Influence of Substrate on the Fermentation Characteristics and Culture-Dependent Microbial Composition of Water Kefir
Abstract
1. Introduction
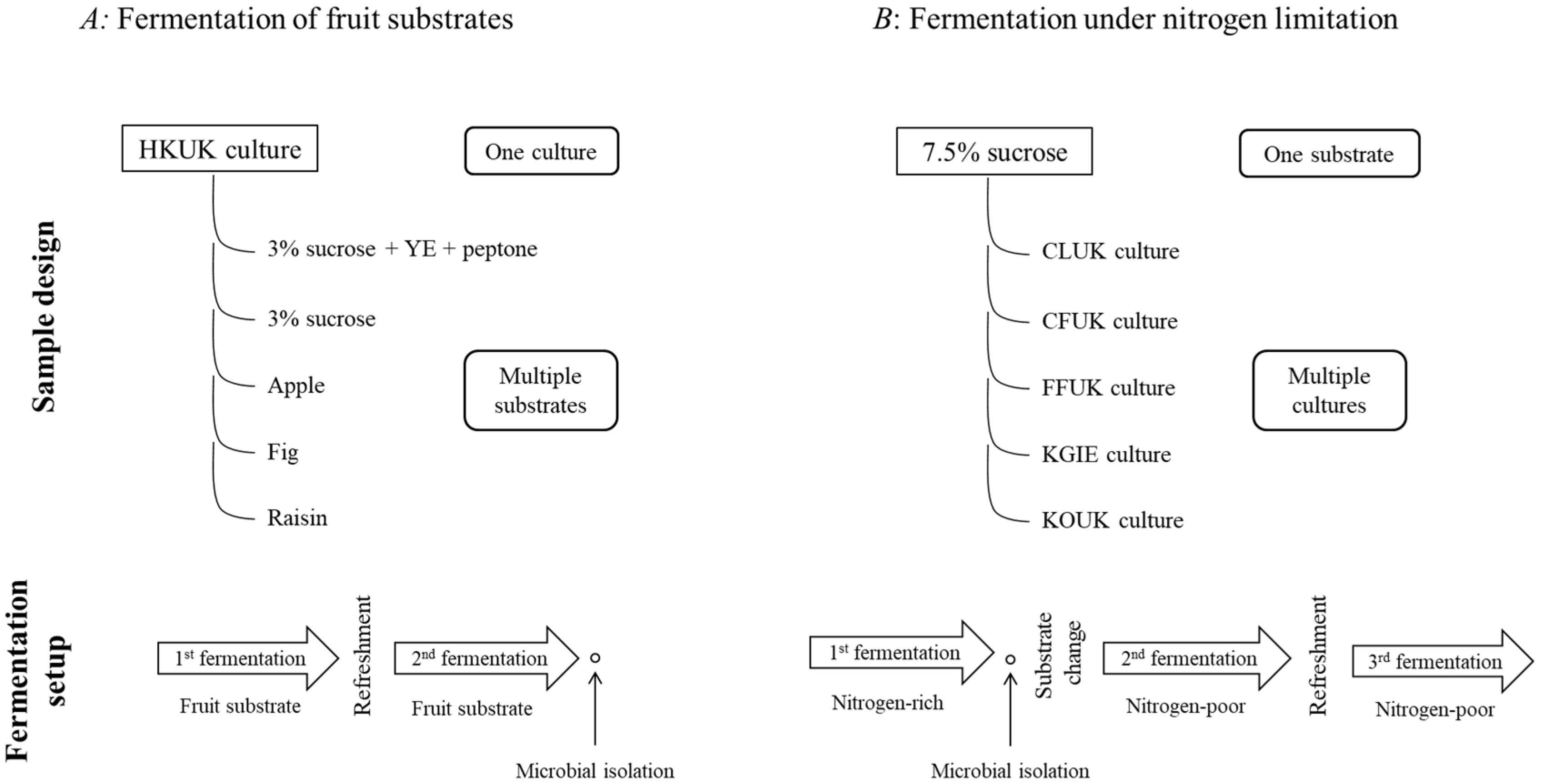
2. Materials and Methods
2.1. Preparation of Water Kefir Grain Cultures
2.2. Fruit-Based Water Kefir Fermentations
2.3. Nitrogen-Limited Water Kefir Fermentations
2.4. Analysis of Water Kefirs
2.5. Microbial Isolation
2.6. Species Identification
2.7. Statistical Analysis
3. Results and Discussion
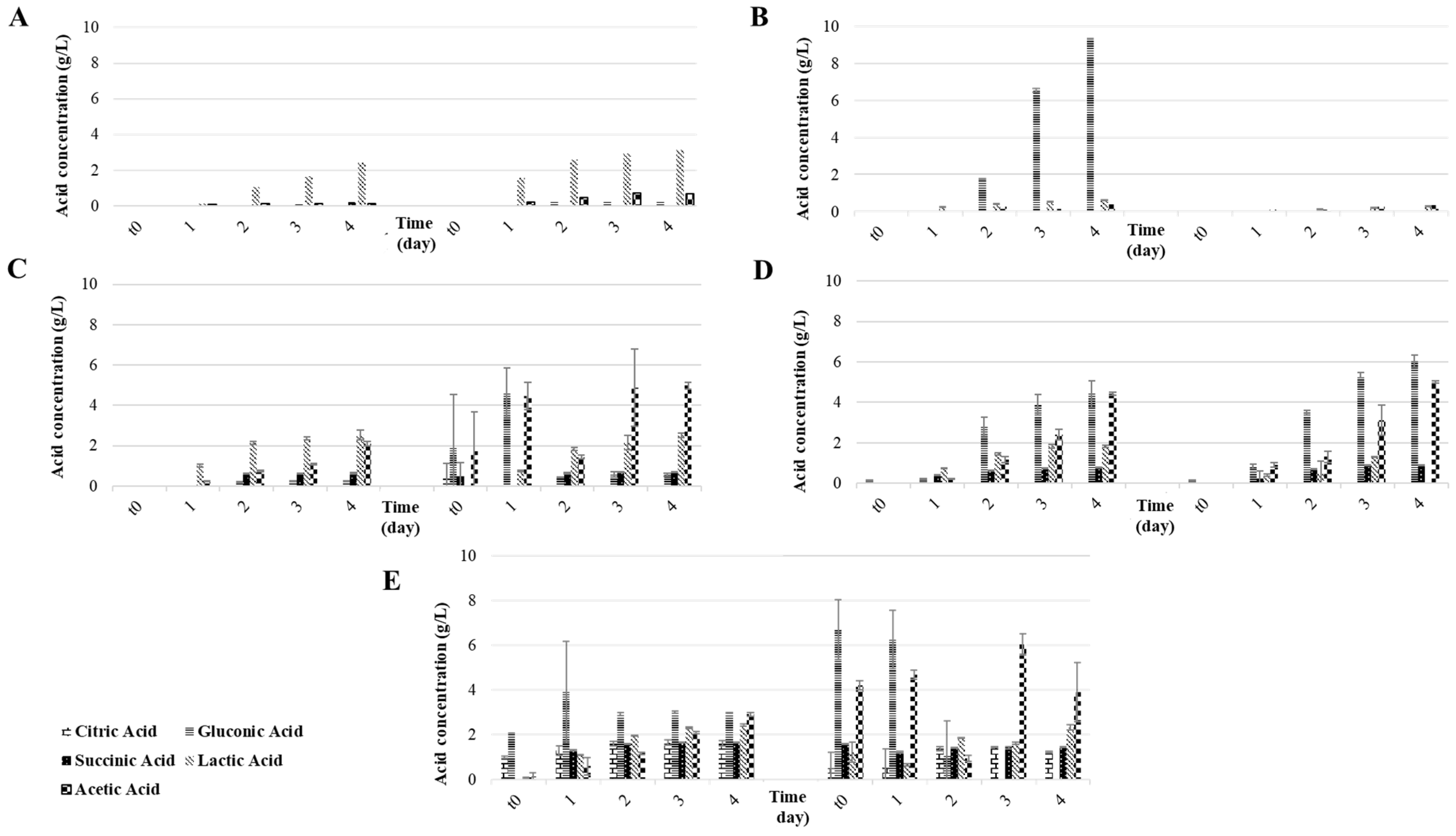
3.1. Part A: Influence of Fruit Substrate on the Water Kefir Fermentation
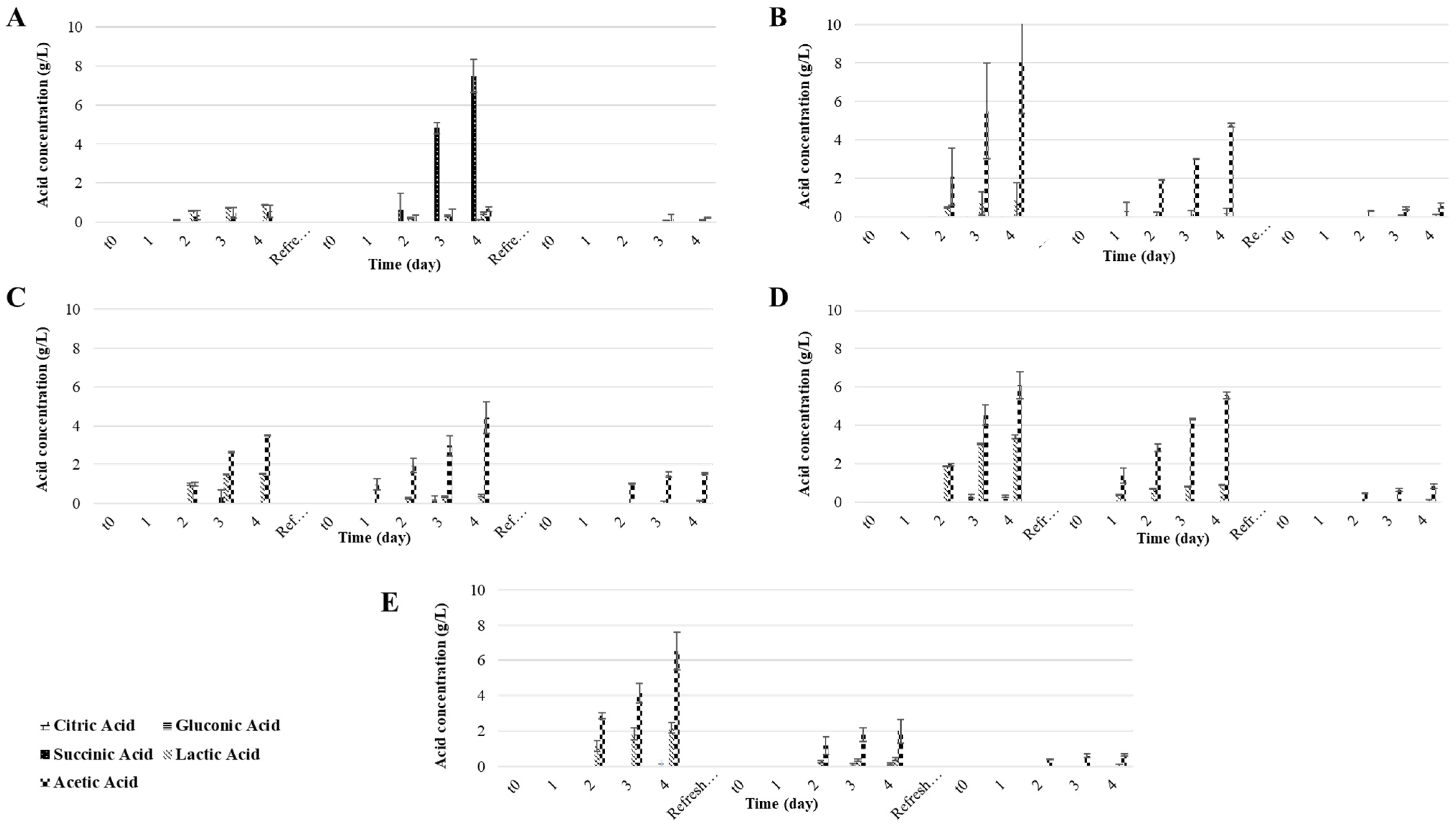
3.2. Part B: Effect of Nitrogen Limitation on the Water Kefir Fermentation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gulitz, A.; Stadie, J.; Wenning, M.; Ehrmann, M.A.; Vogel, R.F. The Microbial Diversity of Water Kefir. Int. J. Food Microbiol. 2011, 151, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Torres, A.; Gutiérrez-Ambrocio, S.; Heredia-del-Orbe, P.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Inferring the Role of Microorganisms in Water Kefir Fermentations. Int. J. Food Sci. Technol. 2017, 52, 559–571. [Google Scholar] [CrossRef]
- Fels, L.; Jakob, F.; Vogel, R.F.; Wefers, D. Structural Characterization of the Exopolysaccharides from Water Kefir. Carbohydr. Polym. 2018, 189, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Pidoux, M.; Brillouet, J.M.; Quemener, B. Characterization of the Polysaccharides from a Lactobacillus Brevis and from Sugary Kefir Grains. Biotechnol. Lett. 1988, 10, 415–420. [Google Scholar] [CrossRef]
- Zanirati, D.F.; Abatemarco, M.; de Cicco Sandes, S.H.; Nicoli, J.R.; Nunes, Á.C.; Neumann, E. Selection of Lactic Acid Bacteria from Brazilian Kefir Grains for Potential Use as Starter or Probiotic Cultures. Anaerobe 2015, 32, 70–76. [Google Scholar] [CrossRef]
- Lynch, K.M.; Wilkinson, S.; Daenen, L.; Arendt, E.K. An Update on Water Kefir: Microbiology, Composition and Production. Int. J. Food Microbiol. 2021, 345, 109128. [Google Scholar] [CrossRef]
- Stadie, J.; Gulitz, A.; Ehrmann, M.A.; Vogel, R.F. Metabolic Activity and Symbiotic Interactions of Lactic Acid Bacteria and Yeasts Isolated from Water Kefir. Food Microbiol. 2013, 35, 92–98. [Google Scholar] [CrossRef]
- Xu, D.; Behr, J.; Geißler, A.J.; Bechtner, J.; Ludwig, C.; Vogel, R.F. Label-Free Quantitative Proteomic Analysis Reveals the Lifestyle of Lactobacillus Hordei in the Presence of Sacchromyces Cerevisiae. Int. J. Food Microbiol. 2019, 294, 18–26. [Google Scholar] [CrossRef]
- Xu, D.; Bechtner, J.; Behr, J.; Eisenbach, L.; Geißler, A.J.; Vogel, R.F. Lifestyle of Lactobacillus Hordei Isolated from Water Kefir Based on Genomic, Proteomic and Physiological Characterization. Int. J. Food Microbiol. 2019, 290, 141–149. [Google Scholar] [CrossRef]
- Laureys, D.; Aerts, M.; Vandamme, P.; De Vuyst, L. Oxygen and Diverse Nutrients Influence the Water Kefir Fermentation Process. Food Microbiol. 2018, 73, 351–361. [Google Scholar] [CrossRef]
- Laureys, D.; Cnockaert, M.; De Vuyst, L.; Vandamme, P. Bifidobacterium Aquikefiri Sp. Nov., Isolated from Water Kefir. Int. J. Syst. Evol. Microbiol. 2016, 66, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Laureys, D.; De Vuyst, L. The Water Kefir Grain Inoculum Determines the Characteristics of the Resulting Water Kefir Fermentation Process. J. Appl. Microbiol. 2017, 122, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Laureys, D.; De Vuyst, L. Microbial Species Diversity, Community Dynamics, and Metabolite Kinetics of Water Kefir Fermentation. Appl. Environ. Microbiol. 2014, 80, 2564–2572. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, K.T.; de Pereira, G.V.M.; Dias, D.R.; Schwan, R.F. Microbial Communities and Chemical Changes during Fermentation of Sugary Brazilian Kefir. World J. Microbiol. Biotechnol. 2010, 26, 1241–1250. [Google Scholar] [CrossRef]
- Fiorda, F.A.; de Melo Pereira, G.V.; Thomaz-Soccol, V.; Rakshit, S.K.; Pagnoncelli, M.G.B.; de Souza Vandenberghe, L.P.; Soccol, C.R. Microbiological, Biochemical, and Functional Aspects of Sugary Kefir Fermentation—A Review. Food Microbiol. 2017, 66, 86–95. [Google Scholar] [CrossRef]
- Laureys, D.; Aerts, M.; Vandamme, P.; De Vuyst, L. The Buffer Capacity and Calcium Concentration of Water Influence the Microbial Species Diversity, Grain Growth, and Metabolite Production During Water Kefir Fermentation. Front. Microbiol. 2019, 10, 2876. [Google Scholar] [CrossRef]
- Laureys, D.; Van Jean, A.; Dumont, J.; De Vuyst, L. Investigation of the Instability and Low Water Kefir Grain Growth during an Industrial Water Kefir Fermentation Process. Appl. Microbiol. Biotechnol. 2017, 101, 2811–2819. [Google Scholar] [CrossRef]
- Gullo, M.; Giudici, P. Acetic Acid Bacteria in Traditional Balsamic Vinegar: Phenotypic Traits Relevant for Starter Cultures Selection. Int. J. Food Microbiol. 2008, 125, 46–53. [Google Scholar] [CrossRef]
- Mamlouk, D.; Gullo, M. Acetic Acid Bacteria: Physiology and Carbon Sources Oxidation. Indian J. Microbiol. 2013, 53, 377–384. [Google Scholar] [CrossRef]
- Camu, N.; González, Á.; De Winter, T.; Van Schoor, A.; De Bruyne, K.; Vandamme, P.; Takrama, J.S.; Addo, S.K.; De Vuyst, L. Influence of Turning and Environmental Contamination on the Dynamics of Populations of Lactic Acid and Acetic Acid Bacteria Involved in Spontaneous Cocoa Bean Heap Fermentation in Ghana. Appl. Environ. Microbiol. 2008, 74, 86–98. [Google Scholar] [CrossRef]
- De Roos, J.; Verce, M.; Aerts, M.; Vandamme, P.; De Vuyst, L. Temporal and Spatial Distribution of the Acetic Acid Bacterium Communities throughout the Wooden Casks Used for the Fermentation and Maturation of Lambic Beer. Appl. Environ. Microbiol. 2018, 84, e02846-17. [Google Scholar] [CrossRef] [PubMed]
- Wieme, A.D.; Spitaels, F.; Aerts, M.; De Bruyne, K.; Van Landschoot, A.; Vandamme, P. Effects of Growth Medium on Matrix-Assisted Laser Desorption-Ionization Time of Flight Mass Spectra: A Case Study of Acetic Acid Bacteria. Appl. Environ. Microbiol. 2014, 80, 1528–1538. [Google Scholar] [CrossRef] [PubMed]
- Papalexandratou, Z.; Lefeber, T.; Bahrim, B.; Lee, O.S.; Daniel, H.M.; De Vuyst, L. Hanseniaspora Opuntiae, Saccharomyces Cerevisiae, Lactobacillus Fermentum, and Acetobacter Pasteurianus Predominate during Well-Performed Malaysian Cocoa Bean Box Fermentations, Underlining the Importance of These Microbial Species for a Successful Cocoa. Food Microbiol. 2013, 35, 73–85. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Gardes, M.; Bruns, T.D. ITS Primers with Enhanced Specificity for Basidiomycetes-application to the Identification of Mycorrhizae and Rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN—List of Prokaryotic Names with Standing in Nomenclature. Nucleic Acids Res. 2014, 42, D613–D616. [Google Scholar] [CrossRef]
- De Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. R Package Version 1.2-0. 2014. Available online: http://CRAN.R-project.org/package=agricolae (accessed on 27 November 2022).
- Gulitz, A.; Stadie, J.; Ehrmann, M.A.; Ludwig, W.; Vogel, R.F. Comparative Phylobiomic Analysis of the Bacterial Community of Water Kefir by 16S RRNA Gene Amplicon Sequencing and ARDRA Analysis. J. Appl. Microbiol. 2013, 114, 1082–1091. [Google Scholar] [CrossRef]
- da C. P. Miguel, M.G.; Cardoso, P.G.; Magalhães, K.T.; Schwan, R.F. Profile of Microbial Communities Present in Tibico (Sugary Kefir) Grains from Different Brazilian States. World J. Microbiol. Biotechnol. 2011, 27, 1875–1884. [Google Scholar] [CrossRef]
- Endo, A.; Okada, S. Lactobacillus Satsumensis Sp. Nov., Isolated from Mashes of Shochu, a Traditional Japanese Distilled Spirit Made from Fermented Rice and Other Starchy Materials. Int. J. Syst. Evol. Microbiol. 2005, 55, 83–85. [Google Scholar] [CrossRef]
- Verce, M.; De Vuyst, L.; Weckx, S. Shotgun Metagenomics of a Water Kefir Fermentation Ecosystem Reveals a Novel Oenococcus Species. Front. Microbiol. 2019, 10, 479. [Google Scholar] [CrossRef]
- Rice, T.; Zannini, E.; Arendt, E.K.; Coffey, A. A Review of Polyols–Biotechnological Production, Food Applications, Regulation, Labeling and Health Effects. Crit. Rev. Food Sci. Nutr. 2019, 60, 2034–2051. [Google Scholar] [CrossRef]
- Von Wright, A.; Axelsson, L. Lactic Acid Bacteria: An Introduction. In Lactic Acid Bacteria. Microbiological and Functional Aspects, 4th ed.; Lahtinen, S., Ouwehand, A.C., Salminen, S., von Wright, A., Eds.; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Warriner, K.S.R.; Morris, J.G. The Effects of Aeration on the Bioreductive Abilities of Some Heterofermentative Lactic Acid Bacteria. Lett. Appl. Microbiol. 1995, 20, 323–327. [Google Scholar] [CrossRef]
- Coton, M.; Pawtowski, A.; Taminiau, B.; Burgaud, G.; Deniel, F.; Coulloumme-Labarthe, L.; Fall, A.; Daube, G.; Coton, E. Unraveling Microbial Ecology of Industrial-Scale Kombucha Fermentations by Metabarcoding and Culture-Based Methods. FEMS Microbiol. Ecol. 2017, 93, 1–16. [Google Scholar] [CrossRef] [PubMed]
- USDA. (n.d.). FoodData Central. 2020. Available online: Fdc.nal.usda.gov (accessed on 22 November 2022).
- Kurniawati, M.; Nurliyani, N.; Budhijanto, W.; Widodo, W. Isolation and Identification of Lactose-Degrading Yeasts and Characterisation of Their Fermentation-Related Ability to Produce Ethanol. Fermentation 2022, 8, 183. [Google Scholar] [CrossRef]
- Kalamaki, M.S.; Angelidis, A.S. Isolation and Molecular Identification of Yeasts in Greek Kefir. Int. J. Dairy Technol. 2017, 70, 261–268. [Google Scholar] [CrossRef]
- Reiß, J. Metabolic Activity of Tibi Grains. Z. Lebensm. Unters. Forsch. 1990, 191, 462–465. [Google Scholar] [CrossRef]
- Lynch, K.M.; Zannini, E.; Wilkinson, S.; Daenen, L.; Arendt, E.K. Physiology of Acetic Acid Bacteria and Their Role in Vinegar and Fermented Beverages. Compr. Rev. Food Sci. Food Saf. 2019, 18, 587–625. [Google Scholar] [CrossRef] [PubMed]
- Pendón, M.D.; Bengoa, A.A.; Iraporda, C.; Medrano, M.; Garrote, G.L.; Abraham, A.G. Water Kefir: Factors Affecting Grain Growth and Health-Promoting Properties of the Fermented Beverage. J. Appl. Microbiol. 2022, 133, 162–180. [Google Scholar] [CrossRef]
| Ferm. Substrates | Ferm. n° | Parameters | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Time (h) | pH | TTA (meq/L) | Alcohol (% v/v) | Extract (% w/w) | Sucrose (g/L) | Glucose (g/L) | Fructose (g/L) | ||
| Sucrose, yeast extract, peptone | 1st | 0 | 6.3 ± 0.0 c | 4.3 ± 0.5 i | 9.7 ± 0.1 abc | 0.1 ± 0.0 d | 87.7 ± 2.4 a | 0.0 ± 0.0 c | 0.0 ± 0.0 e |
| 96 | 3.3 ± 0.1 i | 44.5 ± 0.4 e | 3.3 ± 2.1 e | 4.4 ± 1.2 b | 0.5 ± 0.2 f | 6.2 ± 8.1 c | 13.6 ± 13.6 cde | ||
| 2nd | 0 | 6.5 ±0.0 b | 6.2 ± 0.0 i | 10.4 ± 0.0 a | 0.1 ± 0.0 d | 89.9 ± 0.0 a | 0.0 ± 0.0 c | 0.0 ± 0.0 e | |
| 96 | 3.3 ± 0.0 hi | 66.3 ± 0.5 c | 1.2 ± 0.0 e | 5.7 ± 0.1 a | 0.7 ± 0.1 f | 0.0 ± 0.0 c | 0.1 ± 0.1 e | ||
| Sucrose alone | 1st | 0 | 7.0 ± 0.3 a | 0.0 ± 0.0 i | 7.3 ± 0.1 cd | 0.0 ± 0.0 d | 58.8 ± 5.3 c | 0.4 ± 0.6 c | 0.1 ± 0.2 e |
| 96 | 2.5 ± 0.0 k | 58.2 ± 0.1 cd | 7.0 ± 0.4 d | 0.3 ± 0.1 d | 1.4 ± 0.1 f | 25.7 ± 0.4 ab | 35.5 ± 0.4 ab | ||
| 2nd | 0 | 7.0 ± 0.0 a | 0.2 ± 0.0 i | 7.4 ± 0.0 bcd | 0.0 ± 0.0 d | 76.0 ± 0.8 b | 0.0 ± 0.0 c | 0.0 ± 0.0 e | |
| 96 | 2.9 ± 0.1 j | 21.1 ± 0.3 gh | 6.7 ± 1.3 d | 0.1 ± 0.1 d | 0.2 ± 0.3 f | 0.6 ± 0.1 c | 6.9 ± 0.3 de | ||
| Apple | 1st | 0 | 3.5 ± 0.0 fg | 47.9 ± 0.6 de | 7.2 ± 0.0 d | 0.0 ± 0.0 d | 8.1 ± 0.1 e | 17.7 ± 0.1 b | 35.7 ± 0.2 ab |
| 96 | 3.6 ± 0.0 f | 69.3 ± 0.0 c | 1.2 ± 0.0 e | 3.1 ± 0.0 c | 0.0 ± 0.0 f | 0.0 ± 0.0 c | 0.0 ± 0.0 e | ||
| 2nd | 0 | 3.5 ± 0.0 fg | 42.1 ± 0.3 ef | 7.7 ± 0.0 bcd | 0.1 ± 0.1 d | 2.5 ± 1.8 ef | 3.8 ± 4.7 c | 16.1 ± 22.1 bcde | |
| 96 | 3.5 ± 0.0 fg | 93.7 ± 0.5 b | 1.3 ± 0.0 e | 3.3 ± 0.0 bc | 0.0 ± 0.1 f | 0.0 ± 0.0 c | 0.1 ± 0.0 e | ||
| Fig | 1st | 0 | 4.2 ± 0.0 d | 14.2 ± 0.0 ghi | 8.5 ± 0.0 abcd | 0.0 ± 0.0 d | 1.6 ± 0.0 f | 35.4 ± 0.1 a | 32.8 ± 0.1 abc |
| 96 | 3.4 ± 0.0 gh | 102.5 ± 0.2 b | 2.4 ± 0.0 e | 3.3 ± 0.0 bc | 0.0 ± 0.0 f | 0.0 ± 0.0 c | 0.3 ± 0.0 e | ||
| 2nd | 0 | 4.3 ± 0.0 d | 12.5 ± 0.1 hi | 7.9 ± 0.0 bcd | 0.0 ± 0.0 d | 1.8 ± 0.4 f | 2.9 ± 2.4 c | 12.6 ± 4.8 cde | |
| 96 | 3.4 ± 0.0 fg | 92.6 ± 0.6 b | 2.1 ± 0.6 e | 3.1 ± 0.0 c | 16.5 ± 2.6 d | 28.3 ± 1.5 a | 30.0 ± 1.0 abcd | ||
| Raisin | 1st | 0 | 3.9 ± 0.0 e | 26.4 ± 0.8 gh | 8.2 ± 0.0 abcd | 0.0 ± 0.0 d | 0.3 ± 0.0 f | 34.7 ± 0.0 a | 36.3 ± 0.0 ab |
| 96 | 3.2 ± 0.0 i | 138.2 ± 0.3 a | 1.2 ± 0.0 e | 3.0 ± 0.2 c | 0.3 ± 0.0 f | 0.0 ± 0.0 c | 1.0 ± 0.1 e | ||
| 2nd | 0 | 4.0 ± 0.0 e | 27.7 ± 0.3 fg | 10.1 ± 0.1 ab | 0.1 ± 0.0 d | 0.0 ± 0.0 f | 32.8 ± 0.6 a | 44.7 ± 0.6 a | |
| 96 | 3.2 ± 0.0 i | 146.3 ± 0.5 a | 2.3 ± 0.0 e | 3.9 ± 0.1 bc | 0.0 ± 0.0 f | 0.0 ± 0.0 c | 0.1 ± 0.3 e | ||
| Carbon Source | Liquid (L) or Grain (G) Isolation | Isolated Species | ||
|---|---|---|---|---|
| Lactic Acid Bacteria | Acetic Acid Bacteria | Yeast | ||
| Sucrose | L | n.d. | Gluconobacter cerinus | n.d. |
| G | Liquorilactobacillus satsumensis | Acetobacter syzgii, Gluconobacter cerinus | n.d. | |
| Apple | L | Liquorilactobacillus satsumensis | Acetobacter fabarum, Acetobacter suratthaniensis | n.d. |
| G | n.d. | Acetobacter fabarum | n.d. | |
| Fig | L | n.d. | Acetobacter persici | Pichia kudriavzevii |
| G | Lentilactobacillus hilgardii, Liquorilactobacillus satsumensis | Gluconobacter cerinus, Acetobacter fabarum, Acetobacter syzgii, Komagataeibacter saccharivorans | n.d. | |
| Raisin | L | n.d. | Acetobacter orientalis, Acetobacter syzygii, Gluconobacter cerinus | n.d. |
| G | Liquorilactobacillus satsumensis, Liquorilactobacillus oeni | Acetobacter fabarum, Gluconobacter cerinus, Gluconobacter oxydans | Pichia kudriavzevii | |
| Water Kefir Culture | Fermentation Period | Time (h) | pH | TTA (meq/L) | Extract (% w/w) | Alcohol (% v/v) | Sucrose (g/L) | Glucose (g/L) | Fructose (g/L) |
|---|---|---|---|---|---|---|---|---|---|
| CLUK | 1st | 0 | 6.4 ± 0.0 b | 1.0 ± 0.0 h | 10.5 ± 0.0 a | 0.1 ± 0.0 d | 110.5 ± 0.0 a | 0.0 ± 0.0 g | 0.0 ± 0.0 h |
| 96 | 3.8 ± 0.1 c | 15.2 ± 0.4 fgh | 0.9 ± 0.0 c | 5.8 ± 0.1 a | 1.1 ± 1.6 f | 0.0 ± 0.0 g | 0.0 ± 0.0 h | ||
| 2nd | 0 | 3.7 ± 0.0 c | 0.0 ± 0.0 h | 7.5 ± 0.0 b | 0.0 ± 0.0 d | 80.6 ± 0.0 b | 0.0 ± 0.0 g | 0.0 ± 0.0 h | |
| 96 | 2.5 ± 0.0 i | 46.2 ± 0.6 de | 6.9 ± 0.5 b | 0.5 ± 0.3 cd | 6.7 ± 6.0 f | 32.2 ± 1.7 b | 32.2 ± 1.7 ab | ||
| 3rd | 0 | 7.2 ± 0.0 a | 0.1 ± 0.0 h | 7.5 ± 0.0 b | 0.0 ± 0.0 d | 83.6 ± 0.0 b | 0.0 ± 0.0 g | 0.0 ± 0.0 h | |
| 96 | 3.0 ± 0.1 fgh | 10.3 ± 0.2 gh | 7.5 ± 0.4 b | 0.0 ± 0.0 d | 36.1 ± 5.9 d | 20.8 ± 3.9 bc | 20.8 ± 3.9 cde | ||
| CFUK | 1st | 0 | 6.4 ± 0.0 b | 1.0 ± 0.0 h | 10.5 ± 0.0 a | 0.1 ± 0.0 d | 110.5 ± 0.0 a | 0.0 ± 0.0 g | 0.0 ± 0.0 h |
| 96 | 3.2 ± 0.0 de | 123.3 ± 0.8 a | 2.2 ± 0.4 c | 5.0 ± 0.1 a | 2.9 ± 0.0 f | 0.3 ± 0.4 fg | 4.5 ± 2.4 gh | ||
| 2nd | 0 | 7.2 ± 0.0 a | 0.0 ± 0.0 h | 7.7 ± 0.0 b | 0.0 ± 0.0 d | 81.0 ± 0.0 b | 0.0 ± 0.0 g | 0.0 ± 0.0 h | |
| 96 | 2.7 ± 0.0 hi | 88.3 ± 0.2 b | 7.5 ± 0.1 b | 0.1 ± 0.0 d | 2.8 ± 0.1 f | 33.4 ± 0.0 a | 37.1 ± 0.0 a | ||
| 3rd | 0 | 7.2 ± 0.0 a | 0.3 ± 0.0 h | 7.5 ± 0.0 b | 0.0 ± 0.0 d | 84.4 ± 0.0 b | 0.0 ± 0.0 g | 0.0 ± 0.0 h | |
| 96 | 3.1 ± 0.0 defg | 13.5 ± 0.2 fgh | 7.7 ± 0.01 b | 0.0 ± 0.0 d | 34.8 ± 0.8 de | 21.7 ± 0.3 b | 21.9 ± 0.2 cde | ||
| KGIE | 1st | 0 | 6.4 ± 0.0 b | 1.0 ± 0.0 h | 10.5± 0.0 a | 0.1 ± 0.0 d | 110.5 ± 0.0 a | 0.0 ± 0.0 g | 0.0 ± 0.0 h |
| 96 | 3.5 ± 0.0 cd | 67.9 ± 0.1 bc | 1.9 ± 0.1 c | 4.9 ± 0.1 a | 3.0 ± 0.0 f | 0.0 ± 0.0 g | 0.0 ± 0.0 h | ||
| 2nd | 0 | 7.2 ± 0.1 a | 0.0 ± 0.0 h | 7.6 ± 0.8 b | 0.0 ± 0.0 d | 80.8 ± 0.3 b | 0.0 ± 0.0 g | 0.0± 0.0 h | |
| 96 | 3.0 ± 0.4 efgh | 63.3 ± 1.1 cd | 2.3 ± 0.5 c | 2.9 ± 0.1 b | 2.5 ± 0.0 f | 0.0 ± 0.0 g | 10.7 ± 3.6 fg | ||
| 3rd | 0 | 7.2 ± 0.0 a | 0.2 ± 0.0 h | 7.5 ± 0.0 b | 0.0 ± 0.0 d | 84.0 ± 0.6 b | 0.0 ± 0.0 g | 0.0 ± 0.0 h | |
| 96 | 3.1 ± 0.0 defg | 24.9 ± 0.2 efg | 6.2 ± 1.1 b | 0.7 ± 0.5 cd | 11.7 ± 7.1 f | 20.4 ± 4.5 b | 26.87 ± 0.7 bcd | ||
| FFUK | 1st | 0 | 6.4 ± 0.0 b | 1.0 ± 0.0 gh | 10.5 ± 0.0 a | 0.1 ± 0.0 cd | 110.5 ± 0.0 a | 0.0 ± 0.0 g | 0.0 ± 0.0 h |
| 96 | 3.3 ±0.0 def | 119.5 ± 2.0 a | 10.0 ± 0.2 a | 0.5 ± 0.4 d | 24.1 ± 1.2 e | 19.1 ± 2.5 bcd | 29.4 ± 4.4 abc | ||
| 2nd | 0 | 7.3 ± 0.0 a | 0.0 ± 0.0 h | 7.5 ± 0.0 b | 0.0 ± 0.0 d | 80.6 ± 0.0 b | 0.0 ± 0.0 g | 0.0 ± 0.0 h | |
| 96 | 2.9 ± 0.1 ghi | 72.9 ± 0.5 bc | 6.0 ± 1.1 b | 1.0 ± 0.6 cd | 25.3 ± 6.1 de | 10.9 ± 9.4 de | 18.8 ± 8.5 def | ||
| 3rd | 0 | 7.2 ± 0.0 a | 0.1 ± 0.0 h | 7.5 ± 0.0 b | 0.0 ± 0.0 d | 83.6 ± 0.0 b | 0.0 ± 0.0 g | 0.0 ± 0.0 h | |
| 96 | 3.1 ± 0.2 defg | 13.0 ± 0.3 fgh | 6.2 ± 1.6 b | 0.0 ± 0.4 d | 53.7 ± 2.21 c | 10.9 ± 1.0 cde | 11.4 ± 0.9 fg | ||
| KOUK | 1st | 0 | 6.4 ± 0.0 b | 1.0 ± 1.0 h | 10.5 ± 0.0 a | 0.1 ± 0.0 d | 100.5 ± 0.0 a | 0.0 ± 0.0 g | 0.0 ± 0.0 h |
| 96 | 3.3 ± 0.0 def | 120.8 ± 1.1 a | 6.1 ± 1.5 b | 2.8 ± 0.8 b | 3.6 ± 0.9 f | 9.7 ± 5.2 def | 23.7 ± 4.9 bcde | ||
| 2nd | 0 | 7.2 ± 0.0 a | 0.0 ± 0.0 h | 7.7 ± 0.0 b | 0.0 ± 0.0 d | 81.0 ± 0.0 b | 0.0 ± 0.0 g | 0.0 ± 0.0 h | |
| 96 | 3.0 ± 0.1 fgh | 31.4 ± 0.2 ef | 5.9 ± 0.5 b | 1.2 ± 0.3 c | 26.8 ± 5.0 de | 9.4 ± 0.1 efg | 17.4 ± 0.1 ef | ||
| 3rd | 0 | 7.2 ± 0.0 a | 0.3 ± 0.0 h | 7.5 ± 0.0 b | 0.0 ± 0.0 d | 84.4 ± 0.0 b | 0.0 ± 0.0 g | 0.0 ± 0.0 h | |
| 96 | 3.1 ± 0.0 defg | 12.1 ± 0.3 fgh | 7.1 ± 0.7 b | 0.1 ± 0.1 d | 52.8 ± 6.5 c | 8.3 ± 0.0 efg | 9.5 ± 0.3 fg |
| Water Kefir Culture | Liquid (L) or Grain (G) Isolation | Isolated Species | |||
|---|---|---|---|---|---|
| LAB | AAB | Yeast | Other | ||
| CLUK | L | Liquorilactobacillus nagelii | Acetobacter indonesiensis, Acetobacter orientalis, Acetobacter syzygii, Gluconobacter oxydans | Pichia membranifaciens, Saccharomyces cerevisiae | n.d. |
| G | n.d. | Gluconobacter oxydans | Pichia membranifaciens | n.d. | |
| CFUK | L | Liquorilactobacillus nagelii | Acetobacter persici, Acetobacter syzygii | Zygotorulaspora florentina | n.d. |
| G | Liquorilactobacillus nagelii | Acetobacter persici | Zygotorulaspora florentina | n.d. | |
| FFUK | L | Lacticaseibacillus casei, Lacticaseibacillus paracasei | Acetobacter cerevisiae, Acetobacter fabarum, Acetobacter indonesiensis, Acetobacter orientalis, Gluconobacter oxydans | n.d. | n.d. |
| G | Lacticaseibacillus paracasei | n.d. | n.d. | Uncultured bacterium | |
| KGIE | L | Liquorilactobacillus nagelii | Acetobacter fabarum, Acetobacter indonesiensis, Acetobacter orientalis, Acetobacter tropicalis | Pichia membranifaciens, Zygotorulaspora florentina | Uncultured bacterium, uncultured fungus, Aureobasidium pullulans, Curtobacterium flaccumfaciens, Deinococcus xingiangensis, Paenibacillus humicus |
| G | Liquorilactobacillus nagelii | n.d. | n.d. | - | |
| KOUK | L | Liquorilactobacillus nagelii | Acetobacter cerevisiae, Acetobacter fabarum, Acetobacter papayae, Acetobacter persici, Acetobacter suratthaniensis | Issatchenkia orientalis | Uncultured bacterium |
| G | n.d. | n.d. | n.d. | n.d. | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zannini, E.; Lynch, K.M.; Nyhan, L.; Sahin, A.W.; O’ Riordan, P.; Luk, D.; Arendt, E.K. Influence of Substrate on the Fermentation Characteristics and Culture-Dependent Microbial Composition of Water Kefir. Fermentation 2023, 9, 28. https://doi.org/10.3390/fermentation9010028
Zannini E, Lynch KM, Nyhan L, Sahin AW, O’ Riordan P, Luk D, Arendt EK. Influence of Substrate on the Fermentation Characteristics and Culture-Dependent Microbial Composition of Water Kefir. Fermentation. 2023; 9(1):28. https://doi.org/10.3390/fermentation9010028
Chicago/Turabian StyleZannini, Emanuele, Kieran M. Lynch, Laura Nyhan, Aylin W. Sahin, Patrick O’ Riordan, Daenen Luk, and Elke K. Arendt. 2023. "Influence of Substrate on the Fermentation Characteristics and Culture-Dependent Microbial Composition of Water Kefir" Fermentation 9, no. 1: 28. https://doi.org/10.3390/fermentation9010028
APA StyleZannini, E., Lynch, K. M., Nyhan, L., Sahin, A. W., O’ Riordan, P., Luk, D., & Arendt, E. K. (2023). Influence of Substrate on the Fermentation Characteristics and Culture-Dependent Microbial Composition of Water Kefir. Fermentation, 9(1), 28. https://doi.org/10.3390/fermentation9010028






