Enhancement of Triterpenoid Synthesis in Antrodia cinnamomea through Homologous Expression of the Key Synthetic Pathway Genes AcLSS and AcERG4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Reagents
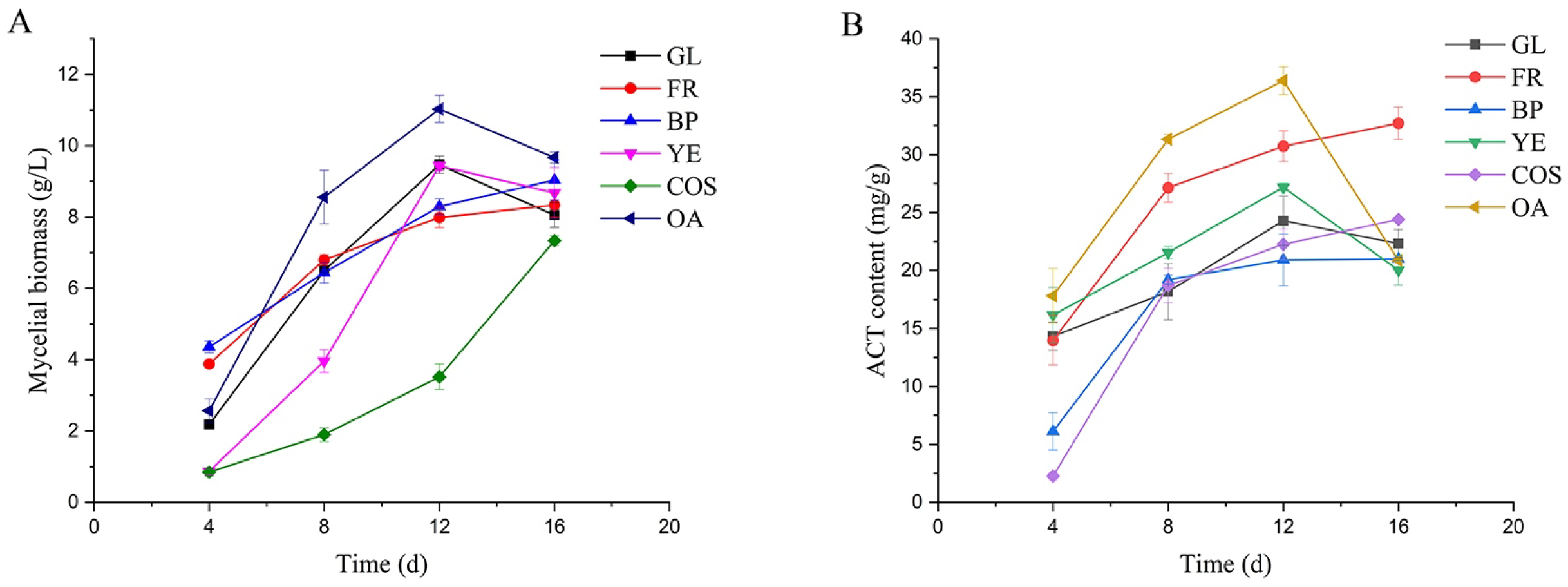
2.2. Determination of Mycelial Biomass and ACT Content under Various Culture Conditions
2.3. Correlation Analysis of ACT Content and Enzyme Expression Levels in Synthetic Pathway
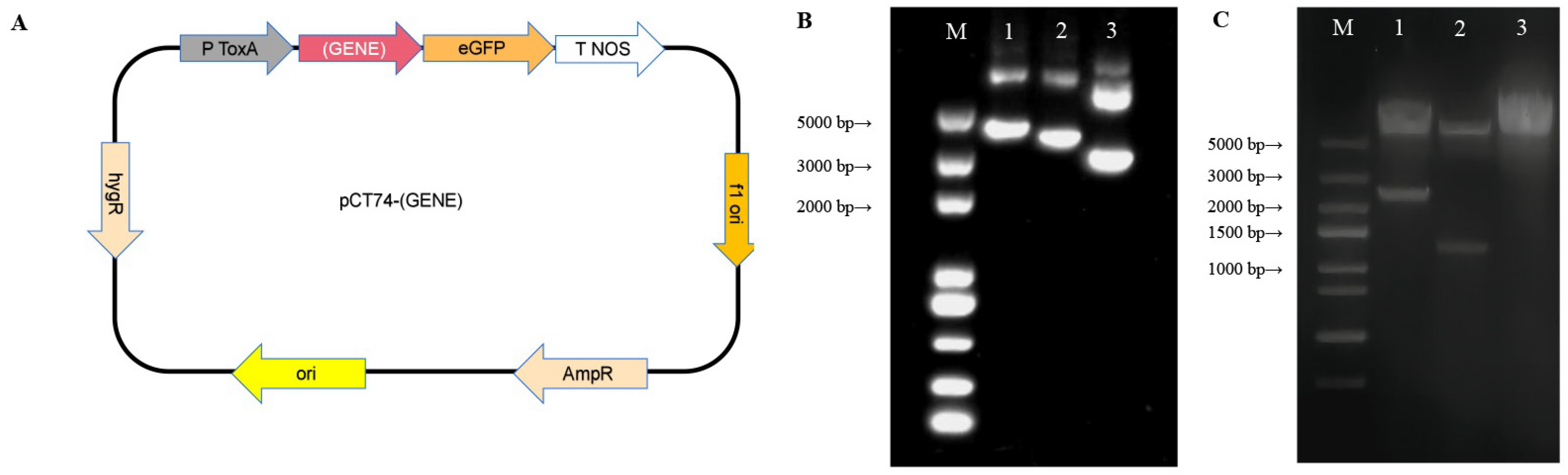
2.4. Construction of Plasmids pCT74-AcLSS and pCT74-AcERG4
2.5. Preparation of AC Protoplasts
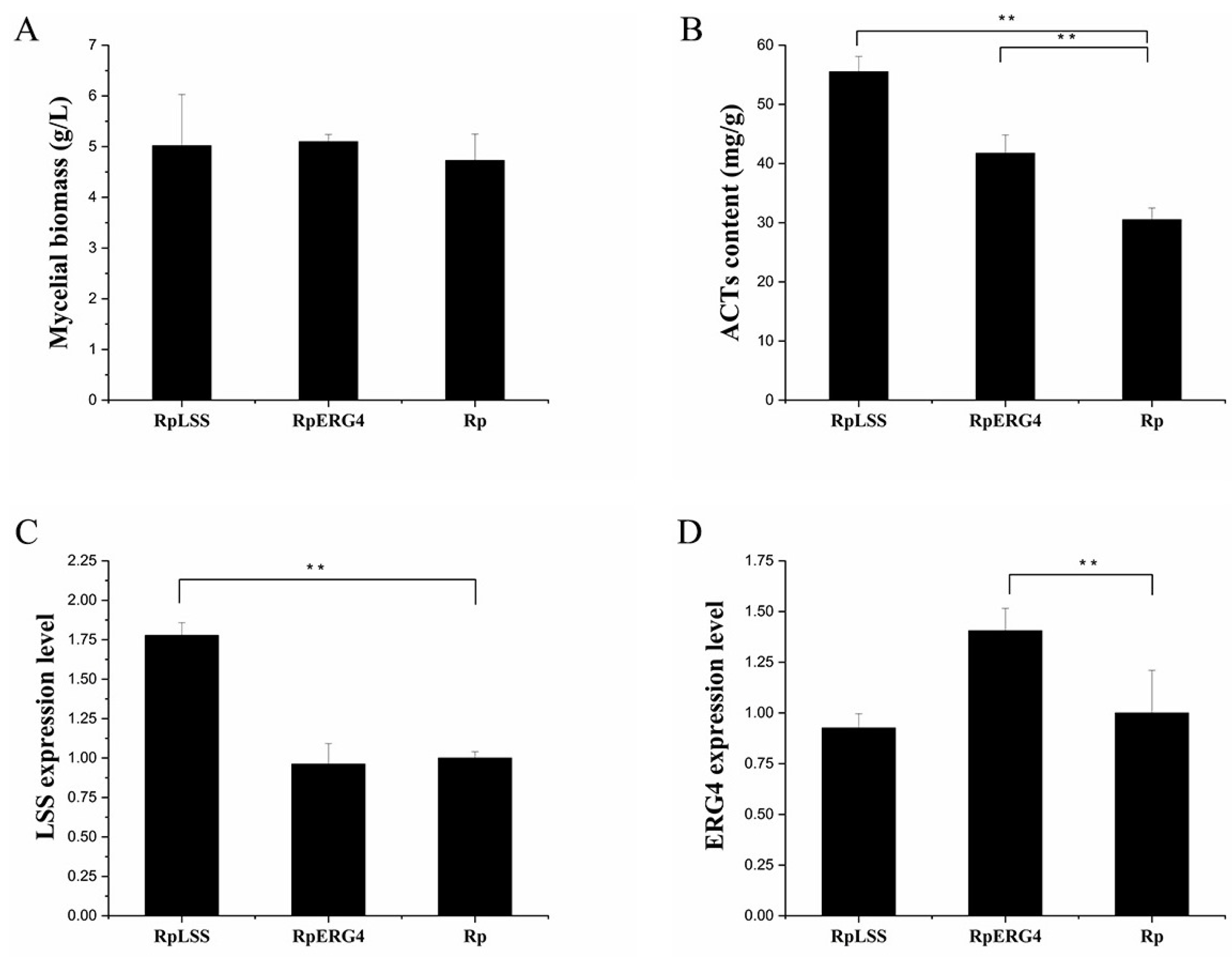
2.6. Homologous Expression of AcLSS and AcERG4
2.7. Statistical Analysis
3. Results
3.1. Changes in Mycelial Biomass and ACT Content under Various Fermentation Conditions
3.2. Expression Levels of Key Genes in ACT Synthetic Pathways
3.3. Correlation Analysis of ACT Content and Synthesis Gene Expression
3.4. Construction of Plasmid for Homologous Transformation
3.5. Protoplast Preparation and PEG-CaCl-Mediated Transformation
3.6. Enhanced Target Gene Expression Levels and ACT Content in Recombinant Strains
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chang, T.; Chou, W. Antrodia cinnamomea sp. nov. on Cinnamomum kanehirai in Taiwan. Mycol. Res. 1995, 99, 756–758. [Google Scholar] [CrossRef]
- Chen, J.J.; Lin, W.J.; Liao, C.H.; Shieh, P.C. Anti-inflammatory benzenoids from Antrodia camphorata. J. Nat. Prod. 2007, 70, 989–992. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, G.; Shen, M.Y.; Lin, K.H.; Lan, M.H.; Wu, L.Y.; Chou, D.S.; Lin, C.H.; Su, C.H.; Sheu, J.R. Antioxidative and hepatoprotective effects of Antrodia camphorata extract. J. Agr. Food Chem. 2003, 51, 3302–3308. [Google Scholar] [CrossRef] [PubMed]
- Song, T.Y.; Yen, G.C. Antioxidant properties of Antrodia camphorata in submerged culture. J. Agr. Food Chem. 2002, 50, 3322–3327. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Shiao, Y.J.; Lin, R.D.; Shao, Y.Y.; Lai, M.N.; Lin, C.C.; Ng, L.T.; Kuo, Y.H. Neuroprotective Diterpenes from the Fruiting Body of Antrodia camphorata. J. Nat. Prod. 2006, 69, 689–691. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.L.; Kuo, Y.C.; Kuo, P.L.; Ng, L.T.; Kuo, Y.H.; Lin, C.C. Apoptotic effects of extract from Antrodia camphorata fruiting bodies in human hepatocellular carcinoma cell lines. Cancer Lett. 2005, 221, 77–89. [Google Scholar] [CrossRef]
- Li, Y.; Ge, J.; Li, Y.; Zheng, S.; Liu, Y.; Liang, Y.; Mei, Y. Isolation, Purification, and Antitumor Activity of a Novel Active Protein from Antrodia cinnamomea Liquid Fermentation Mycelia. Fermentation 2023, 9, 185. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, D.; Wang, Z.; Zang, W.; Rao, P.; Liang, Y.; Mei, Y. Alpha-terpineol affects synthesis and antitumor activity of triterpenoids from Antrodia cinnamomea mycelia in solid-state culture. Food Funct. 2018, 9, 6517–6525. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Li, D.; Zang, W.; Zhu, H.; Wu, P.; Mei, Y.; Liang, Y. A polysaccharide from Antrodia cinnamomea mycelia exerts antitumor activity through blocking of TOP1/TDP1-mediated DNA repair pathway. Int. J. Biol. Macromol. 2018, 120, 1551–1560. [Google Scholar] [CrossRef]
- Kuang, Y.; Li, B.; Wang, Z.; Qiao, X.; Ye, M. Terpenoids from the medicinal mushroom Antrodia camphorata: Chemistry and medicinal potential. Nat. Prod. Rep. 2021, 38, 83–102. [Google Scholar] [CrossRef]
- Yang, C.M.; Zhou, Y.J.; Wang, R.J.; Hu, M.L. Anti-angiogenic effects and mechanisms of polysaccharides from Antrodia cinnamomea with different molecular weights. J. Ethnopharmacol. 2009, 123, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Hirakawa, A.; Gao, J.J.; Kakuda, H.; Shiro, M.; Komatsu, Y.; Sheu, C.c.; Hattori, M. Five New Maleic and Succinic Acid Derivatives from the Mycelium of Antrodia camphorata and Their Cytotoxic Effects on LLC Tumor Cell Line. J. Nat. Prod. 2004, 67, 46–48. [Google Scholar] [CrossRef] [PubMed]
- Geethangili, M.; Tzeng, Y.M. Review of pharmacological effects of Antrodia camphorata and its bioactive compounds. Evid. Based Complement. Altern. Med. 2011, 2011, 212641. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.C.; Wu, T.Y.; Chang, F.R.; Lin, W.Y.; Hsu, Y.M.; Cheng, F.T.; Lu, C.Y.; Yen, M.H.; Tsui, Y.T.; Chen, H.L.; et al. Chemical profiling of the cytotoxic triterpenoid-concentrating fraction and characterization of ergostane stereo-isomer ingredients from Antrodia camphorata. J. Pharm. Biomed. Anal. 2012, 58, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Liao, P.C.; Kuo, D.C.; Lin, C.C.; Ho, K.C.; Lin, T.P.; Hwang, S.Y. Historical spatial range expansion and a very recent bottleneck of Cinnamomum kanehiraeHay.(Lauraceae) in Taiwan inferred from nuclear genes. BMC Evol. Biol. 2010, 10, 124. [Google Scholar] [CrossRef]
- Ho, Y.C.; Lin, M.T.; Duan, K.J.; Chen, Y.S. The hepatoprotective activity against ethanol-induced cytotoxicity by aqueous extract of Antrodia cinnamomea. J. Chin. Inst. Chem. Eng. 2008, 39, 441–447. [Google Scholar] [CrossRef]
- Meng, L.; Luo, B.; Yang, Y.; Faruque, M.O.; Zhang, J.; Li, X.; Hu, X. Addition of vegetable oil to improve triterpenoids production in liquid fermentation of medicinal fungus Antrodia cinnamomea. J. Fungi 2021, 7, 926. [Google Scholar] [CrossRef]
- Lu, M.Y.J.; Fan, W.L.; Wang, W.F.; Chen, T.; Tang, Y.C.; Chu, F.H.; Chang, T.T.; Wang, S.Y.; Li, M.y.; Chen, Y.H.; et al. Genomic and transcriptomic analyses of the medicinal fungus Antrodia cinnamomea for its metabolite biosynthesis and sexual development. Proc. Natl. Acad. Sci. USA 2014, 111, E4743–E4752. [Google Scholar] [CrossRef]
- Fang, X.; Shi, L.; Ren, A.; Jiang, A.L.; Wu, F.L.; Zhao, M.W. The cloning, characterization and functional analysis of a gene encoding an acetyl-CoA acetyltransferase involved in triterpene biosynthesis in Ganoderma lucidum. Mycoscience 2013, 54, 100–105. [Google Scholar] [CrossRef]
- Dai, Z.; Cui, G.; Zhou, S.F.; Zhang, X.; Huang, L. Cloning and characterization of a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase gene from Salvia miltiorrhiza involved in diterpenoid tanshinone accumulation. J. Plant Physiol. 2011, 168, 148–157. [Google Scholar] [CrossRef]
- Shi, L.; Qin, L.; Xu, Y.; Ren, A.; Fang, X.; Mu, D.; Tan, Q.; Zhao, M. Molecular cloning, characterization, and function analysis of a mevalonate pyrophosphate decarboxylase gene from Ganoderma lucidum. Mol. Biol. Rep. 2012, 39, 6149–6159. [Google Scholar] [CrossRef]
- Asadollahi, M.A.; Maury, J.; Schalk, M.; Clark, A.; Nielsen, J. Enhancement of farnesyl diphosphate pool as direct precursor of sesquiterpenes through metabolic engineering of the mevalonate pathway in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2010, 106, 86–96. [Google Scholar]
- Shang, C.H.; Shi, L.; Ren, A.; Qin, L.; Zhao, M.W. Molecular cloning, characterization, and differential expression of a lanosterol synthase gene from Ganoderma lucidum. Biosci. Biotech. Biochem. 2010, 74, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Kristan, K.; Rižner, T.L. Steroid-transforming enzymes in fungi. J. Steroid Biochem. 2012, 129, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.Z.; Zhu, Y.L.; Huang, P.W.; Yang, Q.; Dai, C.C. Strategies for gene disruption and expression in filamentous fungi. Appl. Microbiol. Biotechnol. 2019, 103, 6041–6059. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Lou, H.; Hu, J.; Liu, Z.; Chen, Q. Macrofungi: A review of cultivation strategies, bioactivity, and application of mushrooms. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2333–2356. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, L.; Shen, M.; Liu, J.; Li, Y.; Xu, S.; Chen, L.; Shi, G.; Ding, Z. Establishment of an efficient polyethylene glycol (peg)-mediated transformation system in Pleurotus eryngii var. ferulae using comprehensive optimization and multiple endogenous promoters. J. Fungi 2022, 8, 186. [Google Scholar] [CrossRef]
- Liu, R.; Kim, W.; Paguirigan, J.A.; Jeong, M.H.; Hur, J.S. Establishment of Agrobacterium tumefaciens-mediated transformation of Cladonia macilenta, a model lichen-forming fungus. J. Fungi 2021, 7, 252. [Google Scholar] [CrossRef]
- Dong, Z.; Gao, N.; Deng, B.; Huang, X.; Hu, C.; Chen, P.; Wu, Q.; Lu, C.; Pan, M. Stable transformation of fluorescent proteins into Nosema bombycis by electroporation. Parasites Vectors 2022, 15, 141. [Google Scholar] [CrossRef]
- Schmoll, M.; Zeilinger, S. Resistance marker-and gene gun-mediated transformation of Trichoderma reesei. In Methods in Molecular Biology; Humana: New York, NY, USA, 2021; pp. 55–62. [Google Scholar]
- Attri, C.; Swati; Kulshrestha, S. Restriction enzyme-mediated insertional mutagenesis: An efficient method of Rosellinia necatrix transformation. Arch. Microbiol. 2018, 200, 189–194. [Google Scholar] [CrossRef]
- Liu, X.; Xia, Y.; Zhang, Y.; Liang, L.; Xiong, Z.; Wang, G.; Song, X.; Ai, L. Enhancement of antroquinonol production via the overexpression of 4-hydroxybenzoate polyprenyltransferase biosynthesis-related genes in Antrodia cinnamomea. Phytochemistry 2021, 184, 112677. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lin, X.; Tong, J.; Liu, P.; Lin, Z. ITS sequence analysis of Antrodia cinnamomea and total triterpenoid detection of liquid fermentation mycelium. Southwest China J. Agric. Sci. 2015, 28, 2649–2654. [Google Scholar]
- Li, B.; Kuang, Y.; He, J.B.; Tang, R.; Xu, L.L.; Leung, C.H.; Ma, D.L.; Qiao, X.; Ye, M. Antcamphorols A–K, cytotoxic and ROS scavenging triterpenoids from Antrodia camphorata. J. Nat. Prod. 2019, 83, 45–54. [Google Scholar] [CrossRef]
- Huang, Y.; Lin, X.; Qiao, X.; Ji, S.; Liu, K.; Yeh, C.t.; Tzeng, Y.m.; Guo, D.; Ye, M. Antcamphins A–L, ergostanoids from Antrodia camphorata. J. Nat. Prod. 2014, 77, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Qiao, X.; An, R.; Huang, Y.; Ji, S.; Li, L.; Tzeng, Y.m.; Guo, D.a.; Ye, M. Separation of 25R/S-ergostane triterpenoids in the medicinal mushroom Antrodia camphorata using analytical supercritical-fluid chromatography. J. Chromatogr. A 2014, 1358, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.Y.; Chen, C.Y.; Chien, S.C.; Hsiao, W.W.; Chu, F.H.; Li, W.H.; Lin, C.C.; Shaw, J.F.; Wang, S.Y. Metabolite profiles for Antrodia cinnamomea fruiting bodies harvested at different culture ages and from different wood substrates. J. Agr. Food Chem. 2011, 59, 7626–7635. [Google Scholar] [CrossRef]
- Luo, Z.S.; Lu, Z.M.; Hu, M.M.; Li, X.Y.; Xu, G.Q.; Gong, J.S.; Shi, J.S.; Xu, Z.H. Characterization of Cinnamomum kanehirae Extract-Stimulated Triterpenoids Synthesis in Submerged Fermentation of Antrodia camphorata via Untargeted Metabolomics. J. Agr. Food Chem. 2023, 71, 9175–9186. [Google Scholar] [CrossRef]
- You, H.; Sun, B.; Li, N.; Xu, J.W. Efficient expression of heterologous genes by the introduction of the endogenous glyceraldehyde-3-phosphate dehydrogenase gene intron 1 in Ganoderma lucidum. Microb. Cell Fact. 2021, 20, 164. [Google Scholar] [CrossRef]
- Lin, Y.L.; Lee, Y.R.; Tsao, N.W.; Wang, S.Y.; Shaw, J.F.; Chu, F.H. Characterization of the 2, 3-oxidosqualene cyclase gene from Antrodia cinnamomea and enhancement of cytotoxic triterpenoid compound production. J. Nat. Prod. 2015, 78, 1556–1562. [Google Scholar] [CrossRef]
- Liu, H.; Xing, H.; Jin, Y.; Liu, J.; Tzeng, Y.M.; Deng, L.; Wang, F. Application of multiple strategies to improve the production of the potential cancer drug 4-acetylantroquinonol B (4-aaqb) by the rare fungus Antrodia cinnamomea. Appl. Biochem. Biotechnol. 2022, 194, 2720–2730. [Google Scholar] [CrossRef]
- Li, J.; Lin, X.J.; Shao, E.S.; Lin, Z.X. Molecular cloning and expression analysis of mevalonate pyrophosphate decarboxylase in Antrodia cinnamomea. MATEC Web Conf. 2016, 64, 03001. [Google Scholar] [CrossRef]
- He, H.; Li, Y.; Fang, M.; Li, T.; Liang, Y.; Mei, Y. Carbon source affects synthesis, structures, and activities of mycelial polysaccharides from medicinal fungus Inonotus obliquus. J. Microbiol. Biotechnol. 2021, 31, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.C.F.; Su, Y.H.; Kanagarajan, S.; Agrawal, D.C.; Tsay, H.S. Development of an activation tagging system for the basidiomycetous medicinal fungus Antrodia cinnamomea. Mycol. Res. 2009, 113, 290–297. [Google Scholar] [CrossRef] [PubMed]


| Primer Name | Primer Sequence – |
|---|---|
| QAACT-F | ATCCTCGGCAAGTTCGTC |
| QAACT-R | GCCTCGTTGATCTCGAAAAA |
| QHMGR-F | TATTGTTGCGGAAGCTGTTG |
| QHMGR-R | ATAAGCGTGATGCACTGAGA |
| QMVD-F | GCAGGACTCGAACCAATTTC |
| QMVD-R | AGCATCGTAGGTATAAGCGG |
| QIDI-F | TTCTCCGCTTTTGTTTTCCG |
| QIDI-R | AGTTCATGTTCGAGTTTCCG |
| QFPS-F | CTGAACCCTACTACGTCCAC |
| QFPS-R | CTGTAGAAGGAGTGGCAGAG |
| QSQS-F | TGCGATCTACAAATCCACGA |
| QSQS-R | ATACTTGTTGCGTGTTTCCG |
| QSE-F | CTCATGGAAGGAGTCAACGA |
| QSE-R | TAATTCGTACAGCGCAACAC |
| QLSS-F | GTCCAGAGAGTGATGCTTACA |
| QLSS-R | TGTTCTCTTCCTCGTGAGC |
| QERG27-F | AATGCCTAGAATCCCCCTTG |
| QERG27-R | ATAGTGGCCGAAAACATTGC |
| QERG4-F | TAAGATGTGGCTCGAAGTCC |
| QERG4-R | ACATATCCCACGTTTGTGGA |
| QGAPDH-F | GTATTGGCCGTATCGTGCTG |
| QGAPDH-R | TTGGCTTCGACCCTTCCCT |
| IAcLSS-F | ccaactctattcgcagggtcctATGTCGTACTCCCCTCTCG |
| IAcLSS-R | gcgtgtacttacgattctaggacccTTAAACCTGACGGGCTTCGT |
| IAcERG4-F | ccaactctattcgcagggtcctATGTCTAACATCGCTTCACAGC |
| IAcERG4-R | gcgtgtacttacgattctaggacccCTAATAGATACCGGGTAAAAATTTGTATTTGAC |
| hygR-F | CTATTCCTTTGCCCTCGGAC |
| hygR-R | ATGAAAAAGCCTGAACTCACCG |
| AcLSS-F | ATGTCGTACTCCCCTCTCG |
| AcLSS-R | TTAAACCTGACGGGCTTCGT |
| AcERG4-F | ATGTCTAACATCGCTTCACAGC |
| AcERG4-R | AATAGATACCGGGTAAAAATTTGTATTTGAC |
| AACT | HMGR | MVD | IDI | FPS | SQS | SE | LSS | ERG27 | ERG4 | |
|---|---|---|---|---|---|---|---|---|---|---|
| HMGR | 0.490 * | |||||||||
| MVD | 0.296 | 0.449 * | ||||||||
| IDI | 0.175 | 0.522 ** | 0.693 ** | |||||||
| FPS | 0.472 * | 0.679 ** | 0.808 ** | 0.675 ** | ||||||
| SQS | 0.694 ** | 0.496 * | 0.587 ** | 0.431 * | 0.722 ** | |||||
| SE | 0.265 | 0.423 * | 0.112 | −0.037 | 0.452 * | 0.432 * | ||||
| LSS | 0.506 * | 0.536 ** | 0.467 * | 0.104 | 0.704 ** | 0.606 ** | 0.594 ** | |||
| ERG27 | 0.341 | 0.516 ** | 0.612 ** | 0.393 | 0.651 ** | 0.613 ** | 0.436 * | 0.705 ** | ||
| ERG4 | 0.605 ** | 0.556 ** | 0.858 ** | 0.582 ** | 0.899 ** | 0.844 ** | 0.330 | 0.697 ** | 0.709 ** | |
| ACTc 1 | 0.456 * | 0.506 * | 0.283 | 0.124 | 0.505 * | 0.523 ** | 0.504 * | 0.541 ** | 0.374 | 0.493 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Fang, M.; Huang, J.; Li, Y.; Mei, Y. Enhancement of Triterpenoid Synthesis in Antrodia cinnamomea through Homologous Expression of the Key Synthetic Pathway Genes AcLSS and AcERG4. Fermentation 2023, 9, 880. https://doi.org/10.3390/fermentation9100880
Zheng S, Fang M, Huang J, Li Y, Mei Y. Enhancement of Triterpenoid Synthesis in Antrodia cinnamomea through Homologous Expression of the Key Synthetic Pathway Genes AcLSS and AcERG4. Fermentation. 2023; 9(10):880. https://doi.org/10.3390/fermentation9100880
Chicago/Turabian StyleZheng, Siqi, Mingyue Fang, Jiaxin Huang, Yanbin Li, and Yuxia Mei. 2023. "Enhancement of Triterpenoid Synthesis in Antrodia cinnamomea through Homologous Expression of the Key Synthetic Pathway Genes AcLSS and AcERG4" Fermentation 9, no. 10: 880. https://doi.org/10.3390/fermentation9100880






