S.; Ahmed, M.M.M.; Ramadan, H.A.I.; Saini, K.S.; Baeshen, N.A.; Redwan, E.M. Production of Biopharmaceuticals in E. coli: Current Scenario and Future Perspectives. J. Microbiol. Biotechnol. 2015, 25, 953–962. [Google Scholar] [CrossRef]
- Ratelade, J.; Miot, M.-C.; Johnson, E.; Betton, J.-M.; Mazodier, P.; Benaroudj, N. Production of Recombinant Proteins in the lon -Deficient BL21(DE3) Strain of Escherichia coli in the Absence of the DnaK Chaperone. Appl. Environ. Microbiol. 2009, 75, 3803–3807. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Jeong, H.; Kwon, S.-K.; Kim, J.F. Genomics, biological features, and biotechnological applications of Escherichia coli B: “is B for better?!”. In Systems Biology and Biotechnology of Escherichia coli; Lee, S.Y., Ed.; Springer: Dordrecht, The Netherlands, 2009; pp. 1–17. [Google Scholar] [CrossRef]
- Son, Y.-J.; Phue, J.-N.; Trinh, L.B.; Lee, S.J.; Shiloach, J. The role of Cra in regulating acetate excretion and osmotic tolerance in E. coli K-12 and E. coli B at high density growth. Microb. Cell Factories 2011, 10, 52. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, F.; Wang, W.; Yao, X.; Wei, D.; Cheng, H.; Deng, Z. Improving the Expression of Recombinant Proteins in E. coli BL21 (DE3) under Acetate Stress: An Alkaline pH Shift Approach. PLoS ONE 2014, 9, e112777. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Tegel, H.; Ottosson, J.; Hober, S. Enhancing the protein production levels in Escherichia coli with a strong promoter. FEBS J. 2010, 278, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Durani, V.; Sullivan, B.J.; Magliery, T.J. Simplifying protein expression with ligation-free, traceless and tag-switching plasmids. Protein Expr. Purif. 2012, 85, 9–17. [Google Scholar] [CrossRef]
- Briand, L.; Marcion, G.; Kriznik, A.; Heydel, J.-M.; Artur, Y.; Garrido, C.; Seigneuric, R.; Neiers, F. A self-inducible heterologous protein expression system in Escherichia coli. Sci. Rep. 2016, 6, 33037. [Google Scholar] [CrossRef] [PubMed]
- Marbach, A.; Bettenbrock, K. lac operon induction in Escherichia coli: Systematic comparison of IPTG and TMG induction and influence of the transacetylase LacA. J. Biotechnol. 2012, 157, 82–88. [Google Scholar] [CrossRef]
- Khani, M.-H.; Bagheri, M. Skimmed milk as an alternative for IPTG in induction of recombinant protein expression. Protein Expr. Purif. 2020, 170, 105593. [Google Scholar] [CrossRef]
- Donovan, R.S.; Robinson, C.W.; Glick, B.R. Review: Optimizing inducer and culture conditions for expression of foreign proteins under the control of thelac promoter. J. Ind. Microbiol. Biotechnol. 1996, 16, 145–154. [Google Scholar] [CrossRef]
- Dvorak, P.; Chrast, L.; Nikel, P.I.; Fedr, R.; Soucek, K.; Sedlackova, M.; Chaloupkova, R.; de Lorenzo, V.; Prokop, Z.; Damborsky, J. Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway. Microb. Cell Factories 2015, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Haddadin, F.T.; Harcum, S.W. Transcriptome profiles for high-cell-density recombinant and wild-type Escherichia coli. Biotechnol. Bioeng. 2005, 90, 127–153. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.-S.; Kruse, J.; Pohl, N.L. Synthesis of Isobutyl-C-galactoside (IBCG) as an Isopropylthiogalactoside (IPTG) Substitute for Increased Induction of Protein Expression. Org. Lett. 2003, 5, 1781–1783. [Google Scholar] [CrossRef] [PubMed]
- Menzella, H.G.; Ceccarelli, E.A.; Gramajo, H.C. Novel escherichia coli strain allows efficient recombinant protein production using lactose as inducer. Biotechnol. Bioeng. 2003, 82, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Woyski, D.; Cupp-Vickery, J.R. Enhanced Expression of Cytochrome P450s from lac-Based Plasmids Using Lactose as the Inducer. Arch. Biochem. Biophys. 2001, 388, 276–280. [Google Scholar] [CrossRef]
- Striedner, G.; Cserjan-Puschmann, M.; Pötschacher, F.; Bayer, K. Tuning the Transcription Rate of Recombinant Protein in Strong Escherichia coli Expression Systems through Repressor Titration. Biotechnol. Prog. 2003, 19, 1427–1432. [Google Scholar] [CrossRef]
- Kotik, M.; Kočanová, M.; Marešová, H.; Kyslík, P. High-level expression of a fungal pyranose oxidase in high cell-density fed-batch cultivations of Escherichia coli using lactose as inducer. Protein Expr. Purif. 2004, 36, 61–69. [Google Scholar] [CrossRef]
- Chandra, P.; Enespa; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Fact. 2020, 19, 169. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Trono, D. Recombinant Lipases and Phospholipases and Their Use as Biocatalysts for Industrial Applications. Int. J. Mol. Sci. 2015, 16, 20774–20840. [Google Scholar] [CrossRef]
- Yao, W.; Liu, K.; Liu, H.; Jiang, Y.; Wang, R.; Wang, W.; Wang, T. A Valuable Product of Microbial Cell Factories: Microbial Lipase. Front. Microbiol. 2021, 12, 743377. [Google Scholar] [CrossRef]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzym. Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Ramnath, L.; Sithole, B.; Govinden, R. Classification of lipolytic enzymes and their biotechnological applications in the pulping industry. Can. J. Microbiol. 2017, 63, 179–192. [Google Scholar] [CrossRef]
- Vanleeuw, E.; Winderickx, S.; Thevissen, K.; Lagrain, B.; Dusselier, M.; Cammue, B.P.A.; Sels, B.F. Substrate-Specificity of Candida rugosa Lipase and Its Industrial Application. ACS Sustain. Chem. Eng. 2019, 7, 15828–15844. [Google Scholar] [CrossRef]
- Kumar, A.; Gudiukaite, R.; Gricajeva, A.; Sadauskas, M.; Malunavicius, V.; Kamyab, H.; Sharma, S.; Sharma, T.; Pant, D. Microbial lipolytic enzymes—promising energy-efficient biocatalysts in bioremediation. Energy 2019, 192, 116674. [Google Scholar] [CrossRef]
- Vishnoi, N.; Dixit, S.; Mishra, J. Microbial Lipases and Their Versatile Applications. In Microbial Enzymes: Roles and Applications in Industries. Microorganisms for Sustainability; Arora, N., Mishra, J., Mishra, V., Eds.; Springer: Singapore, 2020; Volume 11, pp. 207–230. [Google Scholar] [CrossRef]
- Contesini, F.J.; Davanço, M.G.; Borin, G.P.; Vanegas, K.G.; Cirino, J.P.G.; De Melo, R.R.; Mortensen, U.H.; Hildén, K.; Campos, D.R.; Carvalho, P.D.O. Advances in Recombinant Lipases: Production, Engineering, Immobilization and Application in the Pharmaceutical Industry. Catalysts 2020, 10, 1032. [Google Scholar] [CrossRef]
- Oh, C.; Kim, T.D.; Kim, K.K. Carboxylic Ester Hydrolases in Bacteria: Active Site, Structure, Function and Application. Crystals 2019, 9, 597. [Google Scholar] [CrossRef]
- Johan, U.U.M.; Rahman, R.N.Z.R.A.; Kamarudin, N.H.A.; Ali, M.S.M. An integrated overview of bacterial carboxylesterase: Structure, function and biocatalytic applications. Colloids Surfaces B Biointerfaces 2021, 205, 111882. [Google Scholar] [CrossRef] [PubMed]
- Baba, A.; Yoshioka, T. Characterization of chemo- and regioselectivity in enzyme-catalyzed consecutive hydrolytic deprotection of methyl acetyl derivatives of 1-β-O-acyl glucuronides. J. Mol. Catal. B Enzym. 2011, 69, 74–82. [Google Scholar] [CrossRef]
- Jeon, J.H.; Kim, S.-J.; Lee, H.S.; Cha, S.-S.; Lee, J.H.; Yoon, S.-H.; Koo, B.-S.; Lee, C.-M.; Choi, S.H.; Lee, S.H.; et al. Novel Metagenome-Derived Carboxylesterase That Hydrolyzes β-Lactam Antibiotics. Appl. Environ. Microbiol. 2011, 77, 7830–7836. [Google Scholar] [CrossRef]
- Romano, D.; Bonomi, F.; de Mattos, M.C.; Fonseca, T.D.S.; Oliveira, M.D.C.F.D.; Molinari, F. Esterases as stereoselective biocatalysts. Biotechnol. Adv. 2015, 33, 547–565. [Google Scholar] [CrossRef]
- Bollinger, A.; Molitor, R.; Thies, S.; Koch, R.; Coscolín, C.; Ferrer, M.; Jaeger, K.-E. Organic-Solvent-Tolerant Carboxylic Ester Hydrolases for Organic Synthesis. Appl. Environ. Microbiol. 2020, 86, e00106-20. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Jeong, G.S.; Lee, H.W.; Kim, H. Biochemical characterization of a family IV esterase with R-form enantioselectivity from a compost metagenomic library. Appl. Biol. Chem. 2021, 64, 1–16. [Google Scholar] [CrossRef]
- Hajighasemi, M.; Nocek, B.P.; Tchigvintsev, A.; Brown, G.; Flick, R.; Xu, X.; Cui, H.; Hai, T.; Joachimiak, A.; Golyshin, P.N.; et al. Biochemical and Structural Insights into Enzymatic Depolymerization of Polylactic Acid and Other Polyesters by Microbial Carboxylesterases. Biomacromolecules 2016, 17, 2027–2039. [Google Scholar] [CrossRef] [PubMed]
- Singh, B. Review on Microbial Carboxylesterase: General Properties and Role in Organophosphate Pesticides Degradation. Biochem. Mol. Biol. 2014, 2, 1. [Google Scholar] [CrossRef]
- Mustafa, S.A. The Development of Bacterial Carboxylesterase Biological Recognition Elements for Cocaine Detection. Mol. Biotechnol. 2018, 60, 601–607. [Google Scholar] [CrossRef]
- Kaushal, J.; Khatri, M.; Arya, S.K. Recent insight into enzymatic degradation of plastics prevalent in the environment: A mini—review. Clean. Eng. Technol. 2021, 2, 100083. [Google Scholar] [CrossRef]
- Ghodke, V.M.; Punekar, N.S. Environmental role of aromatic carboxylesterases. Environ. Microbiol. 2021, 24, 2657–2668. [Google Scholar] [CrossRef]
- Gricajeva, A.; Nadda, A.K.; Gudiukaite, R. Insights into polyester plastic biodegradation by carboxyl ester hydrolases. J. Chem. Technol. Biotechnol. 2021, 97, 359–380. [Google Scholar] [CrossRef]
- Carvalho, C.M.L.; Aires-Barros, M.R.; Cabral, J.M.S. Cutinase structure, function and biocatalytic applications. Electron. J. Biotechnol. 1998, 1, 160–173. [Google Scholar] [CrossRef]
- Dutta, K.; Sen, S.; Veeranki, V.D. Production, characterization and applications of microbial cutinases. Process. Biochem. 2009, 44, 127–134. [Google Scholar] [CrossRef]
- Martínez, A.; Maicas, S. Cutinases: Characteristics and Insights in Industrial Production. Catalysts 2021, 11, 1194. [Google Scholar] [CrossRef]
- Danso, D.; Schmeisser, C.; Chow, J.; Zimmermann, W.; Wei, R.; Leggewie, C.; Li, X.; Hazen, T.; Streit, W.R. New Insights into the Function and Global Distribution of Polyethylene Terephthalate (PET)-Degrading Bacteria and Enzymes in Marine and Terrestrial Metagenomes. Appl. Environ. Microbiol. 2018, 84, e02773-17. [Google Scholar] [CrossRef] [PubMed]
- Nikolaivits, E.; Kanelli, M.; Dimarogona, M.; Topakas, E. A Middle-Aged Enzyme Still in Its Prime: Recent Advances in the Field of Cutinases. Catalysts 2018, 8, 612. [Google Scholar] [CrossRef]
- Kawai, F.; Kawabata, T.; Oda, M. Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields. Appl. Microbiol. Biotechnol. 2019, 103, 4253–4268. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Alcántara, L.; Oliart-Ros, R.M.; García-Bórquez, A.; Peña-Montes, C. Expression of a Cutinase of Moniliophthora roreri with Polyester and PET-Plastic Residues Degradation Activity. Microbiol. Spectr. 2021, 9, e00976-21. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zou, H. Biotechnological Application of Cutinase: A Powerful Tool in Synthetic Biology. SynBio 2022, 1, 54–64. [Google Scholar] [CrossRef]
- Gudiukaite, R.; Sadauskas, M.; Gegeckas, A.; Malunavicius, V.; Citavicius, D. Construction of a novel lipolytic fusion biocatalyst GDEst-lip for industrial application. J. Ind. Microbiol. Biotechnol. 2017, 44, 799–815. [Google Scholar] [CrossRef] [PubMed]
- Druteika, G.; Sadauskas, M.; Malunavicius, V.; Lastauskiene, E.; Statkeviciute, R.; Savickaite, A.; Gudiukaite, R. New engineered Geobacillus lipase GD-95RM for industry focusing on the cleaner production of fatty esters and household washing product formulations. World J. Microbiol. Biotechnol. 2020, 36, 1–15. [Google Scholar] [CrossRef]
- Savickaitė, A.; Gudiukaitė, R. In silico analysis of cutinase from Streptomyces scabiei 87.22. In Proceedings of the 64th International Conference for Students of Physics and Natural Sciences Open Readings 2021, Vilnius, Lithuania, 16–19 March 2021. [Google Scholar]
- Greičius, A.; Savickaitė, A.; Gudiukaitė, R. Analysis of site-directed mutant (Asp94Ala) of Streptomyces scabiei 87.22 cutinase. In Proceedings of the 65th International Conference for Students of Physics and Natural Sciences Open Readings 2022, Vilnius, Lithuania, 15–18 March 2022. [Google Scholar]
- Bignell, D.R.D.; Seipke, R.F.; Huguet-Tapia, J.C.; Chambers, A.H.; Parry, R.J.; Loria, R. Streptomyces scabies 87-22 Contains a Coronafacic Acid-Like Biosynthetic Cluster That Contributes to Plant–Microbe Interactions. Mol. Plant-Microbe Interact. 2010, 23, 161–175. [Google Scholar] [CrossRef]
- Sambrook, J.; Rusell, D.W. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Lab Press: New York, NY, USA, 2001. [Google Scholar]
- Cigno, E.; Magagnoli, C.; Pierce, M.; Iglesias, P. Lubricating ability of two phosphonium-based ionic liquids as additives of a bio-oil for use in wind turbines gearboxes. Wear 2017, 376–377, 756–765. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H. Tricine–SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.R.; Reid, H.J.; Sharp, B.L. Tricine-SDS-PAGE. In Electrophoretic Separation of Proteins; Methods in Molecular Biology, 1855; Kurien, B., Scofield, R., Eds.; Humana Press: New York, NY, USA, 2019; pp. 151–161. [Google Scholar] [CrossRef]
- Levisson, M.; van der Oost, J.; Kengen, S.W.M. Characterization and structural modeling of a new type of thermostable esterase from Thermotoga maritima. FEBS J. 2007, 274, 2832–2842. [Google Scholar] [CrossRef] [PubMed]
- Savickaite, A.; Sadauskas, M.; Gudiukaite, R. Immobilized GDEst-95, GDEst-lip and GD-95RM lipolytic enzymes for continuous flow hydrolysis and transesterification reactions. Int. J. Biol. Macromol. 2021, 173, 421–434. [Google Scholar] [CrossRef]
- Hannig, G.; Makrides, S.C. Strategies for optimizing heterologous protein expression in Escherichia coli. Trends Biotechnol. 1998, 16, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Gombert, A.; Kilikian, B. Recombinant gene expression in Escherichia coli cultivation using lactose as inducer. J. Biotechnol. 1998, 60, 47–54. [Google Scholar] [CrossRef]
- Lim, H.K.; Lee, S.U.; Chung, S.I.; Jung, K.H. Induction of the T7 promoter using lactose for production of recombinant plasminogen kringle 1-3 in Escherichia coli. J. Microb. Biotechnol. 2004, 14, 225–230. [Google Scholar]
- Aucoin, M.G.; McMurray-Beaulieu, V.; Poulin, F.; Boivin, E.B.; Chen, J.; Ardelean, F.M.; Cloutier, M.; Choi, Y.J.; Miguez, C.B.; Jolicoeur, M. Identifying conditions for inducible protein production in E. coli: Combining a fed-batch and multiple induction approach. Microb. Cell Factories 2006, 5, 27. [Google Scholar] [CrossRef]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef]
- Li, Z.; Kessler, W.; Heuvel, J.V.D.; Rinas, U. Simple defined autoinduction medium for high-level recombinant protein production using T7-based Escherichia coli expression systems. Appl. Microbiol. Biotechnol. 2011, 91, 1203–1213. [Google Scholar] [CrossRef]
- Tahara, N.; Tachibana, I.; Takeo, K.; Yamashita, S.; Shimada, A.; Hashimoto, M.; Ohno, S.; Yokogawa, T.; Nakagawa, T.; Suzuki, F.; et al. Boosting Auto-Induction of Recombinant Proteins in Escherichia coli with Glucose and Lactose Additives. Protein Pept. Lett. 2021, 28, 1180–1190. [Google Scholar] [CrossRef] [PubMed]
- Menacho-Melgar, R.; Ye, Z.; Moreb, E.A.; Yang, T.; Efromson, J.P.; Decker, J.S.; Wang, R.; Lynch, M.D. Scalable, two-stage, autoinduction of recombinant protein expression in E. coli utilizing phosphate depletion. Biotechnol. Bioeng. 2020, 117, 2715–2727. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W. Stable Expression Clones and Auto-Induction for Protein Production in E. coli. Methods Mol. Biol. 2013, 1091, 17–32. [Google Scholar] [CrossRef]
- Leone, S.; Sannino, F.; Tutino, M.L.; Parrilli, E.; Picone, D. Acetate: Friend or foe? Efficient production of a sweet protein in Escherichia coli BL21 using acetate as a carbon source. Microb. Cell Factories 2015, 14, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Vyas, H.; Tong, P. Process for Calcium Retention During Skim Milk Ultrafiltration. J. Dairy Sci. 2003, 86, 2761–2766. [Google Scholar] [CrossRef]
- Menchik, P.; Zuber, T.; Zuber, A.; Moraru, C.I. Short communication: Composition of coproduct streams from dairy processing: Acid whey and milk permeate. J. Dairy Sci. 2019, 102, 3978–3984. [Google Scholar] [CrossRef]
- Krefft, D.; Prusinowski, M.; Maciszka, P.; Skokowska, A.; Zebrowska, J.; Skowron, P.M. T7-lac promoter vectors spontaneous derepression caused by plant-derived growth media may lead to serious expression problems: A systematic evaluation. Microb. Cell Factories 2022, 21, 1–13. [Google Scholar] [CrossRef]
- Su, L.; Woodard, R.; Chen, J.; Wu, J. Extracellular Location of Thermobifida fusca Cutinase Expressed in Escherichia coli BL21(DE3) without Mediation of a Signal Peptide. Appl. Environ. Microbiol. 2013, 79, 4192–4198. [Google Scholar] [CrossRef]
- Kawai, F.; Oda, M.; Tamashiro, T.; Waku, T.; Tanaka, N.; Yamamoto, M.; Mizushima, H.; Miyakawa, T.; Tanokura, M. A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 2014, 98, 10053–10064. [Google Scholar] [CrossRef]
- Miyakawa, T.; Mizushima, H.; Ohtsuka, J.; Oda, M.; Kawai, F.; Tanokura, M. Structural basis for the Ca2+-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 2014, 99, 4297–4307. [Google Scholar] [CrossRef]
- Numoto, N.; Kamiya, N.; Bekker, G.-J.; Yamagami, Y.; Inaba, S.; Ishii, K.; Uchiyama, S.; Kawai, F.; Ito, N.; Oda, M. Structural Dynamics of the PET-Degrading Cutinase-like Enzyme from Saccharomonospora viridis AHK190 in Substrate-Bound States Elucidates the Ca2+-Driven Catalytic Cycle. Biochemistry 2018, 57, 5289–5300. [Google Scholar] [CrossRef]
- Oda, M.; Yamagami, Y.; Inaba, S.; Oida, T.; Yamamoto, M.; Kitajima, S.; Kawai, F. Enzymatic hydrolysis of PET: Functional roles of three Ca2+ ions bound to a cutinase-like enzyme, Cut190*, and its engineering for improved activity. Appl. Microbiol. Biotechnol. 2018, 102, 10067–10077. [Google Scholar] [CrossRef] [PubMed]
- Oda, M. Structural basis for Ca2+-dependent catalysis of a cutinase-like enzyme and its engineering: Application to enzymatic PET depolymerization. Biophys. Psysicobiology 2021, 18, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Komeil, D.; Simao-Beaunoir, A.-M.; Beaulieu, C. Detection of potential suberinase-encoding genes in Streptomyces scabiei strains and other actinobacteria. Can. J. Microbiol. 2013, 59, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Jabloune, R.; Khalil, M.; Ben Moussa, I.E.; Simao-Beaunoir, A.-M.; Lerat, S.; Brzezinski, R.; Beaulieu, C. Enzymatic Degradation of p-Nitrophenyl Esters, Polyethylene Terephthalate, Cutin, and Suberin by Sub1, a Suberinase Encoded by the Plant Pathogen Streptomyces scabies. Microbes Environ. 2020, 35, ME19086. [Google Scholar] [CrossRef]
- Feizollahzadeh, S.; Kouhpayeh, S.; Rahimmansh, I.; Khanahmad, H.; Sabzehei, F.; Ganjalikhani-Hakemi, M.; Andalib, A.; Hejazi, Z.; Rezaei, A. The Increase in Protein and Plasmid Yields of E. coli with Optimized Concentration of Ampicillin as Selection Marker. Iran. J. Biotechnol. 2017, 15, 128–134. [Google Scholar] [CrossRef]
- Bahreini, E.; Aghaiypour, K.; Abbasalipourkabir, R.; Goodarzi, M.T.; Saidijam, M.; Safavieh, S.S. An optimized protocol for overproduction of recombinant protein expression in Escherichia coli. Prep. Biochem. Biotechnol. 2014, 44, 510–528. [Google Scholar] [CrossRef]



| Volume of Culture Medium (mL) | Final Ampicillin Concentration in Cultivation Medium (µg/mL) | Tested Inducer and Final Concentration in Cultivation Medium | Cultivation Conditions | Enzymes Produced |
|---|---|---|---|---|
| 50 | 100 | IPTG (1 mM), lactose (2, 4, 6 mM) | 37 °C, 150–180 rpm; Orbicult IBS-R-25-1 incubator benchtop shaker (Kisker Biotech GmbH & Co. KG, Steinfurt, Germany) | Cut+SP, GD-95RM |
| 250 | 100; 50; 25; 10; 0.5 | MP (2, 4, 6 mM) | 37 °C, 150–180 rpm; Orbicult IBS-R-25-1 incubator benchtop shaker | Cut+SP, GD-95RM, GDEst-lip, GDEst-95 |
| 1000 | ~0.5 | MP (4 mM) | 37 °C, magnetic stirrer at 170 rpm; the cultivation was performed in 2000 mL Erlenmeyer flasks in an Enviro-Genie incubator (Scientific Industries, Inc., Bohemia, NY, USA) | Cut+SP, GD-95RM, GDEst-95, GDEst-lip |
| 2000 | ~0.5 | MP (4 mM) | 35–37 °C, magnetic stirrer at 550 rpm; the cultivation was performed in 5000 mL Erlenmeyer flasks using Wisd23 ViseStir MSH-D hotplate magnetic stirrer (TQC bv, Capelle aan den Ijssel, The Netherlands) | Cut+SP, GD-95RM, GDEst-95, GDEst-lip |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greicius, A.; Baliutavicius, T.; Lastauskiene, E.; Gudiukaite, R. Application of Milk Permeate as an Inducer for the Production of Microbial Recombinant Lipolytic Enzymes. Fermentation 2023, 9, 27. https://doi.org/10.3390/fermentation9010027
Greicius A, Baliutavicius T, Lastauskiene E, Gudiukaite R. Application of Milk Permeate as an Inducer for the Production of Microbial Recombinant Lipolytic Enzymes. Fermentation. 2023; 9(1):27. https://doi.org/10.3390/fermentation9010027
Chicago/Turabian StyleGreicius, Aurimas, Tautvydas Baliutavicius, Egle Lastauskiene, and Renata Gudiukaite. 2023. "Application of Milk Permeate as an Inducer for the Production of Microbial Recombinant Lipolytic Enzymes" Fermentation 9, no. 1: 27. https://doi.org/10.3390/fermentation9010027
APA StyleGreicius, A., Baliutavicius, T., Lastauskiene, E., & Gudiukaite, R. (2023). Application of Milk Permeate as an Inducer for the Production of Microbial Recombinant Lipolytic Enzymes. Fermentation, 9(1), 27. https://doi.org/10.3390/fermentation9010027







