Unveiling the Effect of NCgl0580 Gene Deletion on 5-Aminolevulinic Acid Biosynthesis in Corynebacterium glutamicum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Culture Conditions
2.2. Construction of Plasmids
2.3. Analytical Methods
2.4. Transcriptome Analysis
2.5. Real-Time Quantitative PCR (RT-qPCR)
3. Results and Discussion
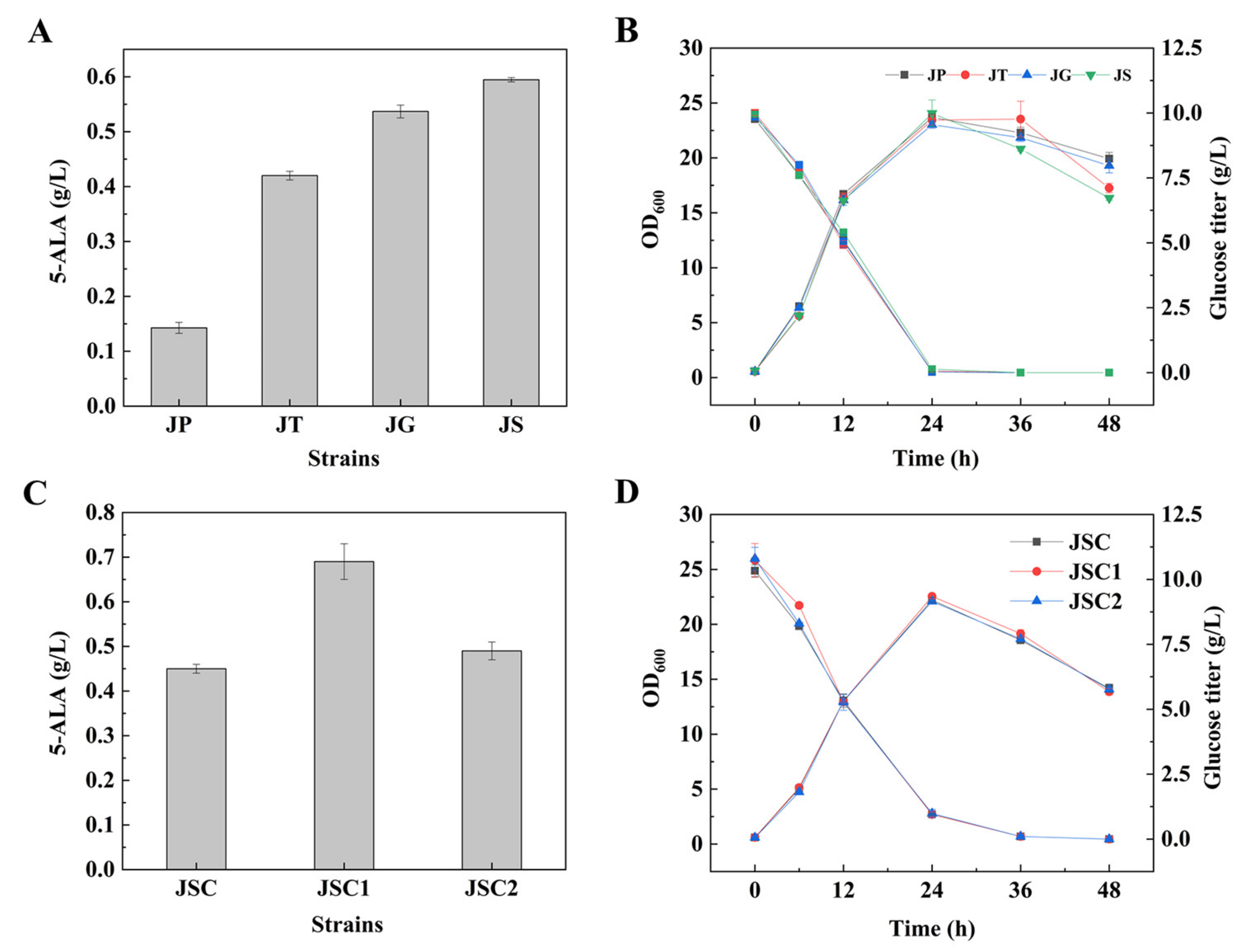
3.1. Enhancement of 5-ALA Synthesis by Overexpressing NCgl0580
3.2. Unexpected Effect of NCgl0580 Deletion on 5-ALA Biosynthesis
3.3. Transcriptomic Analysis of the Effect of NCgl0580 Knockout on 5-ALA Synthesis
3.4. Redistribution of Central Carbon Fluxes toward Succinyl-CoA-Enhanced 5-ALA Biosynthesis
3.5. Enhancement of Iron and Phosphate Uptake Improves 5-ALA Synthesis
3.6. Effect of Multiple Gene Expression Perturbation on 5-ALA Synthesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kang, Z.; Zhang, J.; Zhou, J.; Qi, Q.; Du, G.; Chen, J. Recent advances in microbial production of δ-aminolevulinic acid and vitamin B12. Biotechnol. Adv. 2012, 30, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhao, M.; Wang, W.; Hong, L.; Wu, Z.; Luo, G.; Lu, S.; Tang, Y.; Li, J.; Wang, J.; et al. 5-ALA mediated photodynamic therapy with combined treatment improves anti-tumor efficacy of immunotherapy through boosting immunogenic cell death. Cancer Lett. 2022, 554, 216032. [Google Scholar] [CrossRef] [PubMed]
- McCracken, D.J.; Schupper, A.J.; Lakomkin, N.; Malcolm, J.; Painton Bray, D.; Hadjipanayis, C.G. Turning on the light for brain tumor surgery: A 5-aminolevulinic acid story. Neuro. Oncol. 2022, 24, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Suero Molina, E.; Kaneko, S.; Black, D.; Stummer, W. 5-Aminolevulinic acid-induced porphyrin contents in various brain tumors: Implications regarding imaging device design and their validation. Neurosurgery 2021, 89, 1132–1140. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tian, Y.; Liao, X.; Tang, Y.; Ni, Q.; Sun, J.; Zhao, Y.; Zhang, J.; Teng, Z.; Lu, G. Enhancing selective photosensitizer accumulation and oxygen supply for high-efficacy photodynamic therapy toward glioma by 5-aminolevulinic acid loaded nanoplatform. J. Colloid Interf. Sci. 2020, 565, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Ngwe Tun, M.M.; Sakura, T.; Sakurai, Y.; Kurosaki, Y.; Inaoka, D.K.; Shioda, N.; Yasuda, J.; Kita, K.; Morita, K. Antiviral activity of 5-aminolevulinic acid against variants of severe acute respiratory syndrome coronavirus 2. Trop. Med. Health 2022, 50, 6. [Google Scholar] [CrossRef]
- Sakurai, Y.; Ngwe Tun, M.M.; Kurosaki, Y.; Sakura, T.; Inaoka, D.K.; Fujine, K.; Kita, K.; Morita, K.; Yasuda, J. 5-amino levulinic acid inhibits SARS-CoV-2 infection in vitro. Biochem. Biophys. Res. Commun. 2021, 545, 203–207. [Google Scholar] [CrossRef]
- Yang, L.; Wu, Y.; Wang, X.; Lv, J.; Tang, Z.; Hu, L.; Luo, S.; Wang, R.; Ali, B.; Yu, J. Physiological mechanism of exogenous 5-aminolevulinic acid improved the tolerance of chinese cabbage (Brassica pekinensis L.) to cadmium stress. Front. Plant Sci. 2022, 13, 427. [Google Scholar] [CrossRef]
- Dayan, F.E.; Duke, S.O. Natural compounds as next-generation herbicides. Plant Physiol. 2014, 166, 1090–1105. [Google Scholar] [CrossRef] [Green Version]
- Global 5-aminolevulinic Acid Hydrochloride Market Growth 2022–2028. Available online: https://www.360researchreports.com/global-5-aminolevulinic-acid-hydrochloride-market-19884300 (accessed on 30 December 2022).
- Zhang, J.; Cui, Z.; Zhu, Y.; Zhu, Z.; Qi, Q.; Wang, Q. Recent advances in microbial production of high-value compounds in the tetrapyrrole biosynthesis pathway. Biotechnol. Adv. 2022, 55, 107904. [Google Scholar] [CrossRef]
- Shemin, D.; Russell, C.S. δ-aminolevulinic acid, its role in the biosynthesis of porphyrins and purines1. J. Am. Chem. Soc. 1953, 75, 4873–4874. [Google Scholar] [CrossRef]
- Wang, L.Y.; Brown, L.; Elliott, M.; Elliott, T. Regulation of heme biosynthesis in Salmonella typhimurium: Activity of glutamyl-tRNA reductase (HemA) is greatly elevated during heme limitation by a mechanism which increases abundance of the protein. J. Bacteriol. 1997, 179, 2907–2914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woodard, S.I.; Dailey, H.A. Regulation of heme biosynthesis in Escherichia coli. Arch. Biochem. Biophys. 1995, 316, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Hong, K.; Mao, Y.; Ma, H.; Chen, T.; Wang, Z. Natural 5-aminolevulinic acid: Sources, biosynthesis, detection and applications. Front. Bioeng Biotechnol. 2022, 10, 841443. [Google Scholar] [CrossRef]
- Jørgensen, M.G.; Nielsen, J.S.; Boysen, A.; Franch, T.; Møller-Jensen, J.; Valentin-Hansen, P. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol. Microbiol. 2012, 84, 36–50. [Google Scholar] [CrossRef] [Green Version]
- Livshits, V.A.; Zakataeva, N.P.; Aleshin, V.V.; Vitushkina, M.V. Identification and characterization of the new gene rhtA involved in threonine and homoserine efflux in Escherichia coli. Res. Microbiol. 2003, 154, 123–135. [Google Scholar] [CrossRef]
- Kang, Z.; Wang, Y.; Gu, P.; Wang, Q.; Qi, Q. Engineering Escherichia coli for efficient production of 5-aminolevulinic acid from glucose. Metab. Eng. 2011, 13, 492–498. [Google Scholar] [CrossRef]
- Tan, S.-I.; You, S.-C.; Shih, I.T.; Ng, I.S. Quantification, regulation and production of 5-aminolevulinic acid by green fluorescent protein in recombinant Escherichia coli. J. Biosci. Bioeng. 2020, 129, 387–394. [Google Scholar] [CrossRef]
- Zhang, C.; Li, Y.; Zhu, F.; Li, Z.; Lu, N.; Li, Y.; Xu, Q.; Chen, N. Metabolic engineering of an auto-regulated Corynebacterium glutamicum chassis for biosynthesis of 5-aminolevulinic acid. Bioresour. Technol. 2020, 318, 124064. [Google Scholar] [CrossRef]
- Feng, L.L.; Zhang, Y.; Fu, J.; Mao, Y.F.; Chen, T.; Zhao, X.M.; Wang, Z.W. Metabolic engineering of Corynebacterium glutamicum for efficient production of 5-aminolevulinic acid. Biotechnol. Bioeng. 2016, 113, 1284–1293. [Google Scholar] [CrossRef]
- Laneelle, M.A.; Tropis, M.; Daffe, M. Current knowledge on mycolic acids in Corynebacterium glutamicum and their relevance for biotechnological processes. Appl. Microbiol. Biot. 2013, 97, 9923–9930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.M.; Gao, Y.J.; Chen, Z.W.; Xu, G.Q.; Zhang, X.J.; Li, H.; Shi, J.S.; Koffas, M.A.G.; Xu, Z.H. High-yield production of l-serine through a novel identified exporter combined with synthetic pathway in Corynebacterium glutamicum. Microb. Cell Fact 2020, 19, 115. [Google Scholar] [CrossRef] [PubMed]
- Kishino, M.; Kondoh, M.; Hirasawa, T. Enhanced L-cysteine production by overexpressing potential L-cysteine exporter genes in an L-cysteine-producing recombinant strain of Corynebacterium glutamicum. Biosci. Biotech. Bioch. 2019, 83, 2390–2393. [Google Scholar] [CrossRef] [PubMed]
- Ghiffary, M.R.; Prabowo, C.P.S.; Adidjaja, J.J.; Lee, S.Y.; Kim, H.U. Systems metabolic engineering of Corynebacterium glutamicum for the efficient production of β-alanine. Metab. Eng. 2022, 74, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Mao, Y.; Luo, J.; Liu, P.; Jiang, M.; He, G.; Zhang, Z.; Cao, Q.; Shen, J.; Ma, H.; et al. Global cellular metabolic rewiring adapts Corynebacterium glutamicum to efficient nonnatural xylose utilization. Appl. Environ. Microbiol. 2022, 88, e0151822. [Google Scholar] [CrossRef]
- Sinclair, P.R.; Gorman, N.; Jacobs, J.M. Measurement of heme concentration. Curr. Protoc. Toxicol. 1999, 1, 831–837. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Zheng, P.; Zhao, J.; Tan, Z.J.; Chen, J.Z.; Rao, D.M.; Sun, J.B.; Ma, Y.H. 5-aminolevulinic Acid (ALA) Synthetase Mutant and Host Cells and Application Thereof. China Patent CN108251396B, 1 April 2022. Available online: https://worldwide.espacenet.com/publicationDetails/biblio?CC=CN&NR=108251396B&KC=B&FT=D (accessed on 30 December 2022).
- Shang, X.L.; Chai, X.; Lu, X.M.; Li, Y.; Zhang, Y.; Wang, G.Q.; Zhang, C.; Liu, S.W.; Zhang, Y.; Ma, J.Y.; et al. Native promoters of Corynebacterium glutamicum and its application in l-lysine production. Biotechnol. Lett. 2018, 40, 383–391. [Google Scholar] [CrossRef]
- Dvorak, P.; Chrast, L.; Nikel, P.I.; Fedr, R.; Soucek, K.; Sedlackova, M.; Chaloupkova, R.; de Lorenzo, V.; Prokop, Z.; Damborsky, J. Exacerbation of substrate toxicity by IPTG in Escherichia coli BL21(DE3) carrying a synthetic metabolic pathway. Microb. Cell Fact 2015, 14, 201. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic. Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.M.; Xu, G.Q.; Zhang, X.J.; Shi, J.S.; Xu, Z.H. Integration of ARTP mutagenesis with biosensor-mediated high-throughput screening to improve L-serine yield in Corynebacterium glutamicum. Appl. Microbiol. Biot. 2018, 102, 5939–5951. [Google Scholar] [CrossRef] [PubMed]
- Reese, M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001, 26, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Zakataeva, N.P.; Kutukova, E.A.; Gronskiy, S.V.; Troshin, P.V.; Livshits, V.A.; Aleshin, V.V. Export of metabolites by the proteins of the DMT and RhtB families and its possible role in intercellular communication. Microbiology 2006, 75, 438–448. [Google Scholar] [CrossRef]
- Zavilgelsky, G.B.; Manukhov, I.V. Quorum Sensing, or How Bacteria “Talk” to Each Other. Mol. Biol. 2001, 35, 224–232. [Google Scholar] [CrossRef]
- Banerjee, D.; Eng, T.; Sasaki, Y.; Srinivasan, A.; Oka, A.; Herbert, R.A.; Trinh, J.; Singan, V.R.; Sun, N.; Putnam, D.; et al. Genomics characterization of an engineered Corynebacterium glutamicum in bioreactor cultivation under ionic liquid stress. Front. Bioeng. Biotechnol. 2021, 9, 766674. [Google Scholar] [CrossRef] [PubMed]
- Krings, E.; Krumbach, K.; Bathe, B.; Kelle, R.; Wendisch, V.F.; Sahm, H.; Eggeling, L. Characterization of myo-inositol utilization by Corynebacterium glutamicum: The stimulon, identification of transporters, and influence on L-lysine formation. J. Bacteriol. 2006, 188, 8054–8061. [Google Scholar] [CrossRef] [Green Version]
- Lindner, S.N.; Seibold, G.M.; Henrich, A.; Kramer, R.; Wendisch, V.F. Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases. Appl. Environ. Microb. 2011, 77, 3571–3581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Man, Z.W.; Xu, M.J.; Rao, Z.M.; Guo, J.; Yang, T.W.; Zhang, X.; Xu, Z.H. Systems pathway engineering of Corynebacterium crenatum for improved L-arginine production. Sci. Rep. 2016, 6, 28629. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.Z.; Wu, Z.H.; Gao, S.J.; Zhang, W.G. Rational modification of tricarboxylic acid cycle for improving L-lysine production in Corynebacterium glutamicum. Microb Cell Fact 2018, 17, 105. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.M.; Lai, L.H.; Xu, G.Q.; Zhang, X.J.; Shi, J.S.; Koffas, M.A.G.; Xu, Z.H. Rewiring the central metabolic pathway for high-yield l-serine production in Corynebacterium glutamicum by using glucose. Biotechnol. J. 2019, 14, 1800497. [Google Scholar] [CrossRef]
- Ghiffary, M.R.; Prabowo, C.P.S.; Sharma, K.; Yan, Y.; Lee, S.Y.; Kim, H.U. High-level production of the natural blue pigment indigoidine from metabolically engineered Corynebacterium glutamicum for sustainable fabric dyes. ACS Sustain. Chem. Eng. 2021, 9, 6613–6622. [Google Scholar] [CrossRef]
- Litsanov, B.; Kabus, A.; Brocker, M.; Bott, M. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb. Biotechnol. 2012, 5, 116–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miscevic, D.; Mao, J.Y.; Kefale, T.; Abedi, D.; Moo-Young, M.; Chou, C.P. Strain engineering for high-level 5-aminolevulinic acid production in Escherichia coli. Biotechnol. Bioeng. 2021, 118, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Kim, H.U.; Kim, T.Y.; Lee, S.Y. Systematic engineering of TCA cycle for optimal production of a four-carbon platform chemical 4-hydroxybutyric acid in Escherichia coli. Metab. Eng. 2016, 38, 264–273. [Google Scholar] [CrossRef] [PubMed]
- He, G. Production of 5-Aminolevulinic Acid in the Succinate-Producing Corynebacterium glutamicum by Metabolic Engineering. Master’s Thesis, Tianjin University, Tianjin, China, 28 May 2022. [Google Scholar]
- Rehm, N.; Georgi, T.; Hiery, E.; Degner, U.; Schmiedl, A.; Burkovski, A.; Bott, M. L-glutamine as a nitrogen source for Corynebacterium glutamicum: Derepression of the AmtR regulon and implications for nitrogen sensing. Microbiology 2010, 156, 3180–3193. [Google Scholar] [CrossRef] [Green Version]
- Ge, F.L.; Li, X.K.; Ge, Q.R.; Zhu, D.; Li, W.; Shi, F.H.; Chen, H.J. Modular control of multiple pathways of Corynebacterium glutamicum for 5-aminolevulinic acid production. Amb. Express. 2021, 11, 179. [Google Scholar] [CrossRef]
- Hollenstein, K.; Dawson, R.J.; Locher, K.P. Structure and mechanism of ABC transporter proteins. Curr. Opin. in Struct. Biol. 2007, 17, 412–418. [Google Scholar] [CrossRef]
- Shakoury-Elizeh, M.; Protchenko, O.; Berger, A.; Cox, J.; Gable, K.; Dunn, T.M.; Prinz, W.A.; Bard, M.; Philpott, C.C. Metabolic response to iron deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 14823–14833. [Google Scholar] [CrossRef] [Green Version]
- Ishige, T.; Krause, M.; Bott, M.; Wendisch, V.F.; Sahm, H. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 2003, 185, 4519–4529. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Kang, Z.; Ding, W.; Chen, J.; Du, G. Integrated optimization of the in vivo heme biosynthesis pathway and the in vitro iron concentration for 5-aminolevulinate production. Appl. Biochem. Biotechnol. 2016, 178, 1252–1262. [Google Scholar] [CrossRef]
- Wuttge, S.; Bommer, M.; Jäger, F.; Martins, B.M.; Jacob, S.; Licht, A.; Scheffel, F.; Dobbek, H.; Schneider, E. Determinants of substrate specificity and biochemical properties of the sn-glycerol-3-phosphate ATP binding cassette transporter (UgpB–AEC2) of Escherichia coli. Mol. Microbiol. 2012, 86, 908–920. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Jiang, M.; Kong, S.; Hong, K.; Zhao, J.; Sun, X.; Cui, Z.; Chen, T.; Wang, Z. Unveiling the Effect of NCgl0580 Gene Deletion on 5-Aminolevulinic Acid Biosynthesis in Corynebacterium glutamicum. Fermentation 2023, 9, 213. https://doi.org/10.3390/fermentation9030213
Wu J, Jiang M, Kong S, Hong K, Zhao J, Sun X, Cui Z, Chen T, Wang Z. Unveiling the Effect of NCgl0580 Gene Deletion on 5-Aminolevulinic Acid Biosynthesis in Corynebacterium glutamicum. Fermentation. 2023; 9(3):213. https://doi.org/10.3390/fermentation9030213
Chicago/Turabian StyleWu, Jian, Meiru Jiang, Shutian Kong, Kunqiang Hong, Juntao Zhao, Xi Sun, Zhenzhen Cui, Tao Chen, and Zhiwen Wang. 2023. "Unveiling the Effect of NCgl0580 Gene Deletion on 5-Aminolevulinic Acid Biosynthesis in Corynebacterium glutamicum" Fermentation 9, no. 3: 213. https://doi.org/10.3390/fermentation9030213
APA StyleWu, J., Jiang, M., Kong, S., Hong, K., Zhao, J., Sun, X., Cui, Z., Chen, T., & Wang, Z. (2023). Unveiling the Effect of NCgl0580 Gene Deletion on 5-Aminolevulinic Acid Biosynthesis in Corynebacterium glutamicum. Fermentation, 9(3), 213. https://doi.org/10.3390/fermentation9030213





