Effect of Co-Fermentation of Saccharomyces boulardii CNCM-I745 with Four Different Probiotic Lactobacilli in Coffee Brews on Cell Viabilities and Metabolic Activities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbial Strains, Cultivation, and Enumeration
2.2. Fermentation Conditions and Design
2.3. Non-Volatile Compound Analyses
2.4. Volatile Compound Analyses and Data Processing
2.5. Antioxidant Capacity Assays
2.6. Statistical Analysis
3. Results and Discussion
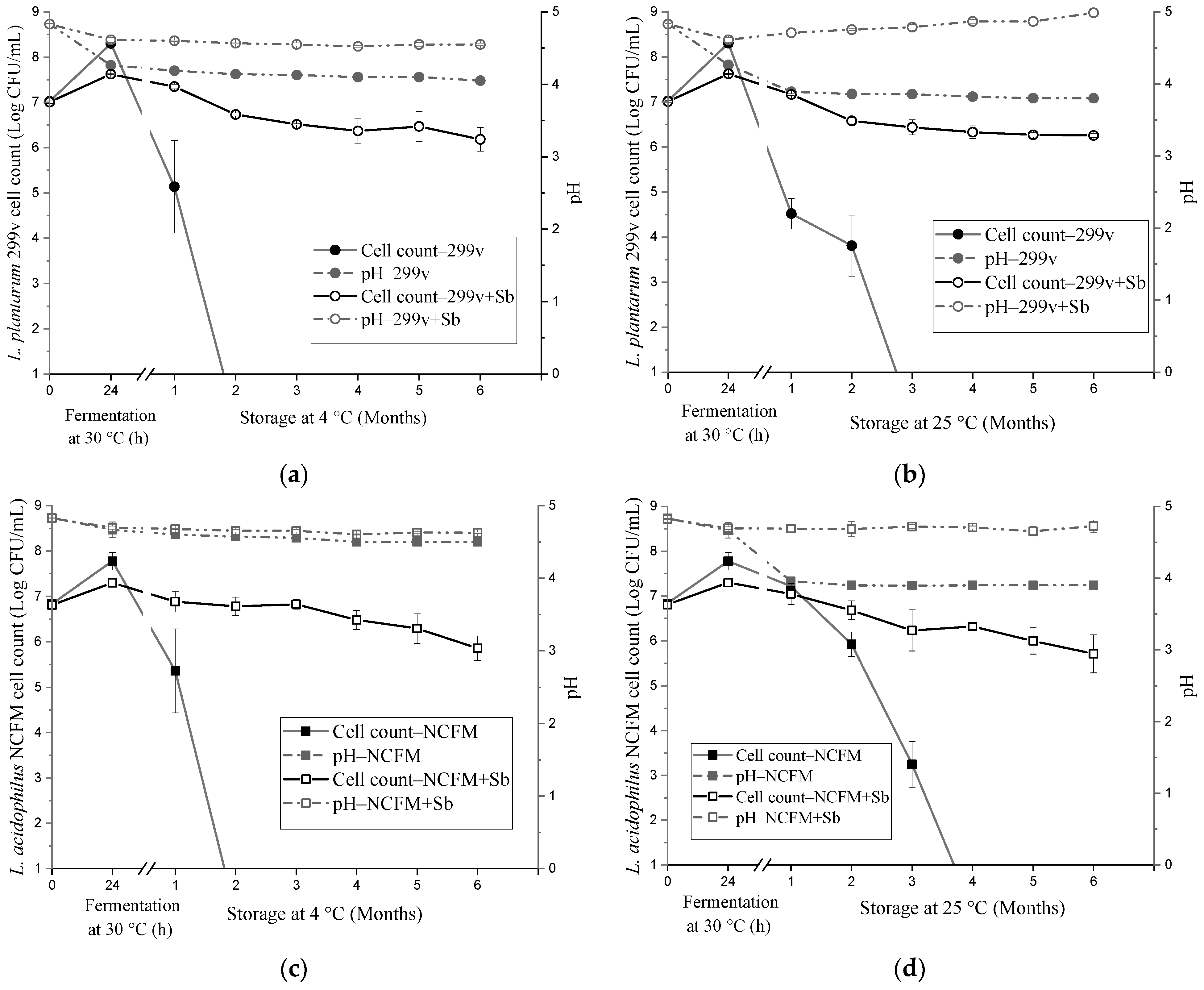
3.1. Probiotic Growth and Survival during Fermentation and Storage in Coffee Brews
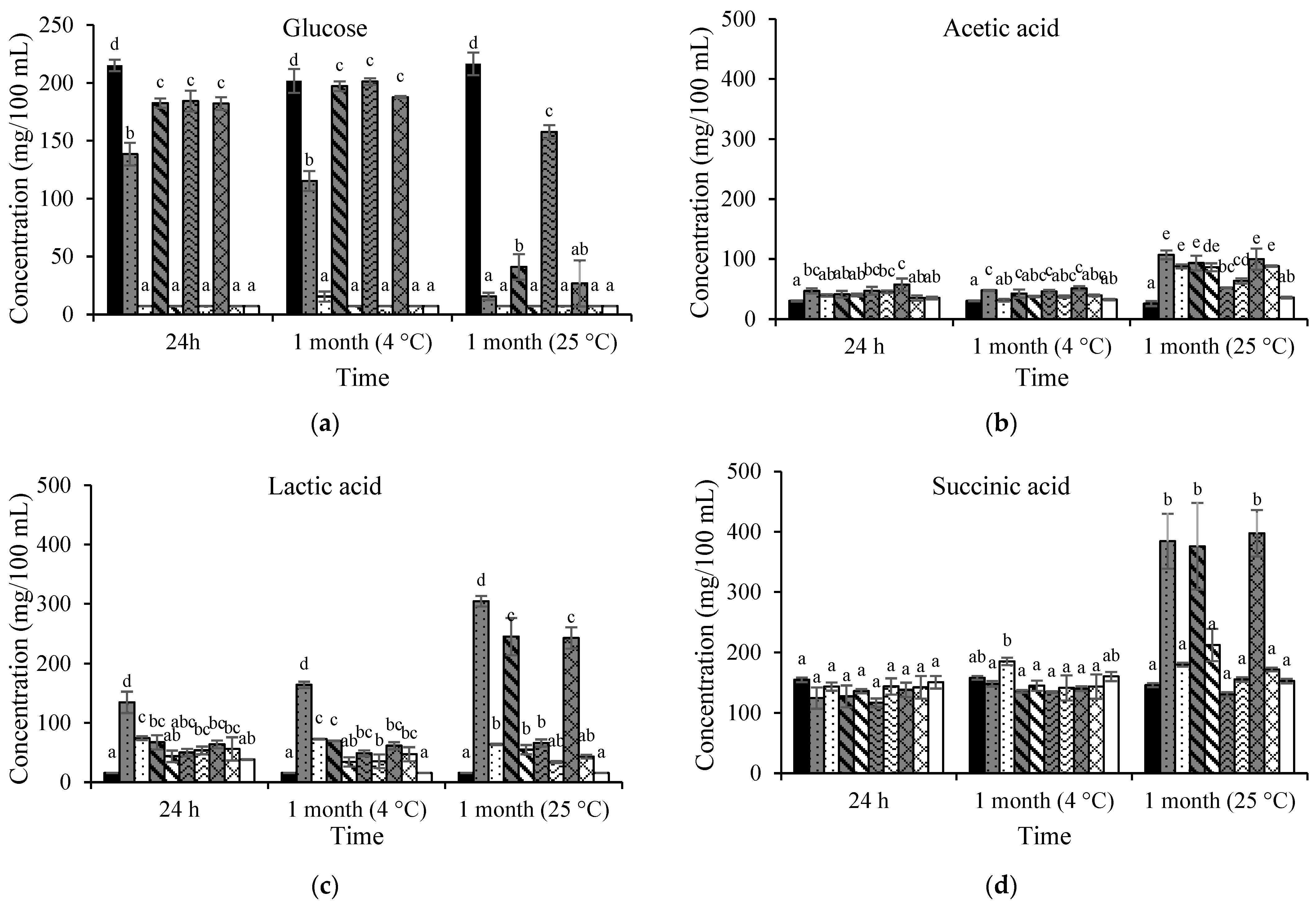
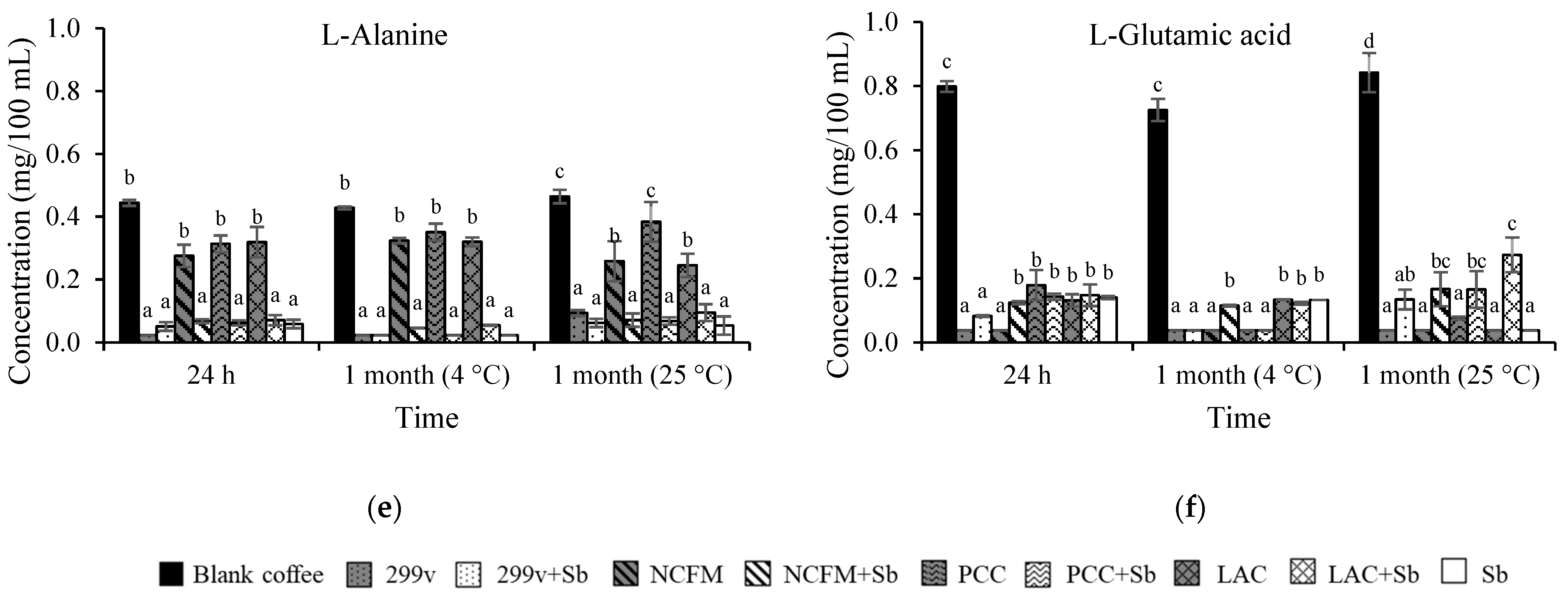
3.2. Changes in Glucose, Organic Acids, and Free Amino Acids
3.3. Changes in Volatile Components
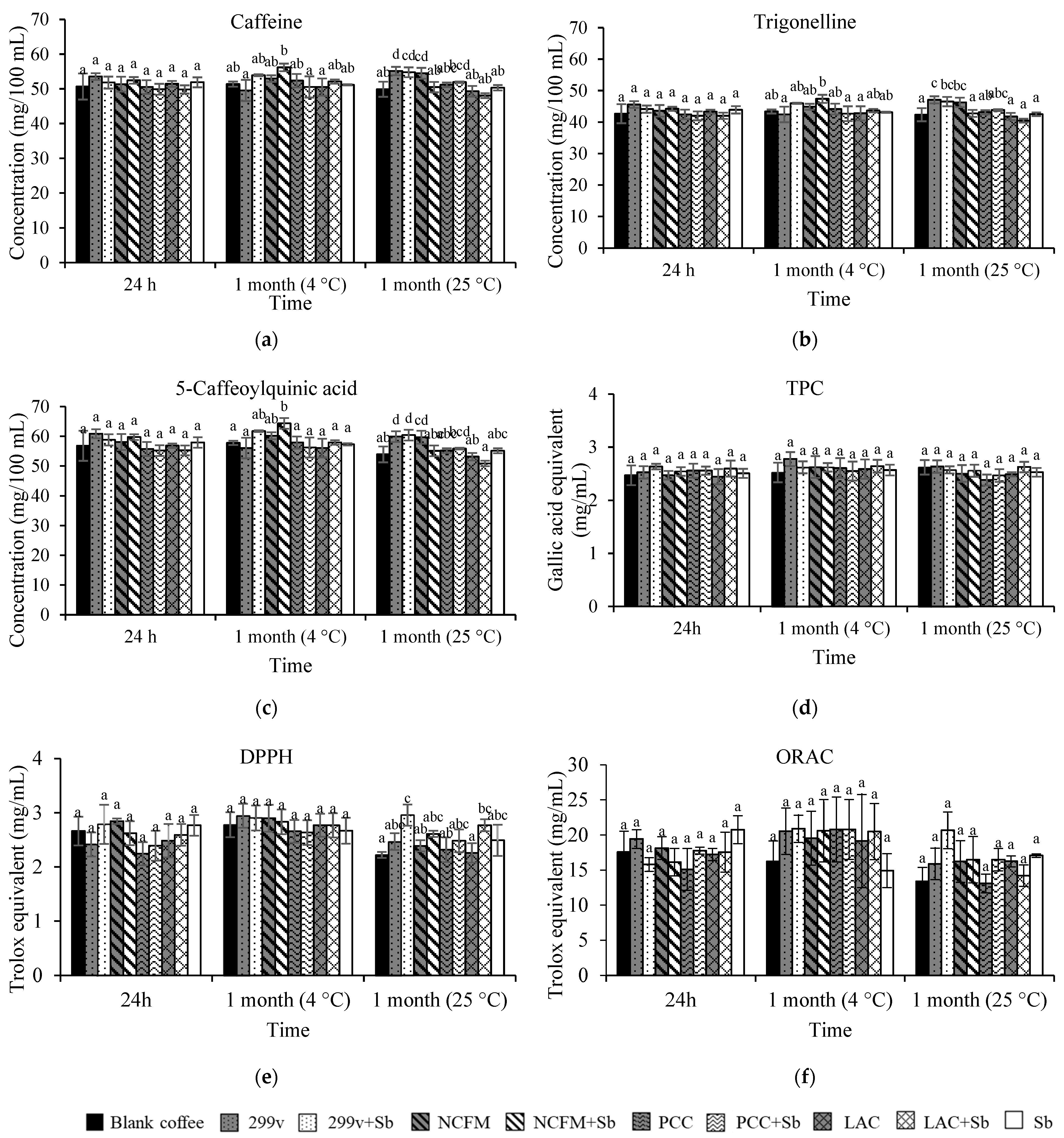
3.4. Changes in Coffee Bioactive Components and Antioxidant Capacities
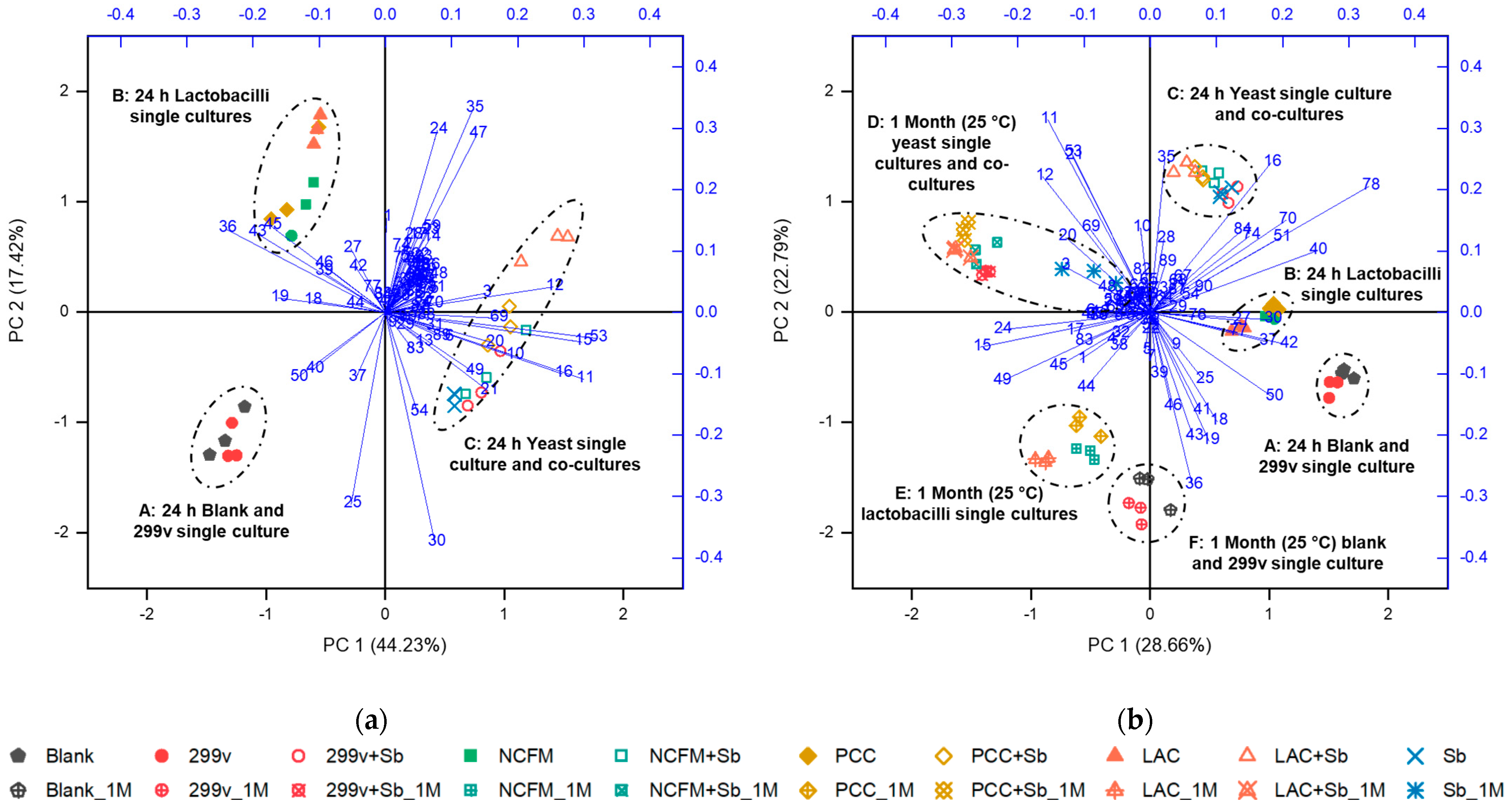
3.5. Other Considerations
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| No | Compound | LRI 1 | m/z2 | Time 3 | Normalised Peak Intensities * | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blank Coffee | 299v | 299v + Sb | NCFM | NCFM + Sb | PCC | PCC + Sb | LAC | LAC + Sb | Sb | |||||
| Acids | ||||||||||||||
| 1 | Acetic acid | 1448 | 43 | 24 h | 114.69 ± 2.47 a | 138.95 ± 22.89 ab | 122.52 ± 42.02 a | 166.92 ± 43.34 b | 145.11 ± 44.68 ab | 203.37 ± 53.64 b | 162.50 ± 8.63 b | 265.31 ± 73.84 b | 232.24 ± 56.50 b | 79.69 ± 20.84 a |
| 1 M | 167.79 ± 48.27 a | 369.32 ± 57.31 abc | 342.21 ± 98.32 abc | 516.11 ± 141.00 bc | 371.46 ± 79.57 abc | 490.19 ± 198.12 bc | 273.14 ± 55.79 ab | 628.89 ± 103.02 c | 408.02 ± 120.81 abc | 108.01 ± 36.62 a | ||||
| 2 | Propanoic acid | 1536 | 74 | 24 h | 5.09 ± 0.65 a | 6.56 ± 0.63 a | 4.78 ± 0.61 a | 6.19 ± 1.50 a | 5.38 ± 0.99 a | 8.97 ± 3.15 ab | 9.59 ± 1.45 b | 10.79 ± 2.38 b | 13.24 ± 1.41 b | 7.35 ± 0.75 ab |
| 1 M | 6.63 ± 3.44 ab | 8.69 ± 1.58 ab | 5.47 ± 0.98 ab | 8.22 ± 2.00 ab | 6.84 ± 1.70 ab | 8.44 ± 1.48 ab | 5.95 ± 0.75 ab | 9.69 ± 2.41 b | 7.71 ± 4.16 ab | 2.99 ± 0.40 a | ||||
| 3 | 2-Methylpropanoic acid (Isobutyric acid) | 1564 | 43 | 24 h | 0.89 ± 0.09 a | 1.05 ± 0.09 a | 2.39 ± 1.32 a | 1.18 ± 0.29 a | 2.66 ± 0.66 b | 1.39 ± 0.42 a | 4.76 ± 1.19 b | 1.92 ± 0.56 a | 6.83 ± 0.37 b | 4.06 ± 0.51 b |
| 1 M | 1.67 ± 0.89 a | 2.97 ± 0.20 ab | 4.50 ± 1.36 bc | 3.49 ± 0.83 abc | 10.37 ± 0.64 c | 3.60 ± 1.44 abc | 4.81 ± 0.68 bc | 3.23 ± 0.76 ab | 9.67 ± 5.41 c | 2.21 ± 0.58 a | ||||
| 4 | Butanoic acid | 1625 | 60 | 24 h | 2.59 ± 0.19 ab | 2.25 ± 0.30 a | 2.27 ± 0.51 a | 2.04 ± 0.11 a | 2.12 ± 0.17 a | 1.95 ± 0.56 a | 4.3 ± 0.59 b | 2.61 ± 0.35 ab | 4.57 ± 1.64 b | 2.52 ± 0.61 ab |
| 1 M | 4.88 ± 2.07 b | 5.22 ± 1.20 b | 3.06 ± 0.93 a | 3.61 ± 0.56 ab | 4.37 ± 0.95 b | 6.52 ± 3.09 b | 4.37 ± 0.92 b | 4.22 ± 0.62 b | 2.52 ± 0.65 a | 3.00 ± 0.14 a | ||||
| 5 | 3-Methyl-2-butenoic acid | 1795 | 82 | 24 h | 4.01 ± 1.45 a | 5.50 ± 0.59 ab | 5.66 ± 1.61 ab | 6.25 ± 1.77 ab | 4.46 ± 1.54 ab | 5.98 ± 1.55 ab | 5.78 ± 0.51 ab | 6.35 ± 1.18 ab | 8.20 ± 1.63 b | 4.84 ± 0.22 ab |
| 1 M | 6.44 ± 1.57 ab | 8.93 ± 2.41 b | 5.79 ± 2.25 ab | 9.80 ± 2.21 b | 3.91 ± 1.55 a | 8.92 ± 3.51 b | 5.14 ± 1.12 ab | 10.24 ± 2.40 b | 4.31 ± 0.94 a | 4.64 ± 1.00 a | ||||
| 6 | Hexanoic acid | 1841 | 42 | 24 h | 0.65 ± 0.21 abc | 0.53 ± 0.06 ab | 1.00 ± 0.07 ab | 0.52 ± 0.06 a | 0.85 ± 0.24 abc | 0.45 ± 0.16 a | 0.80 ± 0.30 abc | 0.52 ± 0.08 ab | 1.30 ± 0.52 c | 0.86 ± 0.18 bc |
| 1 M | 0.57 ± 0.05 a | 1.66 ± 1.05 bc | 1.32 ± 0.12 bc | 1.35 ± 0.16 bc | 1.32 ± 0.30 bc | 1.12 ± 0.13 abc | 1.61 ± 0.21 c | 1.55 ± 0.45 c | 0.80 ± 0.19 a | 1.02 ± 0.03 ab | ||||
| 7 | Heptanoic acid | 1948 | 60 | 24 h | 0.44 ± 0.15 a | 0.32 ± 0.12 a | 0.58 ± 0.15 a | 0.35 ± 0.01 a | 0.34 ± 0.08 a | 0.34 ± 0.13 a | 0.39 ± 0.06 a | 0.39 ± 0.14 a | 0.54 ± 0.04 a | 0.49 ± 0.08 a |
| 1 M | 0.53 ± 0.03 ab | 0.78 ± 0.18 b | 0.41 ± 0.11 ab | 0.99 ± 0.04 b | 0.27 ± 0.09 a | 0.72 ± 0.34 b | 0.30 ± 0.03 a | 0.86 ± 0.39 b | 0.29 ± 0.13 a | 0.31 ± 0.08 a | ||||
| 8 | Octanoic acid | 2055 | 60 | 24 h | 0.67 ± 0.10 a | 1.17 ± 0.14 ab | 1.43 ± 0.42 bc | 1.88 ± 0.20 cd | 1.17 ± 0.22 ab | 1.05 ± 0.12 ab | 1.27 ± 0.16 abc | 1.19 ± 0.09 ab | 2.06 ± 0.17d | 1.23 ± 0.27 ab |
| 1 M | 0.68 ± 0.11 a | 1.69 ± 0.94 ab | 1.66 ± 0.30 ab | 2.35 ± 0.67 ab | 1.05 ± 0.41 ab | 2.46 ± 1.04 ab | 1.76 ± 0.18 ab | 2.88 ± 0.69 b | 1.53 ± 0.83 ab | 1.36 ± 0.62 ab | ||||
| 9 | Nonanoic acid | 2162 | 41 | 24 h | 1.38 ± 0.14 a | 1.23 ± 0.20 a | 1.47 ± 0.37 a | 1.46 ± 0.46 a | 1.15 ± 0.37 a | 1.08 ± 0.14 a | 1.21 ± 0.36 a | 1.08 ± 0.29 a | 1.47 ± 0.54 a | 1.21 ± 0.17 a |
| 1 M | 1.71 ± 0.39 b | 2.19 ± 1.22 b | 0.91 ± 0.07 a | 1.88 ± 0.78 b | 0.63 ± 0.25 a | 1.11 ± 0.39 ab | 1.29 ± 0.13 ab | 1.73 ± 0.55 b | 0.60 ± 0.19 a | 1.36 ± 0.34 ab | ||||
| 10 | Decanoic acid | 2268 | 73 | 24 h | 0.23 ± 0.05 a | 0.26 ± 0.02 a | 2.39 ± 0.32 b | 0.29 ± 0.03 a | 1.87 ± 0.12 b | 0.31 ± 0.03 a | 1.77 ± 0.17 b | 0.28 ± 0.01 a | 1.85 ± 0.29 b | 1.54 ± 0.58 b |
| 1 M | 0.35 ± 0.10 ab | 0.33 ± 0.23 a | 0.47 ± 0.11 ab | 0.21 ± 0.05 a | 0.37 ± 0.18 ab | 0.30 ± 0.10 a | 0.33 ± 0.08 a | 1.58 ± 0.30 b | 1.47 ± 0.90 b | 1.17 ± 0.69 b | ||||
| Alcohols | ||||||||||||||
| 11 | Ethanol | 45 | 24 h | 0.68 ± 0.24 a | 1.62 ± 0.30 a | 62.63 ± 27.17 b | 1.64 ± 0.07 a | 111.13 ± 19.89 b | 0.99 ± 0.33 a | 201.28 ± 65.7 b | 1.56 ± 0.60 a | 206.33 ± 30.30 b | 83.49 ± 27.42 b | |
| 1 M | 0.28 ± 0.21 a | 0.20 ± 0.11 a | 201.13 ± 52.14 b | 4.38 ± 0.61 a | 237.97 ± 75.44 b | 4.68 ± 2.56 a | 83.37 ± 21.58 b | 5.44 ± 1.28 a | 151.96 ± 26.15 b | 124.48 ± 34.69 b | ||||
| 12 | 2/3-Methylbutanol | 1214 | 55 | 24 h | 0.46 ± 0.18 a | 3.04 ± 0.77 a | 37.67 ± 12.30 b | 3.95 ± 0.88 a | 45.94 ± 15.08 b | 5.06 ± 1.71 a | 84.99 ± 36.53 b | 11.10 ± 4.88 a | 113.96 ± 17.29 b | 73.55 ± 9.17 b |
| 1 M | 0.63 ± 0.52 a | 8.55 ± 1.46 ab | 90.15 ± 28.70 c | 12.55 ± 2.78 ab | 213.58 ± 85.00 c | 9.65 ± 0.82 ab | 98.63 ± 36.97 c | 10.66 ± 1.56 ab | 175.52 ± 102.92 c | 27.03 ± 5.77 bc | ||||
| 13 | 2-Heptanol | 1319 | 45 | 24 h | 14.00 ± 8.45 a | 23.79 ± 6.46 b | 29.25 ± 6.00 b | 17.23 ± 5.91 ab | 34.68 ± 17.31 b | 16.82 ± 6.71 ab | 17.46 ± 1.30 ab | 10.19 ± 0.72 a | 19.77 ± 1.03 b | 14.46 ± 1.67 a |
| 1 M | 6.53 ± 1.60 a | 44.34 ± 1.44 ab | 87.75 ± 15.20 b | 59.48 ± 8.41 b | 61.58 ± 24.09 b | 11.33 ± 0.65 a | 9.56 ± 2.69 a | 57.90 ± 18.10 b | 81.92 ± 8.99 b | 5.12 ± 0.30 a | ||||
| 14 | 1-Hexanol | 1357 | 56 | 24 h | 11.05 ± 3.30 a | 15.03 ± 4.46 ab | 17.52 ± 5.79 ab | 19.35 ± 2.48 ab | 17.23 ± 5.74 ab | 27.26 ± 8.24 abc | 28.82 ± 7.48 bc | 27.55 ± 5.90 bc | 40.95 ± 4.68 c | 27.09 ± 5.58 abc |
| 1 M | 12.63 ± 3.64 a | 21.09 ± 2.72 abc | 25.66 ± 6.83 bcd | 30.12 ± 5.57 cd | 27.62 ± 2.22 cd | 27.20 ± 3.15 cd | 24.78 ± 4.98 abcd | 34.44 ± 4.21d | 19.25 ± 4.84 abc | 14.34 ± 0.76 ab | ||||
| 15 | 1-Acetoxy-2-propanol | 1575 | 45 | 24 h | 0.00 ± 0.00 a | 0.04 ± 0.02 a | 1.31 ± 0.40 b | 0.05 ± 0.02 a | 1.92 ± 0.70 b | 0.03 ± 0.02 a | 2.23 ± 0.31 b | 0.05 ± 0.00 a | 4.22 ± 2.31 b | 0.89 ± 0.21 b |
| 1 M | 11.86 ± 4.85 ab | 21.41 ± 4.72 b | 4.89 ± 1.53 a | 19.87 ± 0.48 b | 3.59 ± 0.56 a | 25.82 ± 3.34 b | 4.33 ± 1.40 a | 20.80 ± 4.00 b | 4.77 ± 1.06 a | 10.62 ± 7.21 a | ||||
| Benzoyl derivatives | ||||||||||||||
| 16 | Styrene | 1252 | 104 | 24 h | 0.34 ± 0.07 a | 0.38 ± 0.03 a | 15.22 ± 4.60 b | 0.43 ± 0.04 a | 16.36 ± 5.49 b | 0.46 ± 0.04 a | 14.97 ± 4.29 b | 0.42 ± 0.02 a | 19.15 ± 5.14 b | 9.82 ± 0.69 ab |
| 1 M | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||||
| 17 | 1,3-Di-tert-butylbenzene | 1419 | 57 | 24 h | 11.42 ± 5.00 ab | 6.90 ± 1.52 aab | 12.27 ± 4.41 ab | 12.02 ± 2.75 ab | 12.72 ± 5.11 ab | 12.36 ± 2.55 ab | 18.59 ± 2.94 ab | 19.59 ± 6.51 b | 19.05 ± 7.06 ab | 6.89 ± 1.37 a |
| 1 M | 16.94 ± 9.73 a | 21.93 ± 3.70 ab | 21.13 ± 4.65 a | 24.96 ± 5.84 ab | 37.67 ± 12.71 b | 41.09 ± 17.70 b | 30.34 ± 6.91 b | 29.32 ± 5.45 b | 36.47 ± 8.69 b | 10.10 ± 4.65 a | ||||
| 18 | Benzaldehyde | 1526 | 105 | 24 h | 7.62 ± 2.69 c | 4.96 ± 0.68 bc | 2.56 ± 0.48 ab | 3.74 ± 0.35 bc | 1.52 ± 0.28 a | 4.50 ± 1.52 bc | 1.95 ± 0.39 a | 3.67 ± 1.04 bc | 2.87 ± 0.52 ab | 1.86 ± 0.44 a |
| 1 M | 12.10 ± 5.36 b | 10.95 ± 3.25 b | 0.82 ± 0.24 a | 7.86 ± 2.92 b | 0.79 ± 0.24 a | 12.61 ± 2.32 b | 0.50 ± 0.12 a | 6.73 ± 2.66 b | 0.72 ± 0.09 a | 0.75 ± 0.39 a | ||||
| 19 | 3,4-Dimethylbenzaldehyde | 1817 | 105 | 24 h | 6.20 ± 1.01 b | 4.88 ± 1.73 b | 1.39 ± 0.39 a | 3.01 ± 0.28 b | 0.85 ± 0.13 a | 3.75 ± 1.13 b | 0.96 ± 0.32 a | 2.89 ± 0.67 ab | 1.02 ± 0.05 a | 0.79 ± 0.24 a |
| 1 M | 7.26 ± 2.44 b | 9.25 ± 1.77 b | 0.43 ± 0.07 a | 9.01 ± 1.15 b | 1.02 ± 0.27 ab | 7.98 ± 2.91 b | 0.64 ± 0.20 a | 7.17 ± 2.82 b | 0.26 ± 0.08 a | 0.50 ± 0.08 a | ||||
| 20 | 2-Phenylethyl alcohol | 1925 | 91 | 24 h | 4.20 ± 0.93 a | 5.88 ± 0.68 a | 25.71 ± 6.17 b | 6.10 ± 1.14 a | 19.34 ± 5.25 b | 4.76 ± 1.36 a | 15.43 ± 1.49 b | 4.98 ± 0.87 a | 27.59 ± 4.14 b | 13.11 ± 0.92 ab |
| 1 M | 2.97 ± 1.08 a | 13.58 ± 2.84 a | 50.60 ± 4.35 b | 11.66 ± 4.51 a | 47.03 ± 20.50 b | 6.62 ± 2.10 a | 32.24 ± 6.98 b | 14.29 ± 5.45 a | 32.61 ± 10.75 b | 13.01 ± 3.07 a | ||||
| 21 | 3,4-Dimethoxystyrene (3,4-Dimethoxy-1-vinylbenzene) | 2040 | 164 | 24 h | 3.98 ± 1.30 ab | 1.51 ± 1.00 a | 9.63 ± 0.95 b | 2.29 ± 1.05 a | 7.49 ± 2.24 b | 0.93 ± 0.32 a | 7.10 ± 1.85 b | 1.30 ± 0.46 a | 9.66 ± 2.44 b | 7.68 ± 0.93 b |
| 1 M | 0.10 ± 0.02 a | 0.70 ± 0.40 a | 81.68 ± 10.86 b | 0.48 ± 0.40 a | 48.08 ± 14.76 b | 0.22 ± 0.12 a | 53.09 ± 10.79 b | 0.71 ± 0.38 a | 61.83 ± 13.40 b | 16.55 ± 9.43 b | ||||
| Furans | ||||||||||||||
| 22 | 2-(Methoxymethyl)furan | 1237 | 81 | 24 h | 11.03 ± 2.02 a | 17.13 ± 4.91 ab | 15.09 ± 4.20 ab | 18.89 ± 3.38 ab | 14.61 ± 4.24 ab | 17.01 ± 3.87 ab | 22.33 ± 4.86 bc | 24.38 ± 1.66 bc | 30.19 ± 2.30 c | 23.57 ± 1.97 bc |
| 1 M | 10.99 ± 2.93 a | 32.97 ± 9.74 b | 22.91 ± 1.64 b | 29.46 ± 8.38 b | 21.51 ± 4.94 b | 23.67 ± 5.53 b | 8.58 ± 0.83 a | 30.70 ± 3.58 b | 11.79 ± 2.36 a | 11.46 ± 1.84 a | ||||
| 23 | 2-Methyltetrahydrofuran-3-one (Coffee furanone) | 1265 | 43 | 24 h | 8.00 ± 1.71 a | 11.18 ± 1.70 a | 10.28 ± 4.30 a | 10.34 ± 1.55 a | 12.31 ± 4.85 a | 17.52 ± 6.61 ab | 25.79 ± 11.81 b | 38.89 ± 17.20 b | 35.43 ± 5.29 b | 24.57 ± 2.84 b |
| 1 M | 22.33 ± 5.82 a | 16.12 ± 6.29 a | 18.29 ± 7.77 a | 27.07 ± 6.43 a | 46.06 ± 21.71 a | 22.37 ± 9.74 a | 22.85 ± 10.35 a | 18.69 ± 11.33 a | 36.73 ± 29.86 a | 6.80 ± 1.29 a | ||||
| 24 | Furan-2-carbohydrazide | 1313 | 67 | 24 h | 0.14 ± 0.03 a | 0.16 ± 0.04 a | 0.91 ± 0.12 ab | 2.38 ± 0.47 b | 0.98 ± 0.32 ab | 1.86 ± 0.53 b | 0.91 ± 0.35 ab | 2.77 ± 0.27 b | 2.44 ± 0.72 b | 0.39 ± 0.08 a |
| 1 M | 1.76 ± 0.31 a | 2.85 ± 1.15 a | 10.72 ± 2.76 ab | 10.53 ± 2.41 ab | 26.26 ± 2.48 b | 42.32 ± 18.03 b | 14.93 ± 1.57 b | 5.27 ± 0.92 a | 9.71 ± 4.99 ab | 15.41 ± 0.44 b | ||||
| 25 | Furfural | 1468 | 96 | 24 h | 692.23 ± 41.92 c | 573.18 ± 104.82 c | 97.87 ± 23.77 bc | 15.12 ± 2.17 a | 61.09 ± 14.53 abc | 14.68 ± 4.67 a | 59.87 ± 10.52 abc | 18.26 ± 5.76 a | 54.54 ± 15.7 ab | 83.01 ± 10.71 bc |
| 1 M | 623.60 ± 247.49 b | 567.47 ± 92.09 b | 45.64 ± 13.18 ab | 38.47 ± 5.29 a | 39.37 ± 3.87 a | 53.56 ± 19.17 b | 20.58 ± 4.82 a | 40.05 ± 7.53 a | 40.44 ± 9.60 ab | 37.35 ± 14.91 a | ||||
| 26 | 2-Acetylfuran | 1508 | 95 | 24 h | 139.82 ± 5.81 a | 119.71 ± 15.15 a | 146.74 ± 31.79 a | 146.33 ± 17.34 a | 155.50 ± 52.37 a | 143.11 ± 38.65 a | 164.16 ± 12.77 a | 163.96 ± 12.16 a | 196.25 ± 33.32 a | 132.18 ± 9.61 a |
| 1 M | 134.90 ± 31.82 ab | 166.24 ± 23.37 ab | 188.96 ± 24.69 b | 178.14 ± 19.96 b | 194.03 ± 16.06 b | 186.41 ± 28.50 b | 165.34 ± 25.45 ab | 183.83 ± 10.28 b | 166.12 ± 12.69 ab | 107.74 ± 24.50 a | ||||
| 27 | 1-(2-Furyl)-2-propanone (2-Furfuryl methyl ketone) | 1521 | 81 | 24 h | 5.84 ± 1.07 b | 6.67 ± 1.22 b | 2.14 ± 0.93 a | 7.77 ± 0.71 b | 4.74 ± 1.24 a | 5.55 ± 0.57 ab | 4.61 ± 0.48 a | 8.65 ± 2.18 b | 7.63 ± 1.51 b | 3.86 ± 0.55 a |
| 1 M | 1.42 ± 0.21 a | 4.59 ± 1.52 b | 0.92 ± 0.09 a | 4.09 ± 0.76 b | 1.74 ± 0.56 ab | 2.86 ± 0.65 b | 1.15 ± 0.43 a | 3.43 ± 0.93 b | 0.99 ± 0.18 a | 3.80 ± 2.03 b | ||||
| 28 | Furfuryl acetate | 1537 | 52 | 24 h | 1.98 ± 0.18 a | 2.29 ± 0.74 a | 2.69 ± 0.05 a | 2.80 ± 0.93 a | 2.14 ± 0.39 a | 4.89 ± 1.12 b | 3.90 ± 0.52 b | 3.83 ± 0.29 b | 4.96 ± 0.42 b | 3.27 ± 0.33 ab |
| 1 M | 1.49 ± 0.76 a | 1.20 ± 0.44 a | 3.39 ± 0.75 bc | 1.11 ± 0.22 a | 3.63 ± 0.82 c | 1.19 ± 0.49 a | 2.68 ± 0.44 abc | 1.68 ± 0.75 a | 2.63 ± 0.22 abc | 1.92 ± 0.43 ab | ||||
| 29 | 1-(2-Furyl)-1-propanone (2-Propionylfuran) | 1578 | 95 | 24 h | 35.37 ± 6.83 a | 35.74 ± 9.98 a | 39.67 ± 8.77 a | 32.95 ± 8.59 a | 31.14 ± 2.49 a | 27.19 ± 7.56 a | 33.59 ± 2.01 a | 29.19 ± 1.37 a | 41.61 ± 4.41 a | 30.74 ± 2.36 a |
| 1 M | 29.11 ± 8.04 a | 38.38 ± 8.23 a | 31.08 ± 3.38 a | 31.24 ± 2.39 a | 29.62 ± 7.02 a | 27.56 ± 9.01 a | 25.41 ± 4.42 a | 27.65 ± 4.29 a | 22.01 ± 1.58 a | 23.71 ± 7.30 a | ||||
| 30 | 5-MethyIfurfural | 1578 | 110 | 24 h | 409.03 ± 31.49 b | 345.28 ± 30.13 b | 216.68 ± 43.58 b | 4.38 ± 0.50 a | 136.13 ± 25.90 ab | 4.68 ± 1.49 a | 135.76 ± 8.20 a | 5.85 ± 0.13 a | 143.53 ± 56.33 ab | 154.01 ± 4.55 b |
| 1 M | 340.22 ± 122.84 b | 409.73 ± 63.47 b | 2.45 ± 0.79 a | 8.86 ± 2.52 b | 2.41 ± 0.64 a | 7.04 ± 1.11 ab | 1.86 ± 0.13 a | 4.95 ± 0.82 ab | 1.80 ± 0.52 a | 250.45 ± 131.30 b | ||||
| 31 | Methyl 3-furancarboxylate | 1580 | 126 | 24 h | 0.81 ± 0.22 ab | 0.62 ± 0.09 a | 1.02 ± 0.12 ab | 0.71 ± 0.25 a | 0.90 ± 0.25 ab | 0.56 ± 0.21 a | 0.77 ± 0.04 ab | 0.63 ± 0.10 a | 1.26 ± 0.19 b | 0.79 ± 0.03 ab |
| 1 M | 0.57 ± 0.25 ab | 1.08 ± 0.40 b | 0.76 ± 0.06 b | 0.66 ± 0.04 b | 0.67 ± 0.26 ab | 0.49 ± 0.12 a | 0.65 ± 0.15 ab | 0.45 ± 0.06 a | 0.40 ± 0.07 a | 0.46 ± 0.13 a | ||||
| 32 | 2-Acetyl-5-methylfuran | 1617 | 109 | 24 h | 5.53 ± 0.67 a | 4.46 ± 0.15 a | 6.90 ± 0.96 a | 6.52 ± 1.53 a | 5.75 ± 1.52 a | 4.99 ± 1.50 a | 4.92 ± 0.96 a | 6.51 ± 1.34 a | 7.16 ± 2.12 a | 5.40 ± 0.63 a |
| 1 M | 6.04 ± 1.22 a | 8.21 ± 1.01 b | 7.35 ± 1.74 ab | 9.34 ± 1.59 b | 8.56 ± 1.60 b | 6.88 ± 1.74 ab | 6.87 ± 0.64 ab | 10.93 ± 4.73 b | 5.45 ± 0.74 a | 4.67 ± 0.98 a | ||||
| 33 | 2-Furanmethanol | 1666 | 98 | 24 h | 349.72 ± 8.64 ab | 239.58 ± 25.76 a | 365.44 ± 131.47 ab | 293.10 ± 28.77 a | 362.01 ± 114.35 ab | 385.70 ± 136.71 ab | 488.18 ± 42.42 b | 494.59 ± 34.78 b | 571.43 ± 164.84 b | 331.08 ± 54.81 ab |
| 1 M | 246.20 ± 73.76 a | 303.91 ± 32.46 a | 389.30 ± 86.60 a | 476.61 ± 43.70 a | 403.77 ± 108.71 a | 446.97 ± 102.63 a | 468.34 ± 192.24 a | 455.12 ± 25.27 a | 441.57 ± 94.38 a | 238.93 ± 66.81 a | ||||
| 34 | 2-Methyl-5-propionylfuran | 1684 | 109 | 24 h | 5.54 ± 0.92 a | 4.63 ± 0.45 a | 6.05 ± 1.05 a | 5.87 ± 1.32 a | 5.17 ± 0.45 a | 4.79 ± 1.43 a | 5.18 ± 0.90 a | 5.86 ± 1.72 a | 7.38 ± 0.38 a | 4.21 ± 0.73 a |
| 1 M | 5.79 ± 0.64 a | 6.43 ± 1.38 a | 6.41 ± 0.69 a | 6.39 ± 0.75 a | 6.71 ± 3.12 a | 6.00 ± 2.90 a | 4.26 ± 0.64 a | 6.08 ± 2.11 a | 4.59 ± 1.27 a | 5.17 ± 0.94 a | ||||
| 35 | 5-Methyl-2-furanmethanol (5-Methylfurfuryl alcohol) | 1727 | 95 | 24 h | 0.47 ± 0.10 a | 0.45 ± 0.10 a | 16.50 ± 3.86 ab | 42.56 ± 6.05 b | 13.53 ± 2.11 a | 41.96 ± 16.21 b | 21.88 ± 5.79 ab | 44.95 ± 8.74 b | 32.33 ± 8.89 b | 9.91 ± 2.38 a |
| 1 M | 0.24 ± 0.03 a | 0.25 ± 0.11 a | 7.43 ± 2.00 b | 0.57 ± 0.30 a | 11.16 ± 0.11 b | 5.97 ± 0.76 ab | 6.64 ± 0.23 b | 0.25 ± 0.10 a | 5.80 ± 1.47 ab | 7.48 ± 2.62 b | ||||
| 36 | 3-Ethyl-4-methyl-2,5-furandione | 1745 | 67 | 24 h | 1.52 ± 0.40 ab | 3.47 ± 0.12 b | 0.10 ± 0.07 a | 2.38 ± 0.50 b | 0.06 ± 0.02 a | 2.51 ± 0.93 b | 0.06 ± 0.04 a | 2.22 ± 0.23 b | 0.18 ± 0.10 a | 0.11 ± 0.05 a |
| 1 M | 2.28 ± 0.61 b | 6.61 ± 1.80 b | 0.27 ± 0.12 a | 5.37 ± 1.30 b | 0.64 ± 0.20 ab | 4.98 ± 1.23 b | 0.01 ± 0.01 a | 6.13 ± 1.51 b | 0.26 ± 0.18 a | 0.17 ± 0.04 a | ||||
| 37 | 1-(5-Methyl-2-furyl)-2-propanone | 1781 | 95 | 24 h | 11.31 ± 1.07 b | 15.33 ± 0.29 b | 19.82 ± 1.90 b | 8.78 ± 3.52 ab | 4.69 ± 0.93 a | 5.29 ± 0.82 a | 3.60 ± 0.65 a | 7.22 ± 0.49 ab | 5.65 ± 1.29 a | 13.17 ± 1.23 b |
| 1 M | 3.51 ± 0.95 b | 59.17 ± 10.91 b | 4.13 ± 0.49 b | 2.23 ± 0.59 a | 0.55 ± 0.32 a | 1.84 ± 0.21 a | 0.30 ± 0.14 a | 2.28 ± 0.37 ab | 0.32 ± 0.14 a | 8.17 ± 2.55 b | ||||
| 38 | 4-(2-Furanyl)-3-buten-2-one (Furfural acetone) | 1911 | 121 | 24 h | 0.98 ± 0.31 a | 0.74 ± 0.16 a | 0.56 ± 0.06 a | 0.73 ± 0.07 a | 0.52 ± 0.08 a | 0.68 ± 0.31 a | 0.93 ± 0.06 a | 0.81 ± 0.17 a | 0.95 ± 0.30 a | 0.70 ± 0.12 a |
| 1 M | 0.95 ± 0.26 a | 1.21 ± 0.29 a | 1.17 ± 0.60 a | 1.40 ± 0.16 a | 1.17 ± 0.71 a | 1.79 ± 0.29 a | 1.14 ± 0.29 a | 1.01 ± 0.30 a | 1.11 ± 0.61 a | 0.82 ± 0.41 a | ||||
| Ketones | ||||||||||||||
| 39 | 2,3-Butanedione (Diacetyl) | 43 | 24 h | 18.91 ± 4.57 b | 54.14 ± 11.84 b | 7.49 ± 0.78 a | 53.25 ± 24.04 b | 5.60 ± 0.63 a | 5.15 ± 0.49 a | 8.69 ± 3.33 a | 30.47 ± 6.28 b | 24.83 ± 3.61 b | 3.27 ± 0.38 a | |
| 1 M | 15.70 ± 1.12 a | 18.85 ± 9.43 ab | 32.54 ± 5.61 b | 19.46 ± 2.62 b | 25.74 ± 12.91 b | 38.28 ± 8.31 b | 7.18 ± 3.63 a | 22.23 ± 4.28 b | 6.19 ± 1.48 a | 15.72 ± 0.43 a | ||||
| 40 | 2,3-Pentanedione | 1057 | 43 | 24 h | 32.54 ± 7.48 b | 54.86 ± 8.04 b | 10.10 ± 1.01 b | 81.73 ± 34.35 b | 8.35 ± 0.39 a | 10.29 ± 0.98 b | 7.90 ± 0.59 a | 3.11 ± 0.89 a | 8.95 ± 0.47 ab | 6.54 ± 0.76 a |
| 1 M | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||||
| 41 | 3-Hydroxybutanone (Acetoin) | 1289 | 45 | 24 h | 0.47 ± 0.01 a | 15.20 ± 5.80 b | 4.34 ± 0.50 b | 1.35 ± 0.20 a | 3.43 ± 1.03 b | 0.57 ± 0.15 a | 1.69 ± 0.29 a | 3.46 ± 0.50 b | 3.25 ± 1.54 b | 0.08 ± 0.02 a |
| 1 M | 0.45 ± 0.19 a | 107.29 ± 14.71 b | 1.61 ± 0.25 b | 16.75 ± 1.85 b | 0.13 ± 0.11 a | 0.97 ± 0.84 ab | 0.02 ± 0.01 a | 28.86 ± 1.65 b | 0.41 ± 0.20 a | 0.31 ± 0.16 a | ||||
| 42 | 1-Hydroxy-2-propanone (Hydroxyacetone) | 1307 | 43 | 24 h | 17.80 ± 3.01 b | 13.16 ± 3.30 ab | 9.78 ± 4.64 a | 10.54 ± 1.38 a | 10.62 ± 2.17 a | 9.54 ± 2.40 a | 7.47 ± 5.04 a | 20.62 ± 1.23 b | 17.98 ± 0.98 b | 4.41 ± 1.06 a |
| 1 M | 7.87 ± 3.49 b | 20.86 ± 4.88 b | 0.57 ± 0.42 a | 10.93 ± 3.09 b | 0.33 ± 0.25 a | 6.23 ± 1.28 b | 0.17 ± 0.07 a | 0.67 ± 0.53 a | 0.93 ± 0.87 a | 10.45 ± 6.08 b | ||||
| 43 | 1-Hydroxy-2-propanone acetate (Acetoxyacetone) | 1467 | 43 | 24 h | 80.22 ± 2.63 b | 57.25 ± 9.27 b | 8.13 ± 1.66 a | 66.77 ± 11.67 b | 8.33 ± 2.84 a | 71.56 ± 19.75 b | 6.67 ± 1.14 a | 88.12 ± 16.76 b | 10.21 ± 0.99 ab | 5.95 ± 0.15 a |
| 1 M | 44.74 ± 23.15 b | 78.73 ± 6.22 b | 18.53 ± 2.60 ab | 79.07 ± 9.79 b | 12.15 ± 5.36 a | 85.15 ± 13.92 b | 6.01 ± 0.65 a | 88.52 ± 5.12 b | 12.97 ± 0.97 a | 2.02 ± 0.51 a | ||||
| 44 | 2,5-Hexanedione | 1505 | 43 | 24 h | 1.69 ± 0.87 ab | 1.89 ± 0.14 ab | 2.25 ± 0.22 b | 2.59 ± 0.73 b | 1.35 ± 0.88 a | 1.20 ± 0.62 a | 1.35 ± 0.72 a | 2.56 ± 0.10 b | 1.13 ± 0.78 a | 1.46 ± 0.17 a |
| 1 M | 1.54 ± 0.21 a | 4.64 ± 1.86 b | 2.66 ± 0.88 a | 35.88 ± 3.76 b | 5.49 ± 0.98 b | 4.01 ± 0.27 b | 4.13 ± 0.10 b | 30.78 ± 1.87 b | 2.10 ± 0.11 a | 2.90 ± 0.81 a | ||||
| 45 | 1-Hydroxy-2-butanone acetate | 1534 | 43 | 24 h | 11.43 ± 3.48 b | 13.28 ± 2.30 b | 2.31 ± 0.62 a | 15.63 ± 1.81 b | 1.87 ± 0.43 a | 16.03 ± 5.62 b | 2.11 ± 0.30 a | 16.43 ± 2.36 b | 3.29 ± 0.21 ab | 1.82 ± 0.22 a |
| 1 M | 9.11 ± 1.26 a | 19.94 ± 3.65 ab | 84.41 ± 7.95 b | 18.47 ± 1.17 a | 91.39 ± 15.02 b | 21.06 ± 1.78 ab | 81.12 ± 12.66 b | 18.86 ± 8.69 a | 90.83 ± 16.17 b | 1.35 ± 0.04 a | ||||
| 46 | 4-Cyclopentene-1,3-dione | 1591 | 96 | 24 h | 2.17 ± 0.24 c | 2.10 ± 0.23 c | 0.53 ± 0.14 a | 1.57 ± 0.52 abc | 0.80 ± 0.22 ab | 1.76 ± 0.68 bc | 1.00 ± 0.40 ab | 2.06 ± 0.43 c | 1.52 ± 0.11 abc | 0.85 ± 0.26 ab |
| 1 M | 1.84 ± 0.34 ab | 4.34 ± 0.58 b | 0.64 ± 0.12 a | 3.63 ± 0.69 b | 1.54 ± 0.81 ab | 3.87 ± 1.11 b | 0.46 ± 0.14 a | 4.25 ± 0.98 b | 0.99 ± 0.64 a | 0.32 ± 0.06 a | ||||
| 47 | 3-Hexene-2,5-dione | 1626 | 43 | 24 h | 0.09 ± 0.02 a | 0.09 ± 0.01 a | 2.70 ± 0.78 ab | 3.67 ± 0.74 b | 2.49 ± 0.83 ab | 3.34 ± 0.63 b | 1.92 ± 1.14 a | 3.53 ± 0.08 b | 4.72 ± 0.74 b | 0.77 ± 0.08 a |
| 1 M | 1.58 ± 0.50 a | 3.04 ± 0.41 b | 1.32 ± 0.57 a | 3.63 ± 0.39 b | 1.87 ± 0.74 a | 19.51 ± 3.43 b | 1.68 ± 0.29 a | 3.86 ± 1.39 b | 2.18 ± 0.53 ab | 1.42 ± 0.50 a | ||||
| Lactones | ||||||||||||||
| 48 | Butyrolactone | 1634 | 42 | 24 h | 9.66 ± 1.84 a | 6.47 ± 1.25 a | 10.10 ± 4.29 a | 8.11 ± 1.22 a | 12.79 ± 3.27 ab | 9.47 ± 2.26 a | 13.48 ± 1.97 ab | 11.02 ± 3.02 a | 20.69 ± 4.69 b | 8.59 ± 2.62 a |
| 1 M | 9.52 ± 2.74 a | 9.51 ± 0.35 a | 18.00 ± 5.23 b | 11.80 ± 2.88 ab | 17.33 ± 2.40 b | 16.33 ± 6.22 b | 13.94 ± 2.59 b | 11.12 ± 0.36 ab | 17.91 ± 6.76 b | 6.89 ± 2.23 a | ||||
| Organosulfur compounds | ||||||||||||||
| 49 | 2-Methyl-3-thiolannone | 1527 I | 60 | 24 h | 0.33 ± 0.05 a | 0.73 ± 0.14 ab | 0.99 ± 0.46 b | 0.79 ± 0.16 ab | 1.33 ± 0.07 b | 0.05 ± 0.00 a | 1.64 ± 0.48 b | 0.61 ± 0.27 a | 1.73 ± 0.48 b | 1.46 ± 0.34 b |
| 1 M | 18.17 ± 3.07 a | 24.26 ± 5.35 a | 25.37 ± 2.39 a | 22.52 ± 0.54 a | 20.13 ± 17.22 a | 29.25 ± 3.78 a | 24.38 ± 3.81 a | 23.57 ± 4.54 a | 19.93 ± 16.85 a | 11.30 ± 8.84 a | ||||
| 50 | 2-Thiophenecarboxaldehyde | 1699 | 111 | 24 h | 5.72 ± 0.33 c | 4.24 ± 0.51 b | 0.96 ± 0.14 a | 1.15 ± 0.14 a | 0.57 ± 0.14 a | 1.21 ± 0.36 a | 0.67 ± 0.21 a | 0.85 ± 0.16 a | 0.91 ± 0.29 a | 0.66 ± 0.20 a |
| 1 M | 4.20 ± 1.22 b | 4.00 ± 0.70 b | 0.12 ± 0.03 a | 0.87 ± 0.22 b | 0.15 ± 0.05 a | 1.11 ± 0.09 b | 0.07 ± 0.03 a | 0.41 ± 0.00 ab | 0.11 ± 0.00 a | 0.28 ± 0.18 a | ||||
| 51 | 5-Methyl-2-thiophenecarboxaldehyde | 1717 | 126 | 24 h | 0.37 ± 0.04 a | 0.26 ± 0.04 a | 0.38 ± 0.13 a | 0.44 ± 0.04 a | 0.24 ± 0.08 a | 0.26 ± 0.02 a | 0.39 ± 0.11 a | 0.31 ± 0.06 a | 0.44 ± 0.13 a | 0.31 ± 0.05 a |
| 1 M | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||||
| 52 | 2-Acetylthiophene | 1781 | 111 | 24 h | 1.93 ± 0.26 a | 1.41 ± 0.28 a | 2.38 ± 0.55 a | 1.80 ± 0.43 a | 1.89 ± 0.63 a | 1.60 ± 0.64 a | 1.89 ± 0.19 a | 1.99 ± 0.57 a | 2.88 ± 0.52 a | 2.00 ± 0.09 a |
| 1 M | 2.00 ± 0.23 ab | 2.68 ± 0.75 b | 1.94 ± 0.47 ab | 2.48 ± 0.47 b | 2.17 ± 0.89 ab | 2.46 ± 0.26 b | 1.62 ± 0.40 ab | 2.54 ± 0.60 b | 1.27 ± 0.28 a | 1.36 ± 0.13 a | ||||
| Pyranones | ||||||||||||||
| 53 | α-pyrone-6-carboxylic acid (2-Pyrone-6-carboxylic acid) | 1360 | 95 | 24 h | 0.03 ± 0.01 a | 0.03 ± 0.01 a | 6.03 ± 1.17 b | 0.05 ± 0.01 a | 12.56 ± 3.67 b | 0.18 ± 0.06 a | 6.36 ± 1.61 b | 0.12 ± 0.07 a | 20.97 ± 2.43 b | 2.81 ± 0.86 b |
| 1 M | 0.15 ± 0.03 a | 0.21 ± 0.05 ab | 0.22 ± 0.02 ab | 0.19 ± 0.00 ab | 4.86 ± 7.98 bc | 0.25 ± 0.03 bc | 10.59 ± 2.64 c | 0.20 ± 0.04 ab | 12.80 ± 2.13 c | 9.56 ± 0.23 c | ||||
| 54 | Maltol (3-Hydroxy-2-methyl-4-pyrone) | 1971 | 126 | 24 h | 3.25 ± 0.55 cd | 1.51 ± 0.53 ab | 2.39 ± 0.19 bcd | 3.09 ± 0.92 bcd | 1.84 ± 0.33 bc | 2.15 ± 0.42 bcd | 3.69 ± 0.53d | 0.06 ± 0.00 a | 2.80 ± 0.97 bcd | 1.78 ± 0.59 bc |
| 1 M | 1.17 ± 0.40 ab | 1.74 ± 0.69 ab | 1.95 ± 0.93 b | 2.30 ± 0.64 b | 2.12 ± 0.83 b | 1.32 ± 0.41 ab | 0.74 ± 0.23 ab | 0.73 ± 0.04 ab | 0.19 ± 0.08 a | 1.06 ± 0.40 ab | ||||
| Pyrazines | ||||||||||||||
| 55 | 2,5-Dimethylpyrazine | 1322 | 42 | 24 h | 80.26 ± 10.51 a | 67.43 ± 3.51 a | 94.55 ± 23.50 a | 89.40 ± 14.16 a | 82.78 ± 14.28 a | 83.46 ± 25.46 a | 97.16 ± 2.80 a | 95.22 ± 3.61 a | 113.60 ± 15.43 a | 79.88 ± 1.90 a |
| 1 M | 79.56 ± 17.60 ab | 93.48 ± 16.37 ab | 120.99 ± 13.18 b | 103.62 ± 12.11 ab | 109.90 ± 31.30 ab | 99.40 ± 29.99 ab | 99.14 ± 16.27 ab | 100.22 ± 14.40 ab | 89.57 ± 9.46 ab | 63.86 ± 12.27 a | ||||
| 56 | 2,6-Dimethylpyrazine | 1327 | 108 | 24 h | 120.09 ± 11.66 a | 99.62 ± 8.21 a | 111.09 ± 24.45 a | 132.20 ± 23.81 ab | 153.84 ± 31.14 ab | 150.72 ± 39.12 ab | 149.77 ± 7.34 ab | 140.10 ± 13.24 ab | 189.89 ± 33.27 b | 118.79 ± 7.86 a |
| 1 M | 122.47 ± 29.12 ab | 134.77 ± 20.98 ab | 180.02 ± 23.50 b | 158.80 ± 14.99 ab | 120.00 ± 51.50 ab | 155.33 ± 42.10 ab | 149.02 ± 25.60 ab | 150.88 ± 20.40 ab | 136.47 ± 9.81 ab | 96.89 ± 21.02 a | ||||
| 57 | Ethyl pyrazine | 1332 | 107 | 24 h | 81.90 ± 4.93 a | 64.34 ± 4.86 a | 102.41 ± 19.68 a | 85.14 ± 11.28 a | 95.05 ± 38.61 a | 80.28 ± 28.22 a | 100.81 ± 7.26 a | 97.34 ± 3.94 a | 120.21 ± 23.34 a | 81.05 ± 6.32 a |
| 1 M | 76.14 ± 17.20 ab | 94.93 ± 16.47 ab | 112.01 ± 13.64 b | 101.91 ± 14.46 ab | 106.33 ± 18.49 ab | 99.13 ± 26.13 ab | 93.65 ± 15.72 ab | 104.38 ± 11.98 ab | 84.40 ± 2.68 ab | 61.25 ± 14.78 a | ||||
| 58 | 2,3-Dimethylpyrazine | 1346 | 108 | 24 h | 16.51 ± 0.49 a | 11.52 ± 1.54 a | 21.21 ± 4.62 a | 15.39 ± 2.76 a | 16.04 ± 4.76 a | 19.63 ± 7.19 a | 19.17 ± 2.17 a | 20.34 ± 1.44 a | 25.55 ± 7.12 a | 16.08 ± 3.11 a |
| 1 M | 15.99 ± 5.14 a | 14.38 ± 3.04 a | 24.22 ± 4.29 a | 17.89 ± 3.06 a | 22.96 ± 5.76 a | 20.28 ± 6.81 a | 16.65 ± 2.69 a | 19.41 ± 2.47 a | 16.40 ± 0.63 a | 11.38 ± 2.46 a | ||||
| 59 | Pyrazine | 1215 | 80 | 24 h | 9.07 ± 3.04 a | 10.49 ± 1.59 a | 15.60 ± 5.30 a | 19.00 ± 8.35 a | 22.23 ± 8.73 a | 22.79 ± 8.99 a | 20.24 ± 7.40 a | 26.93 ± 6.21 a | 30.81 ± 10.72 a | 19.29 ± 5.88 a |
| 1 M | 15.22 ± 1.54 ab | 18.16 ± 3.56 ab | 26.64 ± 5.70 abc | 23.88 ± 4.30 abc | 34.62 ± 7.80 c | 24.13 ± 5.38 abc | 21.77 ± 6.10 abc | 29.78 ± 10.62 bc | 16.98 ± 2.92 ab | 11.62 ± 2.74 a | ||||
| 60 | Methyl pyrazine | 1267 | 94 | 24 h | 224.29 ± 29.07 a | 181.42 ± 26.27 a | 232.20 ± 67.29 a | 246.11 ± 28.46 a | 232.00 ± 47.88 a | 235.21 ± 74.64 a | 264.99 ± 84.17 a | 298.04 ± 32.67 a | 370.55 ± 82.46 a | 234.12 ± 34.70 a |
| 1 M | 209.44 ± 62.56 ab | 258.45 ± 41.88 ab | 345.60 ± 72.47 b | 308.22 ± 41.39 ab | 354.40 ± 8.63 b | 342.54 ± 60.45 b | 299.23 ± 63.17 ab | 332.10 ± 29.30 b | 319.95 ± 58.08 ab | 172.34 ± 43.31 a | ||||
| 61 | 2-Ethyl-6-methylpyrazine | 1382 | 121 | 24 h | 109.72 ± 10.61 a | 85.94 ± 2.86 a | 128.83 ± 23.08 a | 117.12 ± 20.55 a | 118.68 ± 40.00 a | 105.76 ± 35.39 a | 118.97 ± 0.77 a | 118.18 ± 14.48 a | 153.08 ± 17.55 a | 101.74 ± 3.55 a |
| 1 M | 100.46 ± 20.15 a | 110.95 ± 26.72 a | 142.23 ± 14.52 a | 127.16 ± 16.22 a | 144.30 ± 32.09 a | 115.76 ± 50.25 a | 115.95 ± 19.69 a | 113.95 ± 26.52 a | 90.99 ± 23.42 a | 79.84 ± 16.57 a | ||||
| 62 | 2-Ethyl-5-methylpyrazine | 1387 | 56 | 24 h | 8.52 ± 1.32 a | 6.41 ± 0.47 a | 8.42 ± 0.67 a | 8.92 ± 1.33 a | 9.43 ± 3.28 a | 8.27 ± 3.30 a | 9.79 ± 0.11 a | 8.71 ± 1.15 a | 12.37 ± 1.06 a | 7.94 ± 0.58 a |
| 1 M | 8.13 ± 1.97 a | 8.66 ± 2.43 a | 11.61 ± 1.28 a | 10.20 ± 1.19 a | 11.04 ± 3.18 a | 11.66 ± 3.63 a | 8.49 ± 1.83 a | 8.23 ± 2.49 a | 6.91 ± 2.39 a | 6.53 ± 1.37 a | ||||
| 63 | 2-Ethyl-3-methylpyrazine | 1401 | 122 | 24 h | 60.52 ± 5.90 a | 44.08 ± 2.35 a | 72.37 ± 14.49 a | 62.45 ± 10.77 a | 65.11 ± 21.82 a | 56.85 ± 18.53 a | 66.71 ± 0.47 a | 64.60 ± 6.68 a | 88.57 ± 11.91 a | 56.50 ± 2.62 a |
| 1 M | 56.14 ± 10.79 a | 51.56 ± 10.77 a | 83.07 ± 9.20 a | 63.48 ± 10.75 a | 82.72 ± 17.87 a | 65.01 ± 27.30 a | 65.92 ± 12.23 a | 57.07 ± 10.62 a | 54.40 ± 10.99 a | 45.33 ± 8.87 a | ||||
| 64 | 2,6-Diethylpyrazine | 1428 | 135 | 24 h | 14.61 ± 1.55 a | 12.22 ± 0.90 a | 16.58 ± 3.98 a | 16.23 ± 2.48 a | 15.56 ± 4.26 a | 13.29 ± 3.54 a | 15.33 ± 0.08 a | 15.50 ± 2.74 a | 19.21 ± 1.89 a | 14.04 ± 1.07 a |
| 1 M | 12.10 ± 2.18 a | 13.47 ± 3.33 a | 16.94 ± 2.70 a | 14.50 ± 2.03 a | 16.58 ± 4.55 a | 17.38 ± 5.31 a | 12.92 ± 2.52 a | 12.76 ± 3.51 a | 8.68 ± 2.63 a | 10.54 ± 2.32 a | ||||
| 65 | 2,5-Dimethyl-3-ethylpyrazine | 1441 | 135 | 24 h | 71.56 ± 11.01 a | 50.10 ± 1.59 a | 82.88 ± 18.68 a | 75.41 ± 14.56 a | 77.21 ± 25.44 a | 65.43 ± 21.47 a | 75.68 ± 0.78 a | 73.67 ± 12.70 a | 99.08 ± 10.17 a | 67.51 ± 4.79 a |
| 1 M | 63.10 ± 11.99 ab | 52.50 ± 12.37 a | 93.42 ± 9.86 b | 64.50 ± 11.17 ab | 91.27 ± 26.37 b | 87.94 ± 26.43 b | 72.70 ± 15.60 ab | 50.69 ± 18.35 a | 55.82 ± 15.58 a | 51.34 ± 9.58 a | ||||
| 66 | 2,3-Dimethyl-5-ethylpyrazine | 1457 | 135 | 24 h | 18.30 ± 2.65 a | 12.49 ± 0.11 a | 16.04 ± 0.78 a | 19.03 ± 4.71 a | 19.05 ± 6.24 a | 16.25 ± 5.20 a | 18.20 ± 1.40 a | 18.53 ± 3.90 a | 23.86 ± 1.33 a | 16.92 ± 1.59 a |
| 1 M | 15.94 ± 2.49 a | 13.39 ± 3.16 a | 23.08 ± 3.04 a | 16.12 ± 2.66 a | 23.08 ± 7.14 a | 22.94 ± 7.91 a | 18.44 ± 3.81 a | 14.03 ± 3.38 a | 13.22 ± 4.11 a | 12.78 ± 2.24 a | ||||
| 67 | 2-Methyl-6-propyl pyrazine | 1461 | 108 | 24 h | 3.01 ± 0.16 a | 2.65 ± 0.12 a | 3.72 ± 1.29 ab | 4.20 ± 0.69 b | 3.05 ± 0.20 a | 2.61 ± 0.19 a | 3.53 ± 1.09 ab | 3.6 ± 0.59 b | 4.75 ± 0.71 b | 3.61 ± 0.15 b |
| 1 M | 1.87 ± 0.42 a | 1.92 ± 0.65 ab | 3.33 ± 0.77 b | 2.57 ± 0.86 b | 3.09 ± 0.68 b | 2.60 ± 0.61 b | 2.73 ± 0.16 b | 2.36 ± 0.57 ab | 1.38 ± 0.42 a | 1.07 ± 0.22 a | ||||
| 68 | 2-Methyl-3,5-diethylpyrazine | 1488 | 149 | 24 h | 16.90 ± 1.96 a | 12.85 ± 0.46 a | 17.72 ± 3.92 a | 16.44 ± 2.66 a | 17.13 ± 4.81 a | 14.20 ± 4.23 a | 15.95 ± 0.50 a | 16.29 ± 3.18 a | 20.31 ± 2.11 a | 15.20 ± 1.11 a |
| 1 M | 12.48 ± 2.48 a | 10.96 ± 2.34 a | 15.92 ± 1.58 a | 12.75 ± 2.13 a | 15.37 ± 4.53 a | 15.37 ± 4.30 a | 13.00 ± 2.86 a | 10.22 ± 2.33 a | 9.48 ± 2.42 a | 10.83 ± 2.60 a | ||||
| Pyridines | ||||||||||||||
| 69 | Pyridine | 1194 | 52 | 24 h | 1.19 ± 0.24 ab | 0.52 ± 0.25 a | 3.03 ± 1.16 b | 1.21 ± 0.53 ab | 9.64 ± 2.26 b | 0.78 ± 0.33 a | 4.64 ± 2.71 b | 0.56 ± 0.08 a | 12.73 ± 6.11 b | 0.62 ± 0.21 a |
| 1 M | 5.62 ± 1.90 b | 0.37 ± 0.02 a | 8.60 ± 2.71 b | 0.62 ± 0.26 a | 9.01 ± 3.27 b | 2.60 ± 0.75 ab | 5.24 ± 2.94 b | 0.66 ± 0.07 a | 3.51 ± 1.21 ab | 3.37 ± 0.86 ab | ||||
| 70 | 1-(5-Hydroxypyridin-2-yl)ethanone | 1639 II | 122 | 24 h | 0.92 ± 0.21 a | 0.88 ± 0.04 a | 1.32 ± 0.19 ab | 0.89 ± 0.21 a | 1.16 ± 0.21 ab | 1.06 ± 0.20 a | 1.22 ± 0.23 ab | 0.86 ± 0.22 a | 1.67 ± 0.15 b | 0.94 ± 0.21 a |
| 1 M | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.01 ± 0.00 ab | 0.82 ± 0.12 b | 1.12 ± 0.51 b | 1.06 ± 0.24 b | 0.01 ± 0.00 ab | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.32 ± 0.55 ab | ||||
| Pyrroles | ||||||||||||||
| 71 | 1-Methyl-1H-pyrrole-2-carboxaldehyde | 1624 | 80 | 24 h | 9.76 ± 2.52 a | 9.23 ± 0.23 a | 11.63 ± 2.82 a | 11.35 ± 1.10 a | 10.19 ± 0.83 a | 13.27 ± 3.68 a | 12.44 ± 0.94 a | 11.73 ± 0.86 a | 15.38 ± 1.95 a | 10.71 ± 0.02 a |
| 1 M | 9.50 ± 2.46 a | 11.09 ± 1.40 a | 13.20 ± 1.43 a | 12.24 ± 1.08 a | 12.15 ± 1.42 a | 11.27 ± 3.42 a | 11.50 ± 2.00 a | 11.99 ± 0.60 a | 10.17 ± 0.86 a | 6.89 ± 0.62 a | ||||
| 72 | 1-Ethyl-2-formyl pyrrole (1-Ethyl-1H-pyrrole-2-carbaldehyde) | 1610 | 123 | 24 h | 3.52 ± 0.67 a | 3.73 ± 0.27 a | 4.24 ± 0.84 a | 4.59 ± 0.28 a | 4.02 ± 0.53 a | 4.78 ± 0.40 a | 4.40 ± 0.04 a | 4.76 ± 0.53 a | 5.11 ± 0.34 a | 4.02 ± 1.15 a |
| 1 M | 3.14 ± 0.26 a | 4.07 ± 0.64 a | 4.29 ± 0.63 a | 4.40 ± 0.41 a | 4.46 ± 0.56 a | 4.19 ± 0.78 a | 4.48 ± 1.36 a | 4.05 ± 0.96 a | 3.31 ± 1.36 a | 2.80 ± 0.56 a | ||||
| 73 | 2-Acetyl-1-methylpyrrole | 1657 | 123 | 24 h | 6.47 ± 1.94 a | 4.55 ± 1.33 a | 7.43 ± 1.62 a | 6.50 ± 1.79 a | 6.53 ± 2.46 a | 7.57 ± 1.12 a | 7.39 ± 1.08 a | 7.16 ± 2.11 a | 10.05 ± 0.51 a | 6.36 ± 0.86 a |
| 1 M | 5.14 ± 2.01 a | 6.10 ± 1.57 a | 7.36 ± 1.72 a | 6.38 ± 0.71 a | 7.43 ± 2.30 a | 8.45 ± 2.12 a | 4.79 ± 1.33 a | 4.84 ± 0.50 a | 5.02 ± 0.60 a | 5.81 ± 1.05 a | ||||
| 74 | 1-(2-Furanylmethyl)-1H-pyrrole | 1831 | 81 | 24 h | 8.79 ± 1.59 a | 7.18 ± 0.59 a | 9.11 ± 1.02 a | 12.54 ± 1.73 b | 11.90 ± 0.99 b | 11.78 ± 2.40 b | 10.63 ± 0.36 b | 11.87 ± 0.94 b | 11.03 ± 2.74 b | 8.84 ± 0.44 a |
| 1 M | 0.94 ± 0.69 a | 1.65 ± 0.36 a | 2.86 ± 0.44 a | 1.53 ± 0.04 a | 1.49 ± 0.22 a | 1.99 ± 0.26 a | 1.79 ± 0.35 a | 1.60 ± 0.31 a | 2.11 ± 0.26 a | 4.27 ± 5.80 a | ||||
| 75 | 2-Acetylpyrrole (1-(1H-pyrrol-2-yl)-ethanone) | 1976 | 94 | 24 h | 8.62 ± 0.83 ab | 6.50 ± 1.54 a | 10.62 ± 1.00 b | 8.43 ± 1.84 ab | 9.27 ± 2.73 ab | 6.71 ± 2.82 a | 11.75 ± 1.02 b | 9.18 ± 2.09 ab | 14.26 ± 3.63 b | 6.26 ± 0.35 a |
| 1 M | 7.82 ± 2.46 ab | 10.81 ± 3.85 ab | 14.17 ± 2.04 b | 9.72 ± 1.57 ab | 12.32 ± 3.59 ab | 10.73 ± 4.24 ab | 10.41 ± 1.44 ab | 9.91 ± 2.19 ab | 9.16 ± 1.15 ab | 6.29 ± 2.15 a | ||||
| 76 | 1H-Pyrrole-2-carboxaldehyde | 2031 | 95 | 24 h | 10.24 ± 3.88 a | 7.44 ± 1.36 a | 16.31 ± 2.98 a | 10.79 ± 1.19 a | 11.70 ± 3.73 a | 12.19 ± 2.89 a | 12.72 ± 1.59 a | 11.22 ± 3.82 a | 18.79 ± 7.01 a | 6.97 ± 0.60 a |
| 1 M | 8.12 ± 0.64 bc | 12.48 ± 3.43 c | 11.15 ± 1.29 c | 10.93 ± 2.59 c | 4.68 ± 2.01 ab | 10.92 ± 1.47 c | 2.50 ± 0.16 a | 9.62 ± 0.80 bc | 5.18 ± 0.4 ab | 7.83 ± 1.47 bc | ||||
| 77 | 1-Furfuryl-2-formyl pyrrole (N-Furfuryl-2-formylpyrrole) | 2255 | 81 | 24 h | 4.82 ± 0.64 a | 4.76 ± 0.67 a | 4.71 ± 1.57 a | 4.74 ± 1.07 a | 3.70 ± 0.37 a | 4.60 ± 1.05 a | 3.63 ± 0.76 a | 4.98 ± 0.85 a | 5.30 ± 0.43 a | 3.59 ± 0.65 a |
| 1 M | 3.11 ± 0.26 b | 4.00 ± 1.41 b | 1.20 ± 0.13 a | 3.78 ± 0.05 b | 1.60 ± 0.37 a | 5.17 ± 0.90 b | 1.18 ± 0.32 a | 2.48 ± 0.83 b | 0.53 ± 0.20 a | 3.32 ± 0.08 b | ||||
| Volatile phenols | ||||||||||||||
| 78 | 2-Hydroxyacetophenone | 1806 | 121 | 24 h | 0.72 ± 0.26 a | 0.63 ± 0.12 a | 0.88 ± 0.19 a | 0.79 ± 0.11 a | 0.88 ± 0.24 a | 0.68 ± 0.23 a | 0.82 ± 0.06 a | 0.69 ± 0.14 a | 0.88 ± 0.26 a | 0.89 ± 0.06 a |
| 1 M | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ||||
| 79 | Guaiacol (2-Methoxyphenol) | 1863 | 124 | 24 h | 8.25 ± 1.74 b | 4.83 ± 1.29 a | 9.63 ± 0.48 b | 7.69 ± 1.38 ab | 6.05 ± 0.95 a | 7.06 ± 2.25 ab | 7.98 ± 0.60 b | 7.08 ± 0.90 ab | 9.62 ± 2.24 b | 6.74 ± 0.23 a |
| 1 M | 5.73 ± 0.94 a | 6.99 ± 1.02 a | 11.11 ± 2.70 a | 7.48 ± 1.38 a | 8.97 ± 3.05 a | 9.50 ± 1.86 a | 8.26 ± 1.77 a | 7.43 ± 1.39 a | 6.06 ± 0.84 a | 7.97 ± 0.89 a | ||||
| 80 | 2-Methylphenol (o-Cresol) | 2006 | 107 | 24 h | 1.46 ± 0.15 ab | 1.17 ± 0.14 a | 1.41 ± 0.29 ab | 1.88 ± 0.20 bc | 1.17 ± 0.22 a | 1.05 ± 0.12 a | 1.27 ± 0.16 a | 1.19 ± 0.09 a | 2.06 ± 0.17 c | 1.23 ± 0.27 a |
| 1 M | 0.82 ± 0.21 a | 1.16 ± 0.17 a | 1.65 ± 0.54 a | 1.78 ± 0.27 a | 1.15 ± 0.48 a | 0.97 ± 0.27 a | 1.36 ± 0.39 a | 1.48 ± 0.33 a | 1.50 ± 0.39 a | 1.11 ± 0.05 a | ||||
| 81 | Phenol | 2009 | 66 | 24 h | 3.84 ± 0.36 ab | 2.78 ± 0.37 a | 5.65 ± 0.68 b | 3.83 ± 0.41 a | 4.22 ± 1.15 ab | 3.59 ± 1.35 a | 4.66 ± 0.41 b | 4.08 ± 0.65 ab | 6.59 ± 1.37 b | 3.64 ± 0.38 a |
| 1 M | 4.21 ± 1.09 a | 4.96 ± 1.26 a | 5.45 ± 1.48 a | 5.10 ± 0.70 a | 5.92 ± 1.37 a | 4.74 ± 1.32 a | 4.66 ± 0.46 a | 5.17 ± 0.98 a | 4.02 ± 0.38 a | 3.39 ± 1.13 a | ||||
| 82 | 4-Ethylguaiacol | 2032 | 137 | 24 h | 5.27 ± 1.27 a | 5.06 ± 0.27 a | 6.38 ± 1.31 a | 6.01 ± 1.00 a | 7.41 ± 3.10 a | 5.20 ± 1.63 a | 7.14 ± 1.91 a | 5.27 ± 0.53 a | 7.15 ± 0.21 a | 5.01 ± 0.72 a |
| 1 M | 2.07 ± 0.59 a | 5.72 ± 3.27 ab | 21.46 ± 2.23 b | 5.75 ± 0.93 b | 9.96 ± 5.77 b | 3.61 ± 0.92 a | 2.17 ± 1.06 a | 3.10 ± 0.83 a | 11.77 ± 2.72 b | 3.09 ± 0.69 a | ||||
| 83 | 4-Ethylphenol | 2178 | 107 | 24 h | 0.83 ± 0.16 a | 3.39 ± 0.38 b | 3.46 ± 1.38 b | 1.40 ± 0.15 ab | 1.66 ± 0.79 ab | 0.74 ± 0.27 a | 1.54 ± 1.87 ab | 0.85 ± 0.19 a | 4.98 ± 3.38 b | 0.96 ± 0.43 ab |
| 1 M | 0.27 ± 0.05 a | 52.51 ± 17.23 b | 23.07 ± 2.87 b | 22.00 ± 10.14 b | 23.61 ± 11.89 b | 0.35 ± 0.22 a | 0.24 ± 0.12 a | 12.22 ± 2.43 ab | 23.81 ± 4.65 b | 0.69 ± 0.03 a | ||||
| 84 | 4-Vinylguaiacol (4-Vinyl-2-methoxy-phenol) | 2202 | 150 | 24 h | 3.90 ± 0.85 a | 6.17 ± 1.46 a | 5.11 ± 1.08 a | 5.45 ± 1.10 a | 5.14 ± 0.72 a | 6.35 ± 0.96 a | 6.05 ± 0.96 a | 4.50 ± 0.16 a | 4.09 ± 0.54 a | 4.20 ± 1.64 a |
| 1 M | 0.34 ± 0.03 a | 1.28 ± 0.68 b | 1.04 ± 0.29 ab | 0.82 ± 0.07 ab | 1.55 ± 0.46 b | 0.84 ± 0.35 ab | 1.94 ± 0.59 b | 0.46 ± 0.15 a | 1.49 ± 0.11 b | 0.51 ± 0.20 a | ||||
| Terpenes and terpenoids | ||||||||||||||
| 85 | trans-Linalool oxide | 1436 III | 59 | 24 h | 17.14 ± 0.03 a | 13.58 ± 0.84 a | 20.20 ± 6.97 a | 18.47 ± 4.48 a | 17.64 ± 3.95 a | 14.94 ± 6.20 a | 19.14 ± 2.25 a | 18.28 ± 2.31 a | 28.29 ± 3.39 a | 17.83 ± 1.44 a |
| 1 M | 16.81 ± 3.87 a | 29.29 ± 5.21 ab | 60.54 ± 8.59 b | 31.38 ± 7.62 b | 40.20 ± 19.39 b | 19.76 ± 7.64 a | 17.13 ± 1.56 a | 35.22 ± 9.05 b | 41.48 ± 1.52 b | 12.30 ± 3.30 a | ||||
| 86 | cis-Linalool oxide | 1465 III | 59 | 24 h | 8.33 ± 0.33 a | 6.25 ± 0.76 a | 10.56 ± 4.20 a | 9.49 ± 2.16 a | 9.52 ± 2.15 a | 10.09 ± 3.27 a | 10.54 ± 1.09 a | 9.70 ± 0.90 a | 15.86 ± 1.91 a | 9.63 ± 0.57 a |
| 1 M | 8.61 ± 2.02 a | 15.18 ± 2.45 ab | 29.42 ± 4.18 b | 16.68 ± 4.26 b | 19.42 ± 9.12 b | 13.37 ± 4.39 a | 8.83 ± 0.96 a | 19.25 ± 4.91 b | 20.05 ± 1.64 b | 4.84 ± 0.30 a | ||||
| 87 | Linalool | 1542 | 71 | 24 h | 17.19 ± 0.30 a | 18.32 ± 2.90 a | 24.40 ± 4.33 a | 19.72 ± 1.16 a | 23.12 ± 7.13 a | 16.85 ± 2.58 a | 19.12 ± 1.62 a | 18.28 ± 0.94 a | 23.42 ± 3.11 a | 17.61 ± 0.60 a |
| 1 M | 11.47 ± 2.00 ab | 18.79 ± 2.26 cd | 19.75 ± 2.85d | 18.23 ± 3.57 cd | 17.66 ± 2.47 bcd | 12.58 ± 2.56 abc | 10.56 ± 1.16 a | 17.60 ± 1.77 bcd | 14.95 ± 1.06 abcd | 11.44 ± 1.92 ab | ||||
| 88 | α-Terpinenol | 1692 | 59 | 24 h | 4.33 ± 0.25 ab | 3.07 ± 0.32 a | 4.87 ± 0.62 b | 3.97 ± 1.29 a | 4.63 ± 1.46 ab | 3.16 ± 0.83 a | 3.82 ± 0.49 a | 3.60 ± 0.78 a | 5.87 ± 0.25 b | 4.19 ± 0.53 ab |
| 1 M | 3.81 ± 0.51 a | 5.89 ± 1.28 a | 5.69 ± 1.39 a | 5.44 ± 1.66 a | 5.96 ± 1.61 a | 4.42 ± 1.79 a | 3.46 ± 0.27 a | 5.76 ± 1.31 a | 3.52 ± 0.66 a | 2.86 ± 0.65 a | ||||
| 89 | Nerol (2,6-Octadien-1-ol, 3,7-dimethyl-, (Z)-) | 1806 | 69 | 24 h | 0.83 ± 0.18 a | 1.64 ± 0.02 b | 2.26 ± 0.42 b | 1.28 ± 0.23 ab | 2.33 ± 0.79 b | 0.78 ± 0.09 a | 1.02 ± 0.11 a | 0.80 ± 0.19 a | 3.10 ± 0.32 b | 0.90 ± 0.16 a |
| 1 M | 0.61 ± 0.09 a | 0.65 ± 0.08 a | 2.31 ± 0.44 b | 1.20 ± 0.25 b | 1.21 ± 0.65 b | 0.64 ± 0.13 a | 0.53 ± 0.09 a | 0.82 ± 0.04 ab | 1.18 ± 0.39 b | 0.57 ± 0.22 a | ||||
| 90 | Geraniol | 1855 | 69 | 24 h | 3.34 ± 0.57 a | 4.09 ± 0.40 a | 4.92 ± 1.00 a | 3.81 ± 0.23 a | 4.57 ± 1.06 a | 3.04 ± 0.79 a | 2.87 ± 0.34 a | 3.46 ± 1.34 a | 4.96 ± 0.56 a | 3.11 ± 0.59 a |
| 1 M | 1.58 ± 0.73 ab | 2.66 ± 0.77 ab | 2.32 ± 0.24 ab | 3.50 ± 0.68 b | 2.69 ± 1.22 ab | 2.04 ± 0.46 ab | 1.48 ± 0.25 a | 2.56 ± 0.25 ab | 1.73 ± 0.58 ab | 2.27 ± 0.28 ab | ||||
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binda, S.; Hill, C.; Johansen, E.; Obis, D.; Pot, B.; Sanders, M.E.; Tremblay, A.; Ouwehand, A.C. Criteria to qualify microorganisms as “probiotic” in foods and dietary supplements. Front. Microbiol. 2020, 11, 1662. [Google Scholar] [CrossRef] [PubMed]
- Muller, J.A.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C. Manufacture of probiotic bacteria. In Prebiotics and Probiotics Science and Technology; Charalampopoulos, D., Rastall, R.A., Eds.; Springer: New York, NY, USA, 2009; pp. 725–759. [Google Scholar]
- Naghmouchi, K.; Belguesmia, Y.; Bendali, F.; Spano, G.; Seal, B.S.; Drider, D. Lactobacillus fermentum: A bacterial species with potential for food preservation and biomedical applications. Crit. Rev. Food Sci. Nutr. 2020, 60, 3387–3399. [Google Scholar] [CrossRef] [PubMed]
- US Food and Drug Administration (U.S. FDA). GRN NO. 685. Lactobacillus plantarum 299v. 2017. Available online: https://www.cfsanappsexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices&id=685&sort=GRN_No&order=DESC&startrow=1&type=basic&search=plantarum (accessed on 6 July 2021).
- US Food and Drug Administration (U.S. FDA). GRN NO. 865. Lactobacillus acidophilus NCFM. 2020. Available online: https://www.cfsanappsexternal.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices&id=865&sort=GRN_No&order=DESC&startrow=1&type=basic&search=865. (accessed on 6 July 2021).
- Chan, M.Z.A.; Liu, S.-Q. Fortifying foods with synbiotic and postbiotic preparations of the probiotic yeast. Saccharomyces boulardii. Curr. Opin. Food Sci. 2022, 43, 216–224. [Google Scholar] [CrossRef]
- Min, M.; Bunt, C.R.; Mason, S.L.; Hussain, M.A. Non-dairy probiotic food products: An emerging group of functional foods. Crit. Rev. Food Sci. Nutr. 2019, 59, 2626–2641. [Google Scholar] [CrossRef]
- Euromonitor International. New Approaches to Functional Coffee. 2019. Available online: https://www.euromonitor.com/new-approaches-to-functional-coffee/report/ (accessed on 25 June 2019).
- Chan, M.Z.A.; Toh, M.; Liu, S.-Q. Growth, survival, and metabolic activities of probiotic lactobacillus spp. in fermented coffee brews supplemented with glucose and inactivated yeast derivatives. Food Res. Int. 2020, 137, 109746. [Google Scholar] [CrossRef]
- Chan, M.Z.A.; Toh, M.; Liu, S.-Q. Growth, survival, and metabolic activities of probiotics Lactobacillus rhamnosus GG and Saccharomyces cerevisiae var. boulardii CNCM-I745 in fermented coffee brews. Int. J. Food Microbiol. 2021, 350, 109229. [Google Scholar] [CrossRef]
- Lim, P.L.; Toh, M.; Liu, S.Q. Saccharomyces cerevisiae EC-1118 enhances the survivability of probiotic Lactobacillus rhamnosus HN001 in an acidic environment. Appl. Microbiol. Biotechnol. 2015, 99, 6803–6811. [Google Scholar] [CrossRef]
- Yeo, A.Y.Y.; Toh, M.Z.; Liu, S.Q. Enhancement of bifidobacteria survival by Williopsis saturnus var. saturnus in milk. Benficial Microbes 2016, 7, 135–144. [Google Scholar] [CrossRef]
- Lu, Y.; Putra, S.D.; Liu, S.-Q. A novel non-dairy beverage from durian pulp fermented with selected probiotics and yeast. Int. J. Food Microbiol. 2018, 265, 1–8. [Google Scholar] [CrossRef]
- Yamasaki-Yashiki, S.; Sawada, H.; Kino-Oka, M.; Katakura, Y. Analysis of gene expression profiles of Lactobacillus paracasei induced by direct contact with Saccharomyces cerevisiae through recognition of yeast mannan. Biosci. Microbiota Food Health 2017, 36, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Zoumpourtikoudi, V.; Pyrgelis, N.; Chatzigrigoriou, M.; Tasakis, R.N.; Touraki, M. Interactions among yeast and probiotic bacteria enhance probiotic properties and metabolism offering augmented protection to Artemia franciscana against Vibrio anguillarum. Microb. Pathog. 2018, 125, 497–506. [Google Scholar] [CrossRef]
- Ponomarova, O.; Gabrielli, N.; Sévin, D.C.; Mülleder, M.; Zirngibl, K.; Bulyha, K.; Andrejev, S.; Kafkia, E.; Typas, A.; Sauer, U.; et al. Yeast creates a niche for symbiotic lactic acid bacteria through nitrogen overflow. Cell Syst. 2017, 5, 345–357.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirai, S.; Kawasumi, T. Enhanced lactic acid bacteria viability with yeast coincubation under acidic conditions. Biosci. Biotechnol. Biochem. 2020, 84, 1706–1713. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-Q.; Tsao, M. Enhancement of survival of probiotic and non-probiotic lactic acid bacteria by yeasts in fermented milk under non-refrigerated conditions. Int. J. Food Microbiol. 2009, 135, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Singleton, V.L.; Rossi, J.A. Colorimetry of total phenolics with phosphomolybdic-phosphotungstic acid reagents. Am. J. Enol. Vitic. 1965, 16, 144–158. [Google Scholar]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT—Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Žuvela, P.; David, J.; Yang, X.; Huang, D.; Wong, M.W. Non-linear quantitative structure−activity relationships modelling, mechanistic study and in-silico design of flavonoids as potent antioxidants. Int. J. Mol. Sci. 2019, 20, 2328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortazavian, A.M.; Ehsani, M.R.; Mousavi, S.M.; Rezaei, K.; Sohrabvandi, S.; Reinheimer, J.A. Effect of refrigerated storage temperature on the viability of probiotic micro-organisms in yogurt. Int. J. Dairy Technol. 2007, 60, 123–127. [Google Scholar] [CrossRef]
- García, C.; Rendueles, M.; Díaz, M. Liquid-phase food fermentations with microbial consortia involving lactic acid bacteria: A review. Food Res. Int. 2019, 119, 207–220. [Google Scholar] [CrossRef]
- Zotta, T.; Parente, E.; Ricciardi, A. Aerobic metabolism in the genus Lactobacillus: Impact on stress response and potential applications in the food industry. J. Appl. Microbiol. 2017, 122, 857–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.-Q. Practical implications of lactate and pyruvate metabolism by lactic acid bacteria in food and beverage fermentations. Int. J. Food Microbiol. 2003, 83, 115–131. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- García-Campusano, F.; Anaya, V.-H.; Robledo-Arratia, L.; Quezada, H.; Hernández, H.; Riego, L.; González, A. ALT1-encoded alanine aminotransferase plays a central role in the metabolism of alanine in Saccharomyces cerevisiae. Can. J. Microbiol. 2009, 55, 368–374. [Google Scholar] [CrossRef]
- Ballester-Tomás, L.; Randez-Gil, F.; Pérez-Torrado, R.; Prieto, J.A. Redox engineering by ectopic expression ofglutamate dehydrogenase genes links NADPH availability and NADH oxidation with cold growth in Saccharomyces cerevisiae. Microb. Cell Fact. 2015, 14, 100. [Google Scholar] [CrossRef] [Green Version]
- Fernández, M.; Zúñiga, M. Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 2006, 32, 155–183. [Google Scholar] [CrossRef]
- Solms, J. Taste of amino acids, peptides, and proteins. J. Agric. Food Chem. 1969, 17, 686–688. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; De Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paczia, N.; Nilgen, A.; Lehmann, T.; Gätgens, J.; Wiechert, W.; Noack, S. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Factories 2012, 11, 122. [Google Scholar] [CrossRef] [Green Version]
- Klug, L.; Daum, G. Yeast lipid metabolism at a glance. FEMS Yeast Res. 2014, 14, 369–388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borrull, A.; López-Martínez, G.; Poblet, M.; Cordero-Otero, R.; Rozès, N. New insights into the toxicity mechanism of octanoic and decanoic acids on Saccharomyces cerevisiae. Yeast 2015, 32, 451–460. [Google Scholar] [CrossRef]
- Murzyn, A.; Krasowska, A.; Stefanowicz, P.; Dziadkowiec, D.; Łukaszewicz, M. Capric acid secreted by S. boulardii inhibits C. Albicans filamentous growth, adhesion and biofilm formation. PLoS ONE 2010, 5, e12050. [Google Scholar] [CrossRef] [PubMed]
- Hazelwood, L.A.; Daran, J.; Van Maris, A.J.A.; Pronk, J.T.; Dickinson, J.R. The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef] [Green Version]
- US Food and Drug Administration (U.S. FDA). CPG Sec 510.400 Dealcoholized Wine and Malt Beverages. 2005. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/cpg-sec-510400-dealcoholized-wine-and-malt-beverages-labeling (accessed on 6 July 2021).
- Regulations (EU). 2017/2119. Establishing the “Prodcom List” of Industrial Products Provided for by Council Regulation (EEC) No 3924/91., p. 25. 2017. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32017R2119 (accessed on 13 April 2022).
- Lentz, M. The impact of simple phenolic compounds on beer aroma and flavor. Fermentation 2018, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Mayer, R.J.; Que, L. 18O Studies of pyrogallol cleavage by catechol 1,2-dioxygenase. J. Biol. Chem. 1984, 259, 13056–13060. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, M.; Fan, J.; Tang, W.; Wang, D.; Ge, H.; Rong, H.; Teng, M.; Niu, L.; Liu, Q.; et al. Crystal structure of 3-hydroxyanthranilic acid 3,4-dioxygenase from Saccharomyces cerevisiae: A special subgroup of the Type III extradiol dioxygenases. Protein Sci. 2006, 15, 761–773. [Google Scholar] [CrossRef] [Green Version]
- Santamaría, L.; Reverón, I.; de Felipe, F.L.; de Las Rivas, B.; Muñoz, R. Ethylphenol formation by Lactobacillus plantarum: Identification of the enzyme involved in the reduction of vinylphenols. Appl. Environ. Microbiol. 2018, 84, e01064-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, I.; Campos, F.M.; Hogg, T.; Couto, J.A. Factors influencing the production of volatile phenols by wine lactic acid bacteria. Int. J. Food Microbiol. 2011, 145, 471–475. [Google Scholar] [CrossRef]
- Kalb, V.; Seewald, T.; Hofmann, T.; Granvogl, M. Studies on the impact of malting and mashing on the free, soluble ester-bound, and insoluble ester-bound forms of desired and undesired phenolic acids aiming at styrene mitigation during wheat beer brewing. J. Agric. Food Chem. 2020, 68, 12421–12432. [Google Scholar] [CrossRef]
- Joint FAO/WHO Expert Committee on Food Additives (JECFA). 3,4-dimethoxy-1-vinylbenzene. 2003. Available online: http://www.fao.org/food/food-safety-quality/scientific-advice/jecfa/jecfa-flav/details/en/c/1260/ (accessed on 13 June 2021).
- Górnaś, P.; Dwiecki, K.; Siger, A.; Tomaszewska-Gras, J.; Michalak, M.; Polewski, K. Contribution of phenolic acids isolated from green and roasted boiled-type coffee brews to total coffee antioxidant capacity. Eur. Food Res. Technol. 2016, 242, 641–653. [Google Scholar] [CrossRef] [Green Version]
- Feng, T.; Wang, J. Oxidative stress tolerance and antioxidant capacity of lactic acid bacteria as probiotic: A systematic review. Gut Microbes 2020, 12, 1801944. [Google Scholar] [CrossRef]
- Ludwig, I.A.; Sánchez, L.; De Peña, M.P.; Cid, C. Contribution of volatile compounds to the antioxidant capacity of coffee. Food Res. Int. 2014, 61, 67–74. [Google Scholar] [CrossRef]
- Fritsch, C.; Jänsch, A.; Ehrmann, M.A.; Toelstede, S.; Vogel, R.F. Characterization of cinnamoyl esterases from different lactobacilli and bifidobacteria. Curr. Microbiol. 2017, 74, 247–256. [Google Scholar] [CrossRef] [PubMed]
- NIST Chemistry WebBook. NIST Standard Reference Database Number 69. 2018. Available online: https://webbook.nist.gov/chemistry/ (accessed on 13 June 2021).
- Castro-Marín, A.; Buglia, A.G.; Riponi, C.; Chinnici, F. Volatile and fixed composition of sulphite-free white wines obtained after fermentation in the presence of chitosan. LWT 2018, 93, 174–180. [Google Scholar] [CrossRef]
- Baek, H.H.; Cadwallader, K.R. Roasted chicory aroma evaluation by gas chromatography/mass spectrometry/olfactometry. J. Food Sci. 1998, 63, 234–237. [Google Scholar] [CrossRef]
- Xiao, Z.; Wang, H.; Niu, Y.; Liu, Q.; Zhu, J.; Chen, H.; Ma, N. Characterization of aroma compositions in different chinese congou black teas using GC–MS and GC–O combined with partial least squares regression. Flavour Fragr. J. 2017, 32, 265–276. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chan, M.Z.A.; Tan, L.T.; Heng, S.W.Q.; Liu, S.Q. Effect of Co-Fermentation of Saccharomyces boulardii CNCM-I745 with Four Different Probiotic Lactobacilli in Coffee Brews on Cell Viabilities and Metabolic Activities. Fermentation 2023, 9, 219. https://doi.org/10.3390/fermentation9030219
Chan MZA, Tan LT, Heng SWQ, Liu SQ. Effect of Co-Fermentation of Saccharomyces boulardii CNCM-I745 with Four Different Probiotic Lactobacilli in Coffee Brews on Cell Viabilities and Metabolic Activities. Fermentation. 2023; 9(3):219. https://doi.org/10.3390/fermentation9030219
Chicago/Turabian StyleChan, Mei Zhi Alcine, Li Ting Tan, Shermaine Wan Qing Heng, and Shao Quan Liu. 2023. "Effect of Co-Fermentation of Saccharomyces boulardii CNCM-I745 with Four Different Probiotic Lactobacilli in Coffee Brews on Cell Viabilities and Metabolic Activities" Fermentation 9, no. 3: 219. https://doi.org/10.3390/fermentation9030219
APA StyleChan, M. Z. A., Tan, L. T., Heng, S. W. Q., & Liu, S. Q. (2023). Effect of Co-Fermentation of Saccharomyces boulardii CNCM-I745 with Four Different Probiotic Lactobacilli in Coffee Brews on Cell Viabilities and Metabolic Activities. Fermentation, 9(3), 219. https://doi.org/10.3390/fermentation9030219





