Abstract
The imbalance of the redox state caused by extra reactive oxygen species is closely related to many diseases. Therefore, it is necessary for people to ingest antioxidants through food. The safety of some synthetic antioxidants has been questioned. In this context, it is worth exploring natural and safe antioxidants from biological sources. Tea has good antioxidant activity, and the antioxidant activity of fermented sour tea is better than that of other types. It is necessary to clarify the antioxidant capacity of sour tea during fermentation, as well as the microbial community and its sources. Nonculture and culture-dependent methods were adopted to track the changes in the microbial population and community structure during the fermentation of sour tea. Sequence analysis of 16S rRNA gene amplification revealed significant differences in community complexity and structure at different fermentation times. The highest proportion of operational taxonomic units (OTU s) in all samples was Latilactobacillus, which was determined to be Lactiplantibacillus plantarum by further analysis. The second highest proportion of OTUs was Enterobacter. With the fermentation of sour tea, the antioxidant capacity increased, and all isolated Lb. plantarum had good DPPH clearance rates. Our findings suggest that Lb. plantarum plays a crucial role in the fermentation process of sour tea. The possibility of discovering new antioxidants was provided by the determination of the antioxidant capacity and bacterial community during the fermentation of sour tea.
1. Introduction
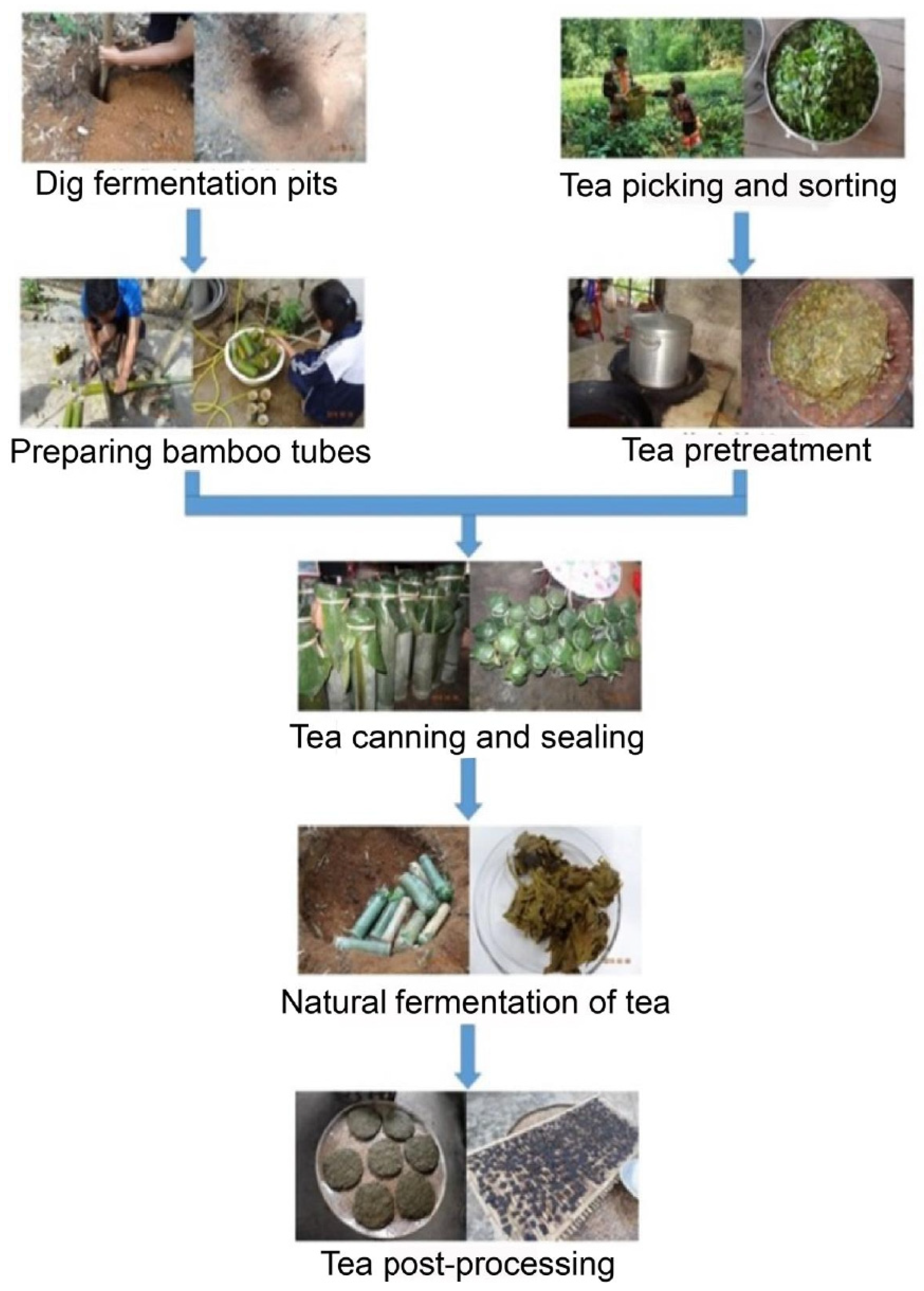
Tea is generally divided into fermented tea (such as black tea) and nonfermented tea (such as green tea) [1]. Traditional fermented tea is produced by microbial fermentation under aerobic and anaerobic conditions using green tea as a raw material through thermal processing [2]. In some areas of Southeast Asia, there is a pickled tea that is significantly different from the traditional fermented tea (in Thailand called Miang, in Myanmar called Laphet, in Japan called Awa-bancha, and in China called sour tea). Miang ferments for several weeks to a year without adding any preservatives [3,4]. Awa-bancha is mainly produced from July to September. The production process of Awa-bancha is to pick tea leaves and boil them until brown and then put them into plastic or wooden containers for fermentation. Awa-bancha takes from two to several weeks to ferment [5,6]. For sour tea, boiled fresh tea leaves are placed into a tight bamboo container without air. The bamboo tube is sealed and buried three meters underground. The fermentation time of sour tea is 60 days, and then the sour tea is removed 30 days post fermentation (Figure 1).
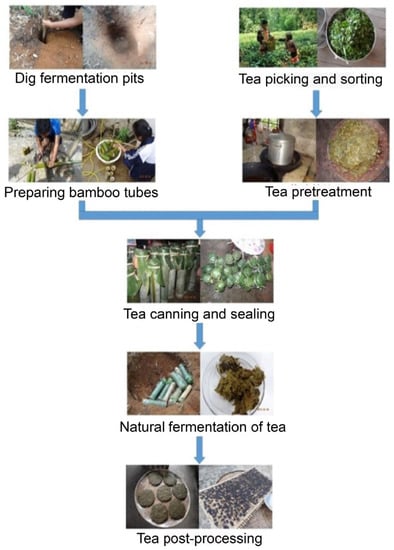
Figure 1.
The sour tea fermentation process. Boiled fresh tea leaves are placed in a bamboo container. The bamboo pipe is sealed and buried three meters underground. The fermentation time of sour tea is 60 days, and then the sour tea is removed from the ground and fermented for 30 days.
Pickled tea is produced by anaerobic microorganism fermentation. Traditional fermented tea is produced by fungal fermentation. For example, Rasamsonia, Thermomyces and Aspergillus are dominant fungi in the middle stage of the fermentation of Pu’er tea, while Aspergillus is the dominant bacteria in other stages [7]. One study found that most of the strains isolated from Miang were Latilactobacillus, among which Lactiplantibacillus plantarum (Lb. plantarum) was dominant (64.6%) [8]. Latilactobacillus and Acetobacter were the most abundant bacteria in Laphet. At the same time, Enterobacteriaceae bacteria were also found in all Laphet samples [9]. In Awa-bancha, Latilactobacillus dominated, followed by Klebsiella. At the species level, Lb. plantarum was the main bacteria, followed by Lb. vaccinalis, Lb. paracolinoides, and Klebsiella variicola (K. variicola) [10]. Among 26 Latilactobacillus isolated from sour tea, 24 were Lb. plantarum, one was Enterococcus faecalis, and one was Lb. acidophilus [11].
Plant-derived food contains many antioxidant substances. Tea is an important source of antioxidants, and its antioxidant substances mainly comprise polyphenols [12]. LAB are also good antioxidants. Many LAB have enzymatic and nonenzymatic antioxidant mechanisms to reduce the production of reactive oxygen species (ROS) to a level harmless to cells [13]. For example, LAB can release and promote the production of glutathione and catalase, which are major nonenzymatic antioxidants and free radical scavengers [14]. LAB can also produce free amino acids and short peptides. Many amino acids, such as histidine, tyrosine, methionine, and cysteine, have antioxidant activities [15]. The physical changes produced by LAB fermentation affect the antioxidant activity of tea. For example, an acidic environment is conducive to the stability of polyphenols [16]. The increase in temperature produced by LAB fermentation increases the content of polyphenols [17]. LAB can improve antioxidant activity by improving the composition of polyphenols in food. For example, the fermentation of apple juice with Lb. plantarum can improve free radical scavenging and antioxidant activities. After fermentation, the contents of 5-o-caffeoylquinic acid, quercetin and phloretin in apple juice increase, and the improved phenolic components are helpful for improving the antioxidant activity [18]. Tea has strong antioxidant activity after microbial fermentation [19,20]. Some studies have shown that LAB fermentation changes the phenolic components extracted from crude tea and enhances the overall antioxidant capacity [21,22]. Research has also shown that Lb. brevis- and Lb. plantarum-fermented black tea extract can effectively alleviate oxidative stress in cells. Fermented black tea has a more obvious protective effect on normal cells, can maintain higher cell vitality, and significantly inhibits the production of ROS. Lb. plantarum is one of the few LAB that can degrade tannin [23]. Tannase produced by Lb. plantarum can catalyze the hydrolysis of ester bonds in tannin and gallate to synthesize effective antioxidant propyl gallate substrates. Lb. plantarum ST and Lb. plantarum STDA10 have good 2,2-diphenyl-1-picrylhydrazyl (DPPH) and 2,2-azino-di-[3-ethylbenzthiazoline sulfonate] (ABTS+) scavenging capacities [11].
However, the role of LAB in sour tea remains unclear. The microbial composition of sour tea during fermentation has not been determined. Therefore, this study combined 16S rRNA gene amplification and culture technology to determine the diversity and dynamics of the bacterial community during the fermentation of sour tea. This study shows that fermentation time has an effect on the microorganisms in sour tea as well as its antioxidant capacity.
2. Materials and Methods
2.1. Sample Collection
De’ang sour tea was selected from fresh tea from Santai Mountain of De’ang Nationality in Yunnan Province, China (Longitude: 98.39, Latitude: 24.33). Three-month fermented sour tea samples were collected (24 samples in total at different time points). Twenty-four Lb. plantarum strains were isolated and identified according to morphology, catalase test, Gram staining, and 16S rDNA sequences as described by Cao [24].
2.2. Analysis of Biochemical Characteristics
2.2.1. pH Determination
During the fermentation of sour tea, the pH value was measured by a pH meter (AS ONE, Osaka, Japan). All analyses were performed in triplicate.
2.2.2. Catechin Detection
Catechin detection was analyzed using a high-performance liquid chromatography system (HPLC, Agilent Technologies Inc., Santa Clara, CA, USA). Briefly, 0.2 g of fermented sour tea was ground and then mixed with 5 mL of 70% methanol solution. After incubation in a water bath at 75 °C for 20 min, the supernatant was collected by centrifugation at 5000 rpm for 10 min and filtered through a 0.45-μm filter (Biosharp, Hefei, Anhui, China). The supernatant was injected into an Agilent ZORBAX SB-C18 column (4.6 × 150 mm, 5 μm), and isocratic elution was used with a mobile phase consisting of acetonitrile-acetic acid-EDTA (45:10:1, v/v/v) and acetonitrile-acetic acid-EDTA (400:10:1, v/v/v) at a flow rate of 1 mL min−1. The catechins were monitored at 278 nm, and their concentrations were determined by integrating the calibration curves obtained from the standards.
2.2.3. Tea Polyphenol Detection
Extract (1 mL) and gallic acid solution (1000 μg/mL), water and 5 mL of 10% folin phenol reagent were mixed. After the mixed solution was left at room temperature for 5 min, 4 mL of 7.5% sodium carbonate was added. The absorbance at 765 nm was recorded. The absorbance of the gallic acid working solution was 10, 20, 30, 40, and 50 μg/mL. The identification method of tea polyphenols is from the Chinese national standard ISO 14502-1:2005.
2.2.4. Caffeine Detection
The tea sample (3 g) was ground, soaked in 450 mL boiling water for 45 min, and then filtered. The sample (10 mL) was added to 4 mL hydrochloric acid (0.01 mol/L) and 1 mL lead acetate alkaline solution, diluted with water to 100 mL and filtered. The filtrate (25 mL) and 0.1 mL sulfuric acid solution (4.5 mol/L) were mixed and diluted to 50 mL. The absorbance at 274 nm was recorded. The caffeine identification method is from the Chinese national standard ISO 10727:1995.
2.2.5. Determination of the Total Amount of Free Amino Acids
The tea sample (3 g) was ground, soaked in 450 mL boiling water for 45 min, and then filtered. The sample was mixed with 0.5 mL phosphate buffer solution (pH 8) and 0.5 mL 2% ninhydrin solution. The mixture was heated in boiling water for 15 min and then cooled, before the volume was stabilized to 25 mL with water. The absorbance of the mixed solution was measured at 570 nm after it was left at room temperature for 10 min.
2.2.6. Determination of the Water Extract
After the tea sample (2 g) was ground, 300 mL boiling water was added. The sample was placed in boiling water for 45 min. After the extraction, the tea residue was filtered by filter paper. The tea residue and filter paper were put into a thermostatic drying oven (120 ± 2 °C), dried for 1 h, cooled for 1 h to room temperature, and weighed after repetition.
2.3. Whole-Genome DNA of Sour Tea Was Extracted, Amplified and Sequenced by PCR
The total DNA in sour tea was extracted by a QIAamp PowerFecal DNA kit (Qiagen, Hilton, Germany). The extracted DNA was amplified by PCR. Primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify V3 and V4 of the 16S rRNA gene [25]. A 10-ng aliquot of each sample was used for PCR with Rapid Taq Master Mix (Vazyme, Nanjing, China), and the annealing temperature was 50 °C. The Illumina MiSeq™ System (Illumina, San Diego, CA, USA) was used for sequencing. The PCR product sequencing data of the sample were uploaded to the National Center for Biotechnology Information (NCBI) under the registration number PRJNA893263.
2.4. Bioinformatics Analysis
All sequences were demultiplexed using each sample barcode. Primer sequences were reduced by using pairwise sequence alignment, and sequences were collected to correct sequencing errors. According to Miseq_SOP, sequence processing was performed by combining the features of mothur v1.44.1 [26]. The SSU rRNA database sequence and taxonomic information from Silva (v138) [27] were downloaded directly from the mothur website. Chimeric sequences were removed using the UCHIME algorithm [28]. According to different analyses, operational taxonomic units (OTUs) were assigned to operational classification units (OTUs) with a 97% similarity threshold [29]. Alpha diversity analysis using QIIME2 (Version 2022.2) software detected Faith’s pd, Shannon’s diversity, observed features, and evenness indices and displayed them using QIIME2 view. Beta diversity was calculated with QIIME2, including the unweighted UniFrac method. Bacterial OTUs were classified using the Silva database and named at the level of domain, phylum, class, order, family, and genus [30].
2.5. Radical Scavenging Activity
The cell-free culture supernatant (CFCS) of the bacterial fermentation broth (109 CFU and 1010 CFU) was mixed with 2 mL DPPH ethanol solution. A mixture of DPPH and normal saline was used as a blank sample. Vitamin C (Vc) was used as a positive control. The absorbance at 517 nm was recorded [11].
The scavenging ability was defined as follows:
Scavenging effect (%) = 1 − [A517(sample)/A517(blank)] × 100%
The CFCS (109 CFU and 1010 CFU) of the bacterial fermentation broth was mixed with 2 mL of Fenton’s reagent (1 mL of ferrous sulfate solution and 1 mL of 3% hydrogen peroxide solution) and 1 mL of salicylic acid. The mixture was placed in an incubator at 37 °C for 15 min. A mixture of Fenton’s reagent, salicylic acid and normal saline was used as a blank sample. The absorbance at 517 nm was recorded [31].
The scavenging ability was defined as follows:
Scavenging effect (%) = 1 − [A510(sample)/A510(blank)] × 100%
The CFCS of the bacterial fermentation broth (109 CFU and 1010 CFU) was mixed with 4.5 mL of Tris-diacetyl pentaacetic acid solution (pH 8.2, Tris concentration 0.012 g/mL, diacetyl pentaacetic acid concentration 0.0008 g/mL) and 300 μL of mixed pyrogallol solution (concentration 0.00025 g/mL). The mixture was incubated at room temperature for 10 min. A mixture of Tris diacetamide pentaacetic acid solution, pyrogallol solution, and normal saline was used as a blank sample. The absorbance at 517 nm was recorded [32].
The scavenging ability was defined as follows:
Scavenging effect (%) = 1 − [A325(sample)/A325(blank)] × 100%
3. Results
3.1. Physicochemical Analysis
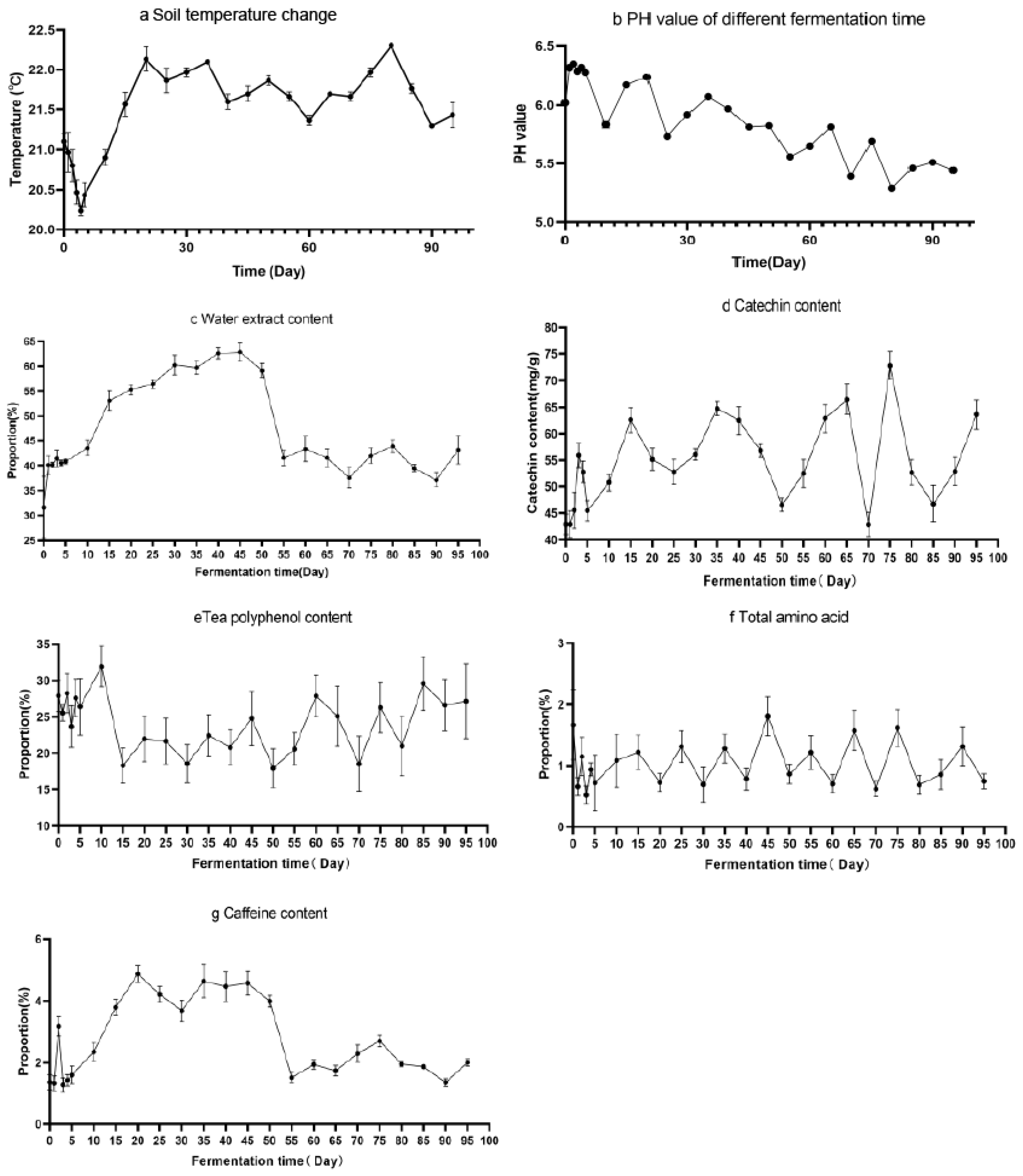
The temperature of fermentation environment is detected during fermentation (Figure 2a). The pH value increased slightly at the beginning of fermentation, but the pH value decreased with fermentation time (Figure 2b). The main antioxidant substances in the fermentation of sour tea were determined (Figure 2c–g). The total amount of water extract gradually increased at the beginning of fermentation, and then decreased until Day 55 (p < 0.01). The contents of polyphenols and catechins increased significantly at the beginning of fermentation and gradually increased with the fermentation process (p < 0.01). The contents of these two substances were basically stable after 60 days of fermentation. The proportion of total soluble sugar increased rapidly in the early stage of fermentation, then decreased slightly, and then remained stable.
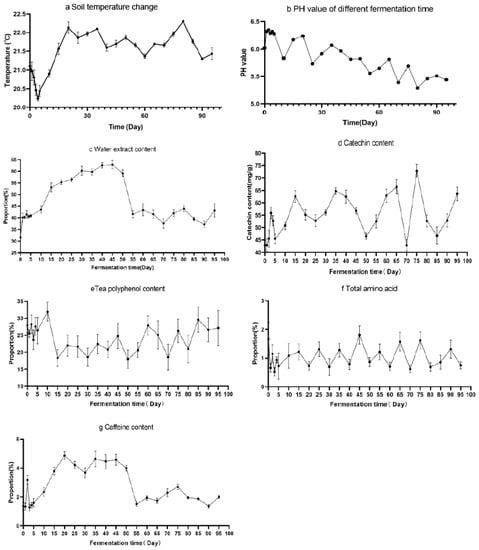
Figure 2.
Physiochemical analysis of sour tea (a) Changes of soil temperature during fermentation (b) pH value changes during the fermentation of sour tea. Contents of the main antioxidant substances in sour tea at different fermentation times. (c) The content of water extract in sour tea fermentation. (d) The catechin content in sour tea fermentation. (e) The tea polyphenol content in sour tea fermentation. (f) The total amino acid content in sour tea fermentation. (g) The caffeine content in sour tea fermentation.
3.2. Free Radical Scavenging Activity of Sour Tea and Lb. plantarum
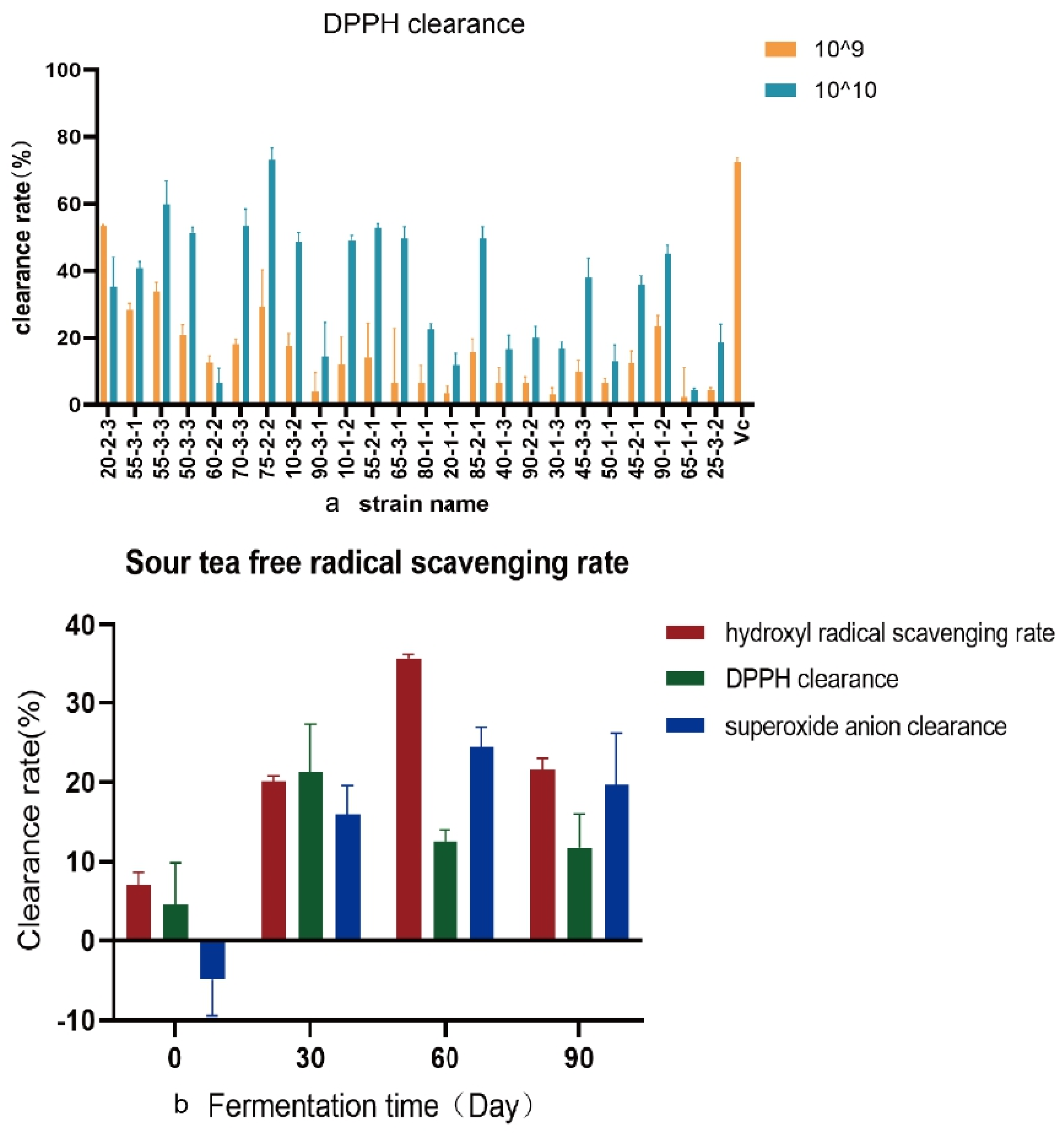
Bacteria in sour tea were isolated by MRS medium. Bacteria were identified by 16S rRNA gene sequencing. The antioxidant capacity of the CFCS of 24 strains of Lb. plantarum was determined (Figure 3a). It was found that the highest DPPH clearance rate of Lb. plantarum 75-2-2 was 73.25% (73.25 ± 3.41), which was similar to that of Vc (0.8 mg/mL; 72.49% ± 1.34). The clearance rates of other CFCS were lower than that of Vc (0.8 mg/mL). The hydroxyl radical scavenging rate and the superoxide anion scavenging rate of these 24 strains of Lb. plantarum were weak. The sour tea was sampled at four time points (0 days of fermentation, one month of fermentation, two months of fermentation, and completion of fermentation) (Figure 3b). The DPPH scavenging rate, hydroxyl radical scavenging rate and superoxide anion scavenging rate of the samples were tested. It was found that the scavenging rate of hydroxyl radicals and superoxide anions increased with increasing fermentation time (p < 0.01). The DPPH removal rate also increased at the beginning of fermentation, reaching the maximum value at one month of fermentation. Post fermentation did not enhance the antioxidant activity of sour tea.
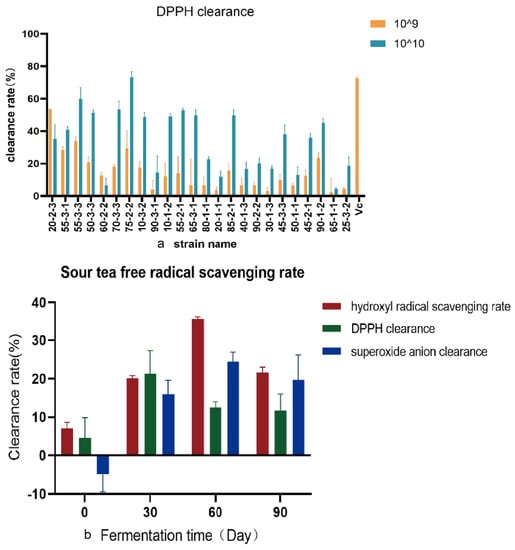
Figure 3.
Antioxidant capacity was measured (a) DPPH free radical scavenging rate of 24 strains of Lb. plantarum isolated from sour tea. Vc (0.8 mg/mL) was the positive control. Orange represents the bacterial concentration of 109 CFU, and blue represents the bacterial concentration of 1010 CFU. (b) Free radical scavenging rate of sour tea fermented at different times.
3.3. Microbial Communities during the Fermentation Process
3.3.1. Alpha Diversity Index
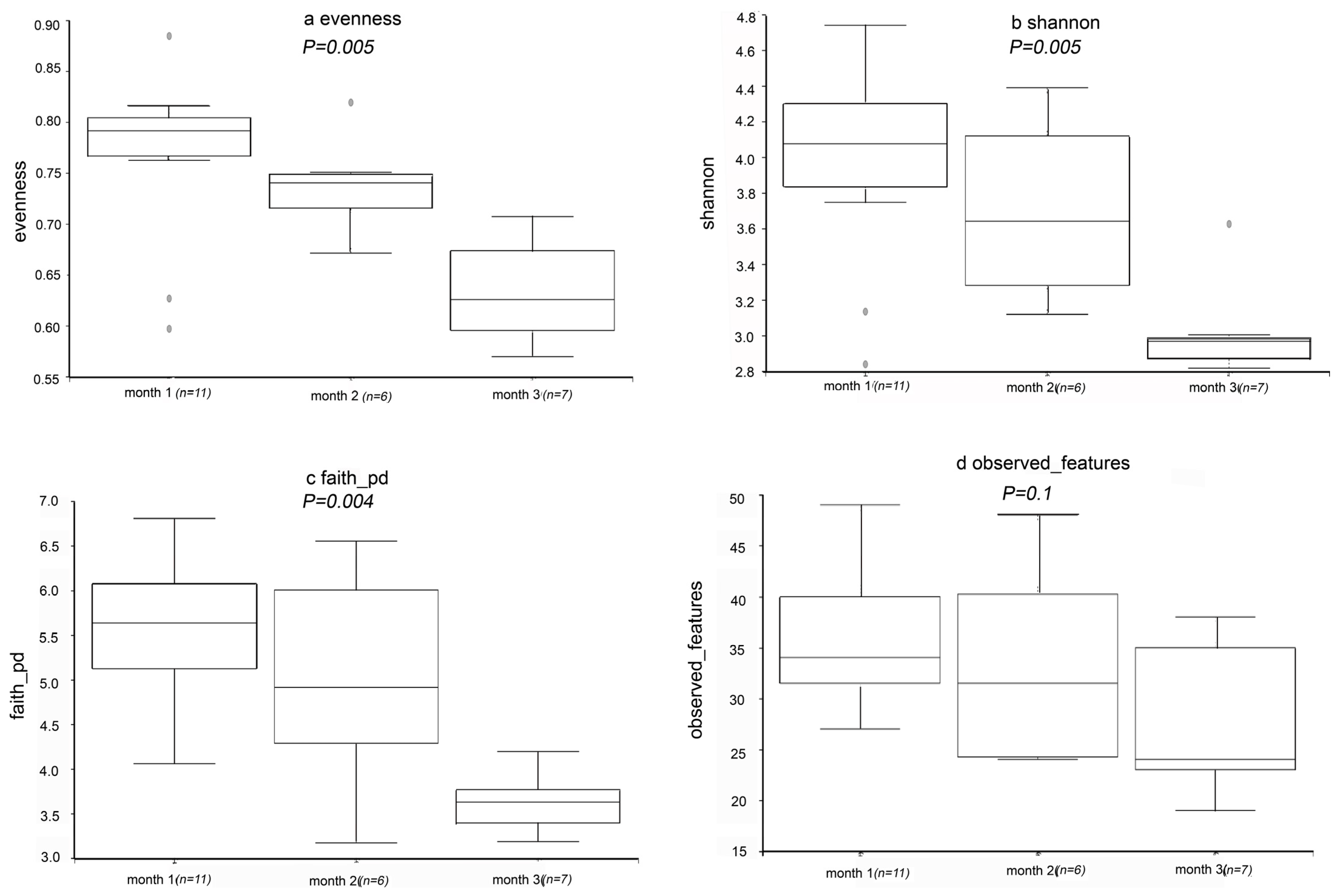
According to the sequencing results, 1596 OTUs were found in the samples. The coverage index of all samples was at least 99%, indicating that the information obtained is sufficient to reveal the existence and relative richness of most components of the bacterial community. The evenness, Shannon’s diversity, Faith’s pd, and observed features indices are represented in the form of box plots (Figure 4a–d). According to Figure 4a–c, evenness, Shannon’s diversity, and Faith’s pd index values decreased significantly during the fermentation process, indicating that microbial diversity decreased significantly. This may be related to the length of fermentation time. The microbial richness was expressed by number of OTUs. In addition, according to Figure 4d, the observed features index did not change significantly. There was no significant change in the observed features index, which may be due to the difference between the algorithm and other indexes [30].
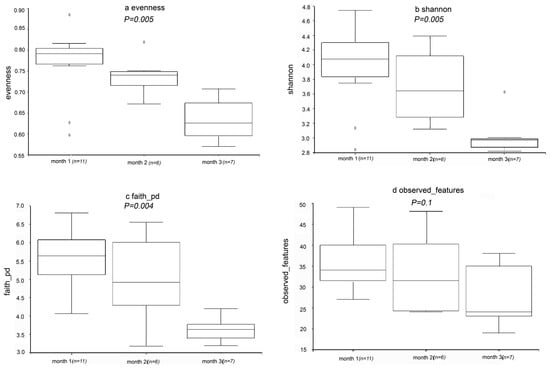
Figure 4.
Alpha diversity analysis. Box plot of index differences between groups of (a) evenness, (b) Shannon’s diversity, (c) Faith’s pd and (d) observed features at different processing steps.
3.3.2. Comparison of Community Composition
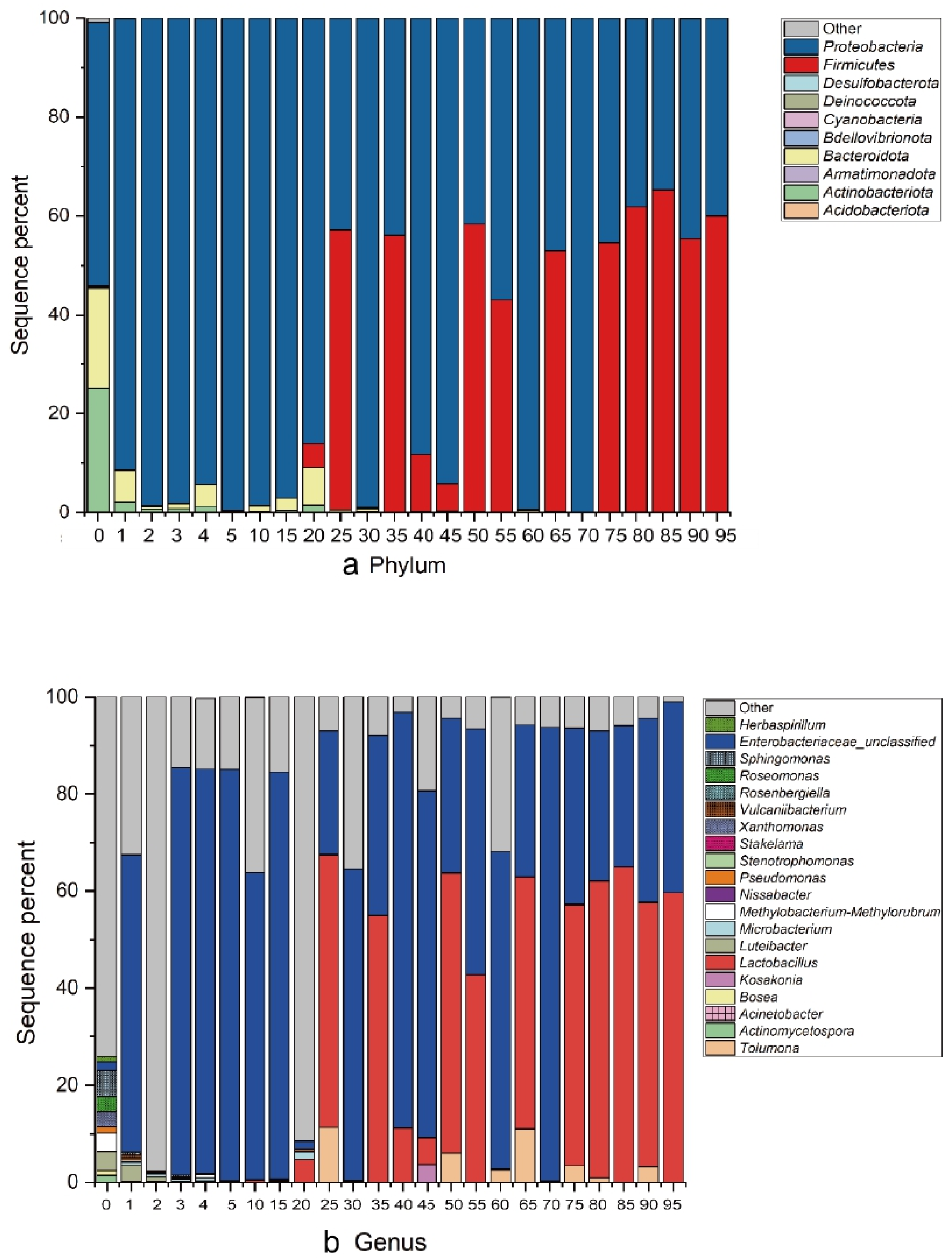
By sequencing and analyzing the samples in the fermentation process, it was detected that the bacteria mainly came from 10 phyla and showed significant taxonomic differences (Figure 5a). Proteobacteria accounted for the largest proportion in the microbiota. With increasing fermentation time, the proportion of Firmicutes gradually increased, and the proportion of actinobacteria gradually decreased.
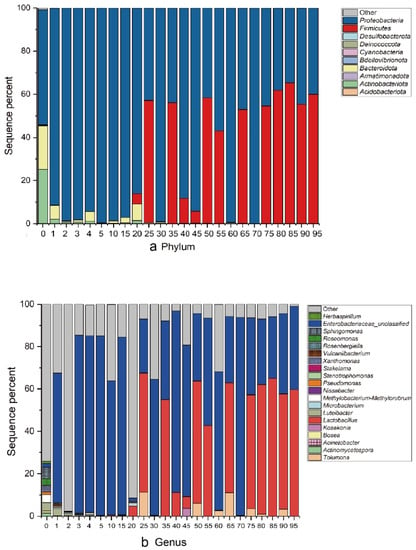
Figure 5.
Changes in the bacterial community structure during different fermentation times. The relative abundance of bacteria at the (a) phylum and (b) genus levels are shown. Each bar represents the relative abundance of each sample. Each color represents a specific phylum or genus.
A total of 68 different bacterial genera were detected, and an overview of the relevant top 20 was presented. As shown in Figure 5b, the proportions of Latilactobacillus, Enterobacteriaceae_unclassified and Tolumona were more abundant in specimens. The highest content of sour tea samples (0 day) was the sequence that could not be determined at the genus level (74%), followed by Sphingomonas (5%), Luteibacter (4%), Methylobacterium-Methylorubrum (4%), Xanthomonas (3%) and Roseomonas (3%). In the process of fermentation, the composition of the microbial flora showed significant fluctuations. In the second and third months of fermentation, the dominant strain became Enterobacteriaceae_unclassified (29–93%) and Latilactobacillus (0–65%). As a less studied fermented food, sour tea may contain a large number of uncultured microbes. On days 60 and 70 of fermentation, the abnormal proportion of Lactiplantibacillus may be due to the change of fermentation environment (sour tea being taken out of the pit for post-fermentation).
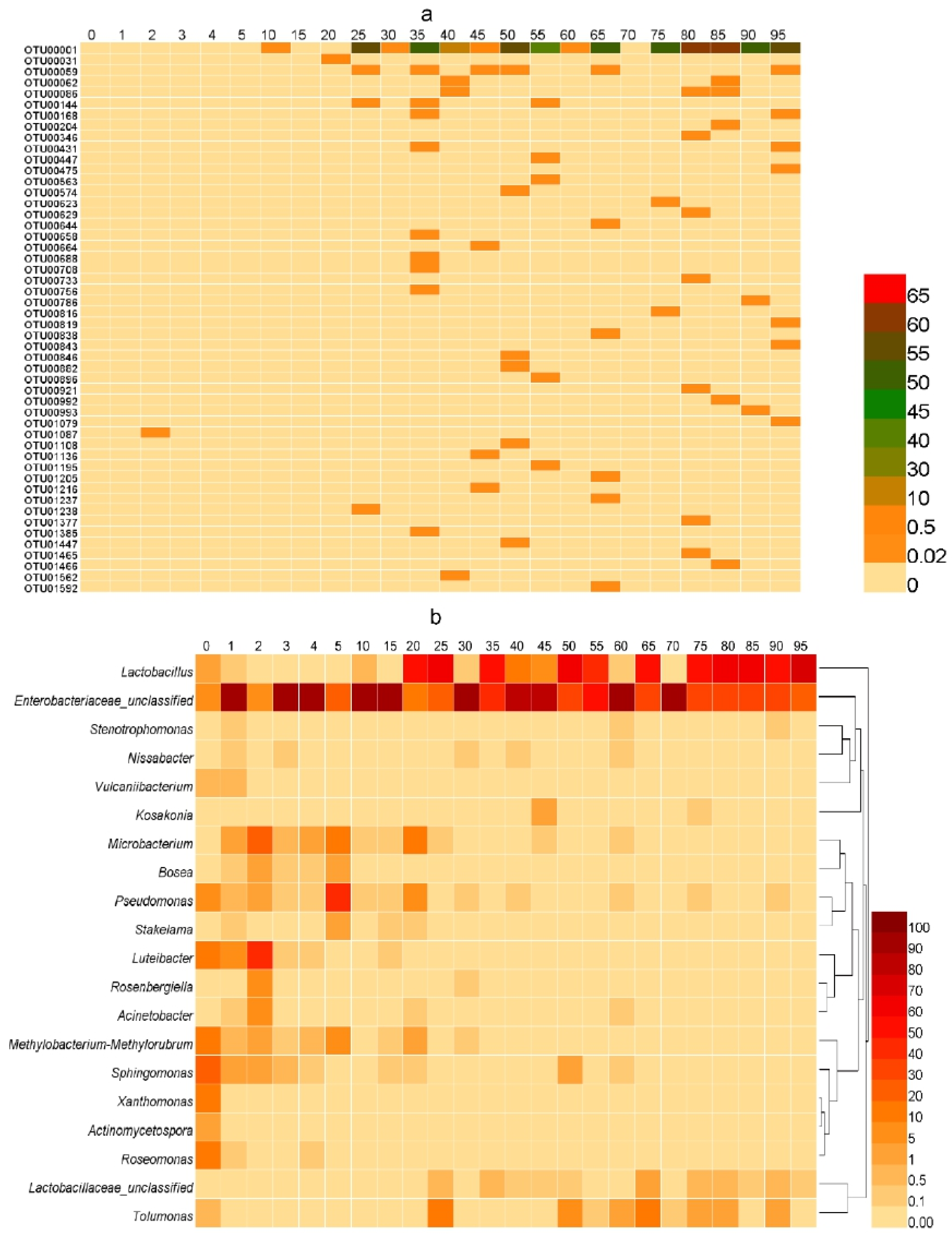
Among all samples, the OTU with the highest sequence proportion was Latilactobacillus (0–65%). After further analysis, it was determined to be the Lb. plantarum sequence. At the same time, a relatively high number of OTUs belonged to Enterobacter, which is consistent with the classification information. This OTU belonged to Pantoea septica after blast comparison, which is a bacterium often detected in plants. The changes in these OTUs during fermentation are shown in Figure 6a (those with low proportions are shown in red). From Day 25, the proportion of Lb. plantarum increased to 56%, and in most samples thereafter, the proportion remained at approximately 50%. The highest proportion of Lb. plantarum was 64% on Day 85 of fermentation. This shows that the main bacteria in the fermentation process of sour tea are Lb. plantarum and Enterobacter. The tea was pretreated before fermentation (stir-fry the tea). Although tea was pretreated before fermentation, some trace bacteria will still be retained. This may be one of the possible sources of Lb. plantarum in the fermentation process. In the 0 day sour tea samples, Roseomonas and Sphingomonas account for the highest proportion. They were very common bacteria in the environment. This was the same as most fermented tea [33].
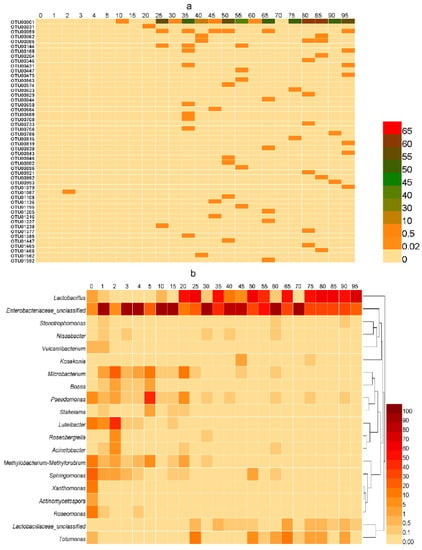
Figure 6.
The proportion of microbial community in sour tea at different fermentation times. (a) The proportion of OTUs belonging to Lb. plantarum in the sour tea fermentation process. The proportion of Lb. plantarum in unfermented sour tea was very low. Lb. plantarum accounted for only 5% 20 days before the fermentation of sour tea. Lb. plantarum accounted for 56% on the 25th day of fermentation. The highest proportion of Lb. plantarum was 64% on the 85th day of fermentation. (b) Heatmap showing changes in the microbial community of sour tea during fermentation. The top 20 bacteria were selected for their relative abundance at the genus level.
3.3.3. Microbial Diversity Changes with Fermentation Time
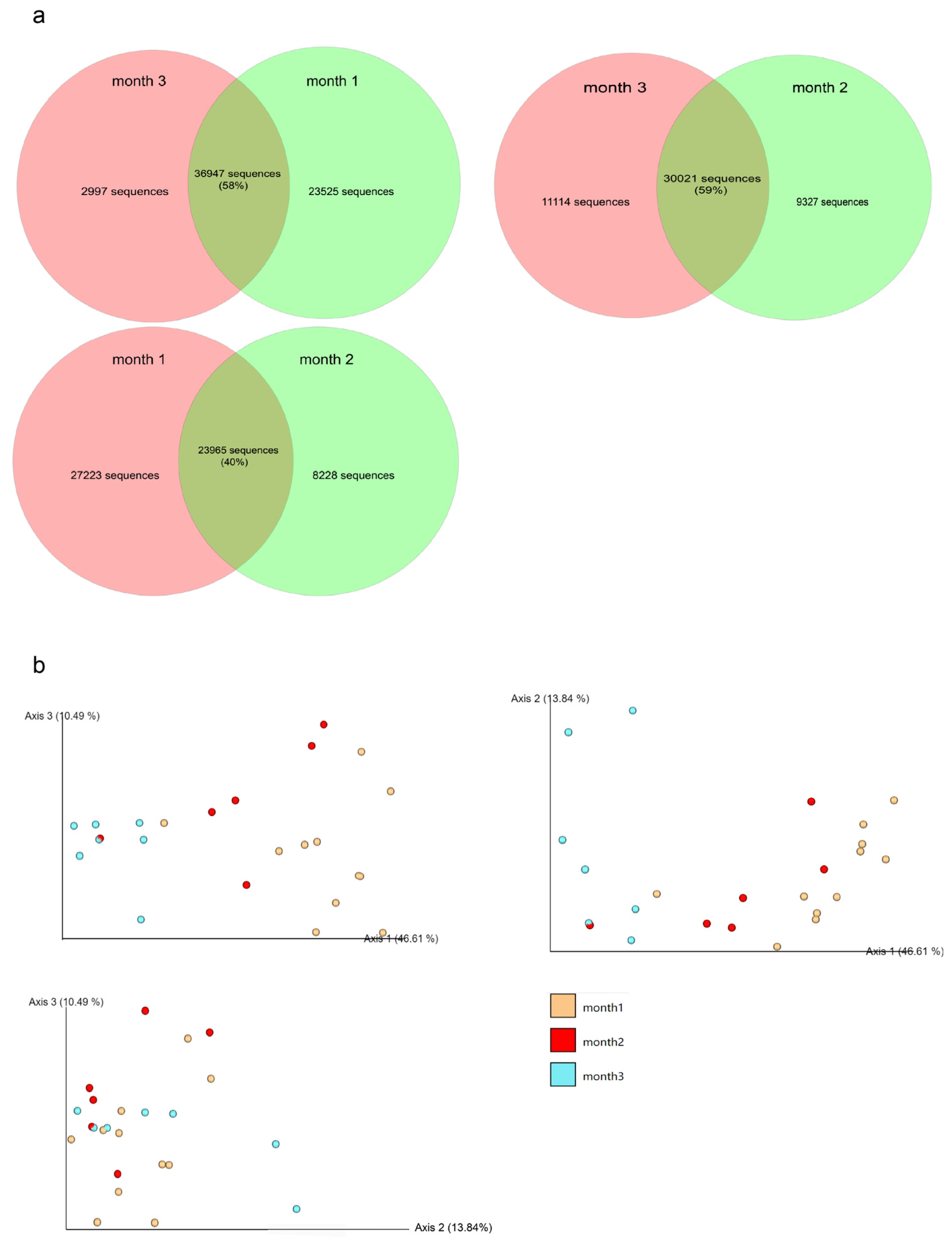
The bacterial sequences shared between the sour teas with different fermentation times are shown by the Venn diagram. Sour tea fermented for three months shared nearly 60% of the sequence with sour tea fermented for one month and two months. However, 40% of bacterial sequences were shared between one-month fermented sour tea and two-month fermented sour tea (Figure 7a).
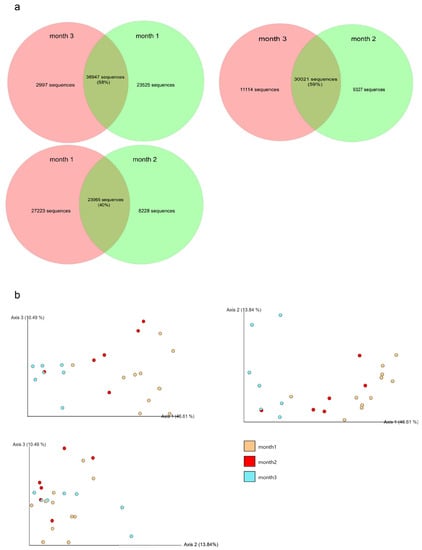
Figure 7.
The effect of ferment times on microbial community composition. (a) Venn diagram showing unique and shared sequences of bacterial communities at different fermentation times. (b) Principal coordinate analysis of the bacterial community during the fermentation of sour tea. The analysis was conducted using the unweighted UniFrac method.
For PCoA, based on the unweighted UniFrac distances, the first principal components accounted for 47% of the total variance, which covered most of the variable information. As shown in Figure 7b, the microbial communities of the three groups of samples were separated from each other, and different fermentation times had a major impact on the bacteria. The larger the difference in fermentation time of samples, the greater the difference in microbial communities.
The differences and similarities of the bacterial community structure between different fermentation times were compared by a heatmap (Figure 6b). There were significant differences in the types of dominant bacteria at different times during the fermentation of sour tea. For unfermented sour tea, the main bacterial species included Actinomycetospora, Bosea, Roseomonas, Xanthomonas, and Sphingomonas. However, their abundance was very low, and the number was negligible. Conversely, the microbial species were significantly altered by one month of fermentation, among which the bacteria included Luteibacter, Vulcaniibacterium, Acinetobacter, Rosenbergiella, Stakelama, Pseudomonas, and Microbacterium. Latilactobacillus, Enterobacteriaceae_unclassified, and Tolumonas predominated when fermentation proceeded to two months. The proportion of Latilactobacillus increased further during the third month of fermentation.
4. Discussion
In this study, the microbial community composition and changes in sour tea during fermentation were analyzed. The antioxidant capacity and antioxidant content of sour tea were tested. The relationship between the microbial community and the antioxidant capacity of sour tea was revealed. This provides a basis for improving the fermentation of sour tea.
According to previous reports, the most abundant bacteria in Japanese Awa-bancha are Lb. pentosus and Lb. plantarum [34,35]. The most abundant bacterial genus in kombucha is Acetobacter, followed by Foalobacter and Gluconobacter [36]. The pH value decreases significantly during the fermentation of sour tea, and the results of our study were consistent with previous reports in this regard [37]. The pH value of the sour tea sample was higher than that of fermented Kombucha (pH 3) and lower than that of fermented black tea (pH 5.9) [21,38]. The difference in pH value in the process of sour tea fermentation is the result of the accumulation of acid secreted by microorganisms and the transformation of tea ingredients [20]. The most abundant bacteria in sour tea were Latilactobacillus and Enterobacteriaceae. Further analysis revealed that K. variicola accounted for the highest proportion of Enterobacteriaceae. K. variicola was not found in unfermented sour tea, but it accounted for the highest proportion in sour tea fermented for two months, while the proportion in sour tea fermented for three months decreased significantly. A large proportion of K. variicola was also found in Awa-bancha [10]. K. variicola is a gram-negative bacterium that is isolated from banana trees [39]. K. variicola is habitually isolated from a wide range of plant ecosystems, where it plays an important role in nitrogen fixation and plant growth promotion [40]. This shows that the main microorganisms in pickled tea are different in different regions. This may also be related to the fermentation mode and fermentation time.
With the progress of fermentation, the proportion of Lb. plantarum in sour tea increased and the antioxidant capacity of sour tea also increased. These findings are similar to those reported for Miang [41], which show that Lb. plantarum plays an important role in the antioxidation of sour tea. This may be caused by an excessively long fermentation time. The DPPH clearance rate of sour tea was lower than that of Lb. plantarum CFCS. This may be because Lb. plantarum has a variety of esterase (tannase) genes, TanaA (tanALp) [42] and TanB (tanBLp) [43], which can hydrolyze the ester bond of methyl gallate to generate gallic acid [44]. Lb. plantarum is the most common LAB in the food fermentation of plants rich in phenolic compounds and is a model bacterium for studying the metabolism of phenolic compounds [44]. Phenolic acids and flavonoids exist in tea. Indeed, thousands of molecules with polyphenol structures have been found in some plants. These molecules are secondary metabolites of plants [45]. The B ring of polyphenol compounds contains a 3′-4′ dihydroxy group, and the C ring contains gallic acid ester, which is an excellent antioxidant [16,46]. Phenolic acids account for almost one third of dietary phenols, mainly including benzoic acid derivatives (such as gallic acid and protocatechin) and cinnamic acid derivatives (such as ferulic acid, coumaric acid, and caffeic acid), among which tea is an important source of gallic acid (3,4,5-trihydroxybenzoic acid). In the reported Lb. plantarum, the transformation of two carbon frameworks follows a similar two-step metabolism, first esterase action and then decarboxylation [44].
It is important to explore natural and safe antioxidants from biological resources. Research on sour tea and Lb. plantarum provided conditions for this exploration.
5. Conclusions
Overall, the complex bacterial structure in the fermentation of sour tea is highly dynamic, showing significant changes at different stages. The dynamic changes in the dominant bacteria in different fermentation stages were identified. During the third month of sour tea fermentation, the bacterial composition changed greatly. The decrease in microbial diversity and the increase in Lb. plantarum and K. variicola during fermentation may be related to the anaerobic fermentation environment and time. The antioxidant capacity of sour tea is closely related to fermentation time and the proportion of Lb. plantarum. It is not the case that the longer the fermentation time of sour tea is, the stronger its antioxidant capacity. The traditional fermentation method of sour tea needs improvement. In conclusion, sour tea can be considered an important source of biological antioxidants. These data provide a useful framework for further improving sour tea fermentation and exploring biological antioxidants.
Author Contributions
S.Z.: data curation, writing—original draft, sequencing data analysis. C.S.: data curation, writing—original draft. C.L.: antioxidant activity determination, statistical analysis. X.Z.: DNA extraction and PCR amplification. F.G.: sample preparation and collection, writing—review & editing, project administration. X.L.: funding acquisition, writing—review & editing, project administration. All authors have read and agreed to the published version of the manuscript.
Funding
Natural Science Foundation of Yunnan Province (202101AT070139).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, D.M.; Lu, J.L.; Miao, A.Q.; Xie, Z.Y.; Yang, D.P. HPLC-DAD-ESI-MS/MS analysis in leaves of 22 tea of polyphenols and purine alkaloids cultivars in China. J. Food Compos. Anal. 2008, 21, 361–369. [Google Scholar] [CrossRef]
- Gong, Z.; Watanabe, N.; Yagi, A.; Etoh, H.; Sakata, K.; Ina, K.; Liu, Q. Compositional Change of Pu-erh Tea during Processing. Biosci. Biotechnol. Biochem. 1993, 57, 1745–1746. [Google Scholar] [CrossRef]
- Kanpiengjai, A.; Chui-Chai, N.; Chaikaew, S.; Khanongnuch, C. Distribution of tannin-’tolerant yeasts isolated from Miang, a traditional fermented tea leaf (Camellia sinensis var. assamica) in northern Thailand. Int. J. Food Microbiol. 2016, 238, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Unban, K.; Khatthongngam, N.; Shetty, K.; Khanongnuch, C. Nutritional biotransformation in traditional fermented tea (Miang) from north Thailand and its impact on antioxidant and antimicrobial activities. J. Food Sci. Technol. 2019, 56, 2687–2699. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Tamura, A.; Mizooti, Y.; Omori, M.; Nanba, A.; Miyagawa, K. Changes of Flavor during Manufacturing Process of Japanese Fermented Tea (Awa-bancha) and Its Characteristic. J. Home Econ. Jpn. 1993, 44, 561–565. [Google Scholar]
- Miyazaki, E.; Nakanishi, K. Various component analysis during manufacturing process of Awa-bancha made in spring. Rep. Tokushima Prefect. Ind. Technol. Cent. 2007, 16, 37–40. [Google Scholar]
- Zhao, M.; Su, X.Q.; Nian, B.; Chen, L.J.; Zhang, D.L.; Duan, S.M.; Wang, L.Y.; Shi, X.Y.; Jiang, B.; Jiang, W.W.; et al. Integrated Meta-omits Approaches To Understand the Microbiome of Spontaneous Fermentation of Traditional Chinese Pu-erh Tea. Msystems 2019, 4, e00680-19. [Google Scholar] [CrossRef]
- Chaikaew, S.; Baipong, S.; Sone, T.; Kanpiengjai, A.; Chui-chai, N.; Asano, K.; Khanongnuch, C. Diversity of lactic acid bacteria from Miang, a traditional fermented tea leaf in northern Thailand and their tannin-tolerant ability in tea extract. J. Microbiol. 2017, 55, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Bo, B.; Kim, S.A.; Han, N.S. Bacterial and fungal diversity in Laphet, traditional fermented tea leaves in Myanmar, analyzed by culturing, DNA amplicon-based sequencing, and PCR-DGGE methods. Int. J. Food Microbiol. 2020, 320, 108508. [Google Scholar] [CrossRef]
- Nishioka, H.; Mizuno, T.; Iwahashi, H.; Horie, M. Changes in lactic acid bacteria and components of Awa-bancha by anaerobic fermentation. Biosci. Biotechnol. Biochem. 2020, 84, 1921–1935. [Google Scholar] [CrossRef]
- Cao, Z.H.; Pan, H.B.; Li, S.J.; Shi, C.Y.; Wang, S.F.; Wang, F.Y.; Ye, P.F.; Jia, J.J.; Ge, C.R.; Lin, Q.Y.; et al. In Vitro Evaluation of Probiotic Potential of Lactic Acid Bacteria Isolated from Yunnan De’ang Pickled Tea. Probiotics Antimicrob. Proteins 2019, 11, 103–112. [Google Scholar] [CrossRef]
- Naczk, M.; Shahidi, F. Extraction and analysis of phenolics in food. J. Chromatogr. A 2004, 1054, 95–111. [Google Scholar] [CrossRef]
- Rodríguez, H.; Curiel, J.A.; Landete, J.M.; de las Rivas, B.; de Felipe, F.L.; Gómez-Cordovés, C.; Mancheno, J.M.; Munoz, R. Food phenolics and lactic acid bacteria. Int. J. Food Microbiol. 2009, 132, 79–90. [Google Scholar] [CrossRef]
- Kachouri, F.; Ksontini, H.; Kraiem, M.; Setti, K.; Mechmeche, M.; Hamdi, M. Involvement of antioxidant activity of Lactobacillus plantarum on functional properties of olive phenolic compounds. J. Food Sci. Technol. 2015, 52, 7924–7933. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Ju, X.R.; Yuan, J.; Wang, L.F.; Girgih, A.T.; Aluko, R.E. Antioxidant activities of rapeseed peptides produced by solid state fermentation. Food Res. Int. 2012, 49, 432–438. [Google Scholar] [CrossRef]
- Chu, S.C.; Chen, C.S. Effects of origins and fermentation time on the antioxidant activities of kombucha. Food Chem. 2006, 98, 502–507. [Google Scholar] [CrossRef]
- Ajila, C.M.; Brar, S.K.; Verma, M.; Tyagi, R.D.; Valero, J.R. Solid-state fermentation of apple pomace using Phanerocheate chrysosporium—Liberation and extraction of phenolic antioxidants. Food Chem. 2011, 126, 1071–1080. [Google Scholar] [CrossRef]
- Li, Z.X.; Teng, J.; Lyu, Y.L.; Hu, X.Q.; Zhao, Y.L.; Wang, M.F. Enhanced Antioxidant Activity for Apple Juice Fermented with Lactobacillus plantarum ATCC14917. Molecules 2019, 24, 51. [Google Scholar] [CrossRef] [PubMed]
- Okada, S.; Daengsubha, W.; Uchimura, T.A.I.; Ohara, N.; Kozaki, M. Flora of lactic acid bacteria in Miang produced in northern Thailand. J. Gen. Appl. Microbiol. 1986, 32, 57–65. [Google Scholar]
- Xiao, P.; Huang, Y.Y.; Yang, W.P.; Zhang, B.W.; Quan, X.X. Screening lactic acid bacteria with high yielding-acid capacity from pickled tea for their potential uses of inoculating to ferment tea products. J. Food Sci. Technol. 2015, 52, 6727–6734. [Google Scholar] [CrossRef]
- Jayabalan, R.; Subathradevi, P.; Marimuthu, S.; Sathishkumar, M.; Swaminathan, K. Changes in free-radical scavenging ability of kombucha tea during fermentation. Food Chem. 2008, 109, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Shah, N.P. Lactic acid bacterial fermentation modified phenolic composition in tea extracts and enhanced their antioxidant activity and cellular uptake of phenolic compounds following in vitro digestion. J. Funct. Foods 2016, 20, 182–194. [Google Scholar] [CrossRef]
- Zhao, D.Y.; Shah, N.P. Synergistic Application of Black Tea Extracts and Lactic Acid Bacteria in Protecting Human Colonocytes against Oxidative Damage. J. Agric. Food Chem. 2016, 64, 2238–2246. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.H.; Pan, H.B.; Tong, H.Q.; Gu, D.H.; Li, S.Y.; Xu, Y.P.; Ge, C.R.; Lin, Q.Y. In vitro evaluation of probiotic potential of Pediococcus pentosaceus L1 isolated from paocai-a Chinese fermented vegetable. Ann. Microbiol. 2016, 66, 963–971. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537. [Google Scholar] [CrossRef]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.G.; Peplies, J.; Glockner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Hall, M.; Beiko, R.G. 16S rRNA Gene Analysis with QIIME2. Methods Mol. Biol. 2018, 1849, 113–129. [Google Scholar] [CrossRef]
- Smirnoff, N.; Cumbes, Q.J. Hyroxyl radical scavenging activity of compatible soluts. Phytochemistry 1989, 28, 1057–1060. [Google Scholar] [CrossRef]
- Fernandez, C.; Kalpowitz, N.; Garcia, R.C.; Colell, A.; Miranda, M.; Mari, M.; Ardite, E.; Morales, A. GSH transport in mitochondria: Defense against TNF-induced oxidative stress and alcohol-induced defect. Am. J. Physiol. 1997, 273, 17–271. [Google Scholar]
- Li, Q.; Chai, S.; Li, Y.D.; Huang, J.A.; Luo, Y.; Xiao, L.Z.H. Biochemical Components Associated With Microbial Community Shift During the Pile-Fermentation of Primary Dark Tea. Front. Microbiol. 2018, 9, 1509. [Google Scholar]
- Okada, S.; Takahashi, N.; Ohara, N.; Uchimura, T.; Kozaki, M. Microorganisms Involving in Fermentation of Awa-bancha, Japanese Fermented Tea Leaves: Microorganisms Involving in Fermentation of Japanese Fermented Tea Leaves. Part I. Nippon Shokuhin Kogyo Gakkai-Shi 1996, 43, 12–20. [Google Scholar]
- Horie, M.; Sato, H.; Tada, A.; Nakamura, S.; Sugino, S.; Tabei, Y.; Katoh, M.; Toyotome, T. Regional characteristics of Lactobacillus plantarum group strains isolated from two kinds of Japanese post-fermented teas, Ishizuchi-kurocha and Awa-bancha. Biosci. Microbiota Food Health 2019, 38, 11–22. [Google Scholar] [CrossRef]
- Jayabalan, R.; Malbasa, R.V.; Loncar, E.S.; Vitas, J.S.; Sathishkumar, M. A Review on Kombucha TeaMicrobiology, Composition, Fermentation, Beneficial Effects, Toxicity, and Tea Fungus. Compr. Rev. Food Sci. Food Saf. 2014, 13, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.D.; Udompornmongkol, P.; Yang, J.H.; Chen, S.Y.; Mau, J.L. Quality and antioxidant property of three types of tea infusions. J. Food Process. Preserv. 2014, 38, 1401–1408. [Google Scholar] [CrossRef]
- Abe, M.; Takaoka, N.; Idemoto, Y.; Takagi, C.; Imai, T.; Nakasaki, K. Characteristic fungi observed in the fermentation process for Puer tea. Int. J. Food Microbiol. 2008, 124, 199–203. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Martinez, L.; Silva, J.; Martinez-Romero, E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst. Appl. Microbiol. 2004, 27, 27–35. [Google Scholar] [CrossRef]
- Duran-Bedolla, J.; Garza-Ramos, U.; Rodriguez-Medina, N.; Vera, A.A.; Barrios-Camacho, H. Exploring the environmental traits and applications of Klebsiella variicola. Braz. J. Microbiol. 2021, 52, 2233–2245. [Google Scholar] [CrossRef]
- Unban, K.; Chaichana, W.; Baipong, S.; Abdullahi, A.D.; Kanpiengjai, A.; Shetty, K.; Khanongnuch, C. Probiotic and Antioxidant Properties of Lactic Acid Bacteria Isolated from Indigenous Fermented Tea Leaves (Miang) of North Thailand and Promising Application in Synbiotic Formulation. Fermentation 2021, 7, 195. [Google Scholar] [CrossRef]
- Jimenez, N.; Esteban-Torres, M.; Mancheno, J.M.; de las Rivas, B.; Munoz, R. Tannin Degradation by a Novel Tannase Enzyme Present in Some Lactobacillus plantarum Strains. Appl. Environ. Microbiol. 2014, 80, 2991–2997. [Google Scholar] [CrossRef] [PubMed]
- Curiel, J.A.; Rodriguez, H.; Acebron, I.; Mancheno, J.M.; De Las Rivas, B.L.; Munoz, R. Production and Physicochemical Properties of Recombinant Lactobacillus plantarum Tannase. J. Agric. Food Chem. 2009, 57, 6224–6230. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; Plaza-Vinuesa, L.; Montenegro, C.; Santamaria, L.; Reveron, I.; de las Rivas, B.; Munoz, R. The use of Lactobacillus plantarum esterase genes: A biotechnological strategy to increase the bioavailability of dietary phenolic compounds in lactic acid bacteria. Int. J. Food Sci. Nutr. 2021, 72, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Manach, C.; Scalbert, A.; Morand, C.; Remesy, C.; Jimenez, L. Polyphenols: Food sources and bioavailability. Am. J. Clin. Nutr. 2004, 79, 727–747. [Google Scholar] [CrossRef] [PubMed]
- Khokhar, S.; Apenten, R.K.O. Iron binding characteristics of phenolic compounds: Some tentative structure-activity relations. Food Chem. 2003, 81, 133–140. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).