Exploring the Inhibitory Activity of Selected Lactic Acid Bacteria against Bread Rope Spoilage Agents
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms Used in This Study
2.1.1. Lactic Acid Bacteria Strains
2.1.2. Spoilage Bacteria Strains
2.2. Screenings for Antibacterial Activity of the LAB Strains
2.2.1. High-Throughput Screening Assay with LAB Cell-Free Supernatants (CFS)
2.2.2. Double-Agar-Layer Screening Assay
2.3. Confirmatory Assay of Antibacterial Activity in Bread Medium (BM)
2.4. Assessment of the LAB Bioactive Compounds
2.4.1. High-Performance Liquid Chromatography (HPLC) Detection of the Main Compounds Produced in MRS Fermentation
2.4.2. Bacteriocins Production
2.4.3. Kinetic Screening Assay
2.5. Phenotypical Characterization of the LAB Strains
2.5.1. API Test
2.5.2. Determination of Acidification Ability in BM USING ICINAC
2.6. Safety Assessment
2.6.1. Hemolytic Activity
2.6.2. LAB Antimicrobial Susceptibility
2.7. Whole-Genome Sequencing Analysis: In Silico Screening for Functional Genes and Virulence Genes
3. Results
3.1. Screening Assays
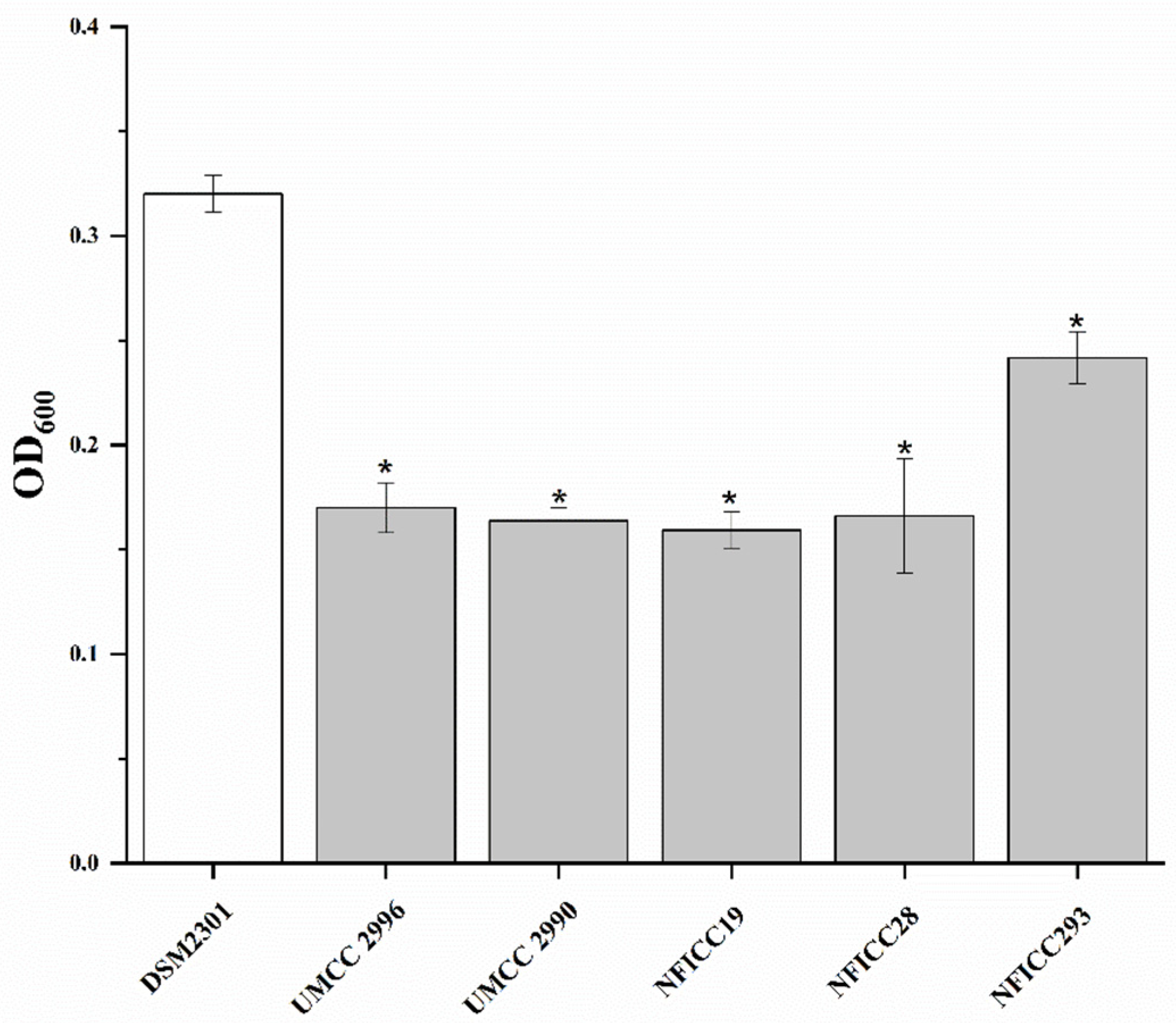
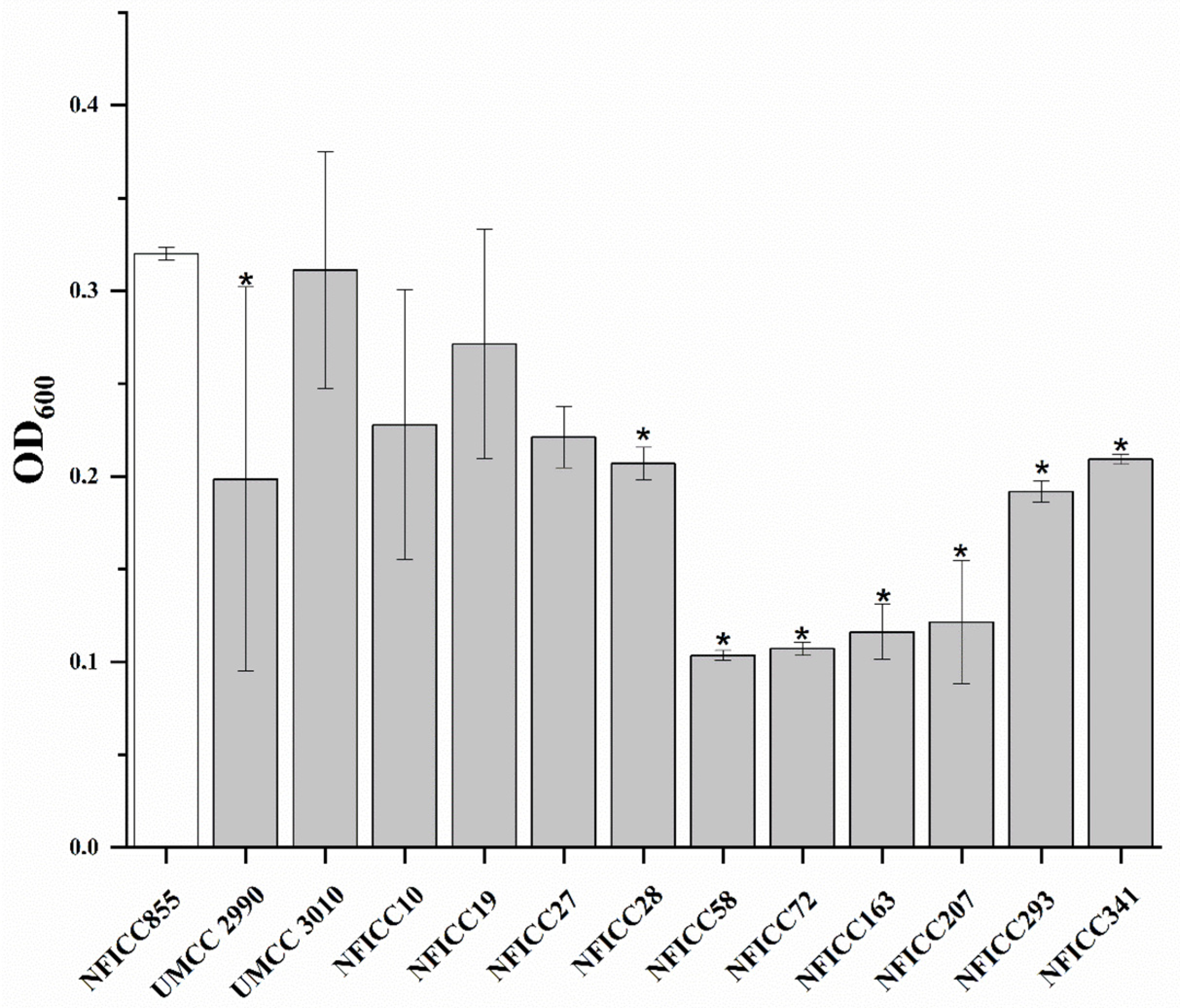
3.1.1. High-Throughput Preliminary Screening
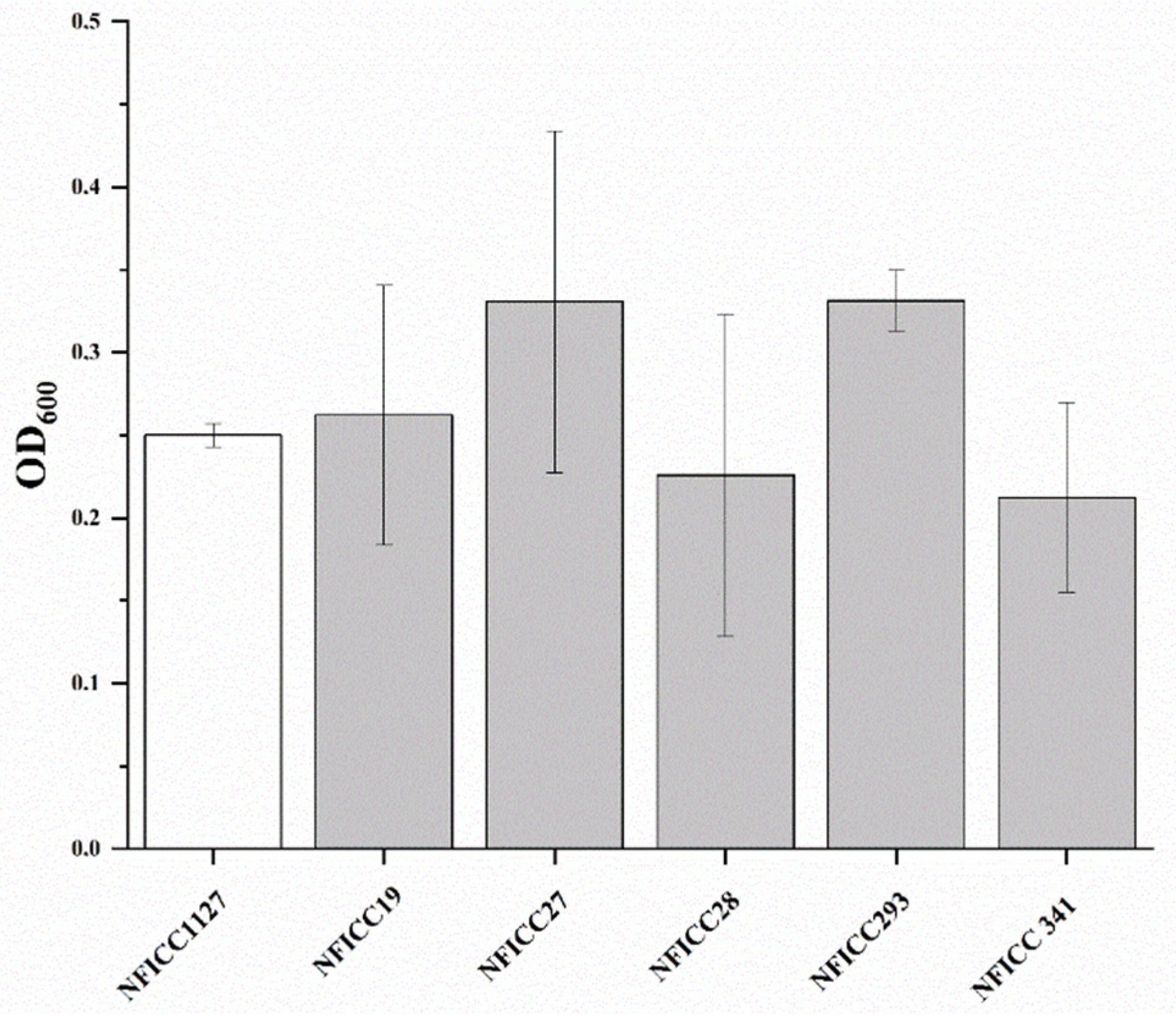
3.1.2. Antibacterial Activity of LAB Assessed by Double-Agar-Layer Screening Assay
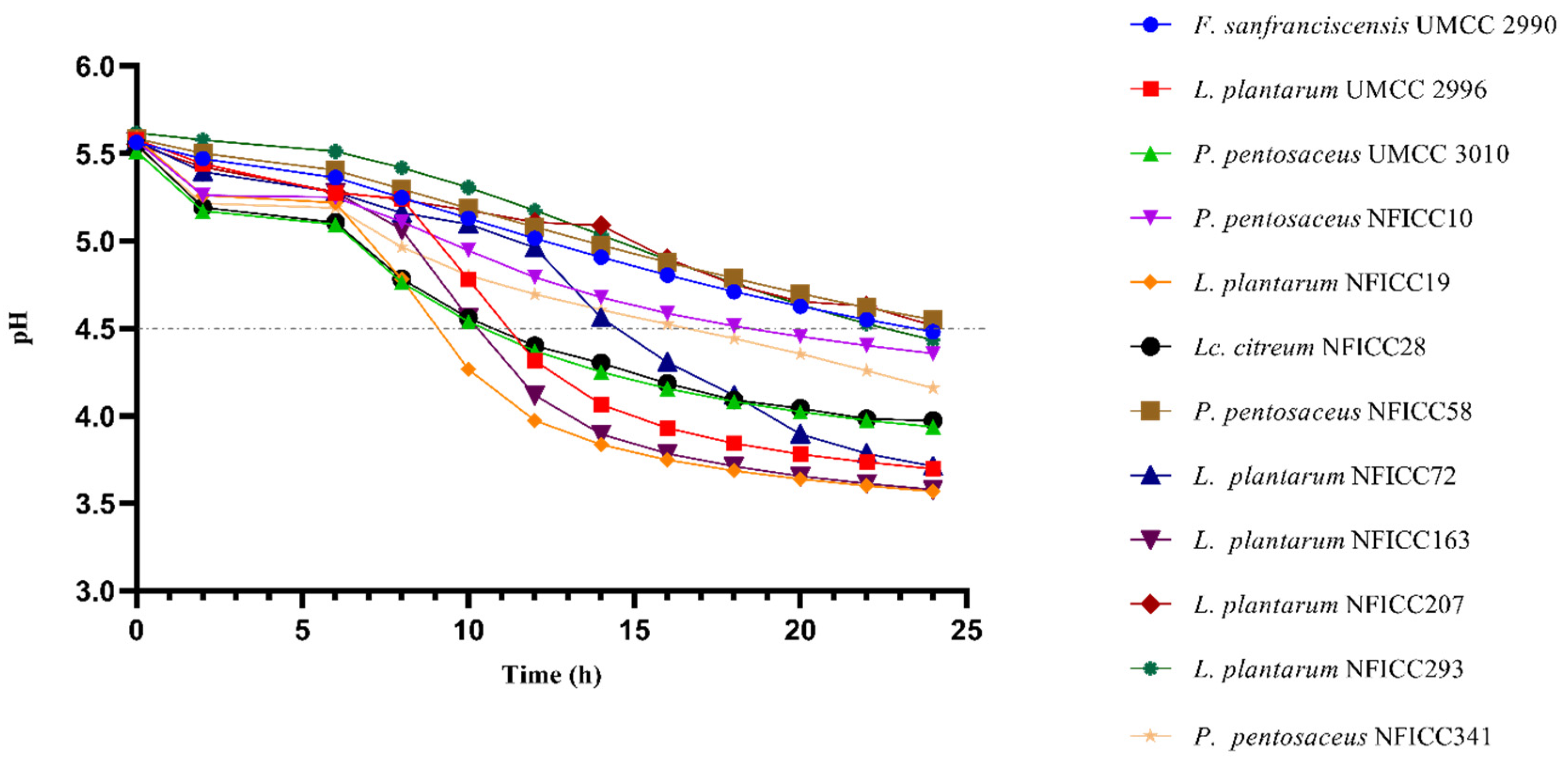
3.2. Antibacterial Activity of LAB by Confirmatory Assay in BM
3.3. Detection of Bioactive Compounds
3.3.1. Metabolites Detected by HPLC during Fermentation in MRS
3.3.2. Evaluation of the Bacteriocins Production and Kinetic Screening Assay
3.4. Assessment of Technological Features of the Candidate LAB Strains
3.4.1. Sugar Fermentation Profiling (API Test)
3.4.2. Acidification Ability in BM
3.5. Safety Profile of the Selected LAB Strains
3.5.1. Hemolytic Activity Profile
3.5.2. Antimicrobial Susceptibility Profiles (AST)
3.5.3. In Silico Screening for Resistance Genes
3.6. Whole-Genome Sequencing Analysis: Functional Gene and Potential Virulence In Silico Screening
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zain, M.Z.M.; Shori, A.B.; Baba, A.S. Potential functional food ingredients in bread and their health benefits. Biointerface Res. Appl. Chem. 2022, 12, 6533–6542. [Google Scholar] [CrossRef]
- Goryńska-Goldmann, E.; Gazdecki, M.; Rejman, K.; Łaba, S.; Kobus-Cisowska, J.; Szczepański, K. Magnitude, causes and scope for reducing food losses in the baking and confectionery industry a multi-method approach. Agriculture 2021, 11, 936. [Google Scholar] [CrossRef]
- Cicatiello, C.; Franco, S.; Pancino, B.; Blasi, E.; Falasconi, L. The dark side of retail food waste: Evidences from in-store data. Resour. Conserv. Recycl. 2017, 125, 273–281. [Google Scholar] [CrossRef]
- Muthappa, D.M.; Lamba, S.; Sivasankaran, S.K.; Naithani, A.; Rogers, N.; Srikumar, S.; Macori, G.; Scannell, A.G.M.; Fanning, S. 16S rRNA Based Profiling of Bacterial Communities Colonizing Bakery-Production Environments. Foodborne Pathog. Dis. 2022, 19, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Pepe, O.; Blaiotta, G.; Moschetti, G.; Greco, T.; Villani, F. Rope-producing strains of Bacillus spp. from wheat bread and strategy for their control by lactic acid bacteria. Appl. Environ. Microbiol. 2003, 69, 2321–2329. [Google Scholar] [CrossRef] [PubMed]
- Valerio, F.; De Bellis, P.; Di Biase, M.; Lonigro, S.L.; Giussani, B.; Visconti, A.; Lavermicocca, P.; Sisto, A. Diversity of spore-forming bacteria and identification of Bacillus amyloliquefaciens as a species frequently associated with the ropy spoilage of bread. Int. J. Food Microbiol. 2012, 156, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Valerio, F.; Di Biase, M.; Huchet, V.; Desriac, N.; Lonigro, S.L.; Lavermicocca, P.; Sohier, D.; Postollec, F. Comparison of three Bacillus amyloliquefaciens strains growth behaviour and evaluation of the spoilage risk during bread shelf-life. Food Microbiol. 2015, 45, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Iurie, R.; Cahul, B.P.H.; Uniersity, S.; Turtoi, M.; Dunarea, U.; Galati, D.J. The occurrence of the rope spoilage of bread at Cahulpan bread-making company. Sci. J. Cahul State Univ. “Bogdan Petriceicu Hasdeu” Econ. Eng. Stud. 2021, 2, 107–116. [Google Scholar]
- Thompson, J.M.; Waites, W.M.; Dodd, C.E.R. Detection of rope spoilage in bread caused by Bacillus species. J. Appl. Microbiol. 1998, 85, 481–486. [Google Scholar] [CrossRef]
- Pereira, A.P.M.; Stradiotto, G.C.; Freire, L.; Alvarenga, V.O.; Crucello, A.; Morassi, L.L.P.; Silva, F.P.; Sant’Ana, A.S. Occurrence and enumeration of rope-producing spore forming bacteria in flour and their spoilage potential in different bread formulations. LWT 2020, 133, 110108. [Google Scholar] [CrossRef]
- De Vero, L.; Iosca, G.; Gullo, M.; Pulvirenti, A. Functional and healthy features of conventional and non-conventional sourdoughs. Appl. Sci. 2021, 11, 3694. [Google Scholar] [CrossRef]
- Ibrahim, S.A.; Ayivi, R.D.; Zimmerman, T.; Siddiqui, S.A.; Altemimi, A.B.; Fidan, H.; Esatbeyoglu, T.; Bakhshayesh, R.V. Lactic acid bacteria as antimicrobial agents: Food safety and microbial food spoilage prevention. Foods 2021, 10, 3131. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Lv, M.; Shao, Z.; Hungwe, M.; Wang, J.; Bai, X.; Xie, J.; Wang, Y.; Geng, W. Metabolism Characteristics of Lactic Acid Bacteria and the Expanding Applications in Food Industry. Front. Bioeng. Biotechnol. 2021, 9, 612285. [Google Scholar] [CrossRef] [PubMed]
- De Vero, L.; Iosca, G.; La China, S.; Licciardello, F.; Gullo, M.; Pulvirenti, A. Yeasts and lactic acid bacteria for panettone production: An assessment of candidate strains. Microorganisms 2021, 9, 1093. [Google Scholar] [CrossRef] [PubMed]
- Iosca, G.; De Vero, L.; Di Rocco, G.; Perrone, G.; Gullo, M.; Pulvirenti, A. Anti-spoilage activity and exopolysaccharides production by selected lactic acid bacteria. Foods 2022, 11, 1914. [Google Scholar] [CrossRef]
- De Vuyst, L.; Leroy, F. Bacteriocins from lactic acid bacteria: Production, purification, and food applications. J. Mol. Microbiol. Biotechnol. 2007, 13, 194–199. [Google Scholar] [CrossRef]
- Iosca, G.; Turetta, M.; De Vero, L.; Bang-berthelsen, C.H.; Gullo, M.; Pulvirenti, A. Valorization of wheat bread waste and cheese whey through cultivation of lactic acid bacteria for bio-preservation of bakery products. LWT 2023, 176, 114524. [Google Scholar] [CrossRef]
- Zotta, T.; Parente, E.; Ricciardi, A. Viability staining and detection of metabolic activity of sourdough lactic acid bacteria under stress conditions. World J. Microbiol. Biotechnol. 2009, 25, 1119–1124. [Google Scholar] [CrossRef]
- Cizeikiene, D.; Juodeikiene, G.; Paskevicius, A.; Bartkiene, E. Antimicrobial activity of lactic acid bacteria against pathogenic and spoilage microorganism isolated from food and their control in wheat bread. Food Control 2013, 31, 539–545. [Google Scholar] [CrossRef]
- Katina, K.; Sauri, M.; Alakomi, H.L.; Mattila-Sandholm, T. Potential of lactic acid bacteria to inhibit rope spoilage in wheat sourdough bread. LWT 2002, 35, 38–45. [Google Scholar] [CrossRef]
- Menteş, Ö.; Ercan, R.; Akçelik, M. Inhibitor activities of two Lactobacillus strains, isolated from sourdough, against rope-forming Bacillus strains. Food Control 2007, 18, 359–363. [Google Scholar] [CrossRef]
- Valerio, F.; De Bellis, P.; Lonigro, S.L.; Visconti, A.; Lavermicocca, P. Use of Lactobacillus plantarum fermentation products in bread-making to prevent Bacillus subtilis ropy spoilage. Int. J. Food Microbiol. 2008, 122, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Madsen, S.K.; Priess, C.; Wätjen, A.P.; Øzmerih, S.; Mohammadifar, M.A.; Heiner Bang-Berthelsen, C. Development of a yoghurt alternative, based on plant-adapted lactic acid bacteria, soy drink and the liquid fraction of brewers’ spent grain. FEMS Microbiol. Lett. 2021, 368, fnab093. [Google Scholar] [CrossRef] [PubMed]
- Saranraj, P.; Gheeta, M. Microbial Spoilage of Bakery Products and Its Control by Preservatives. Int. J. Pharm. Biol. 2012, 3, 38–48. [Google Scholar]
- Inglin, R.C.; Stevens, M.J.A.; Meile, L.; Lacroix, C.; Meile, L. High-throughput screening assays for antibacterial and antifungal activities of Lactobacillus species. J. Microbiol. Methods 2015, 114, 26–29. [Google Scholar] [CrossRef]
- Verni, M.; Minisci, A.; Convertino, S.; Nionelli, L.; Rizzello, C.G. Wasted Bread as Substrate for the Cultivation of Starters for the Food Industry. Front. Microbiol. 2020, 11, 293. [Google Scholar] [CrossRef] [PubMed]
- Hui-Hu, C.; Ren, L.Q.; Zhou, Y.; Ye, B.C. Characterization of antimicrobial activity of three Lactobacillus plantarum strains isolated from Chinese traditional dairy food. Food Sci. Nutr. 2019, 7, 1997–2005. [Google Scholar] [CrossRef]
- Fugaban, J.I.I.; Vazquez Bucheli, J.E.; Holzapfel, W.H.; Todorov, S.D. Characterization of Partially Purified Bacteriocins Produced by Enterococcus faecium Strains Isolated from Soybean Paste Active Against Listeria spp. and Vancomycin-Resistant Enterococci. Microorganisms 2021, 9, 1085. [Google Scholar] [CrossRef]
- Corrieu, G.; Spinnler, H.E.; Picque, D.; Jomier, Y. Automated system to follow up 403 and control the acidification activity of lactic acid starters. Fr. Pat. FR 1988, 2, 612. [Google Scholar]
- Madsen, S.K.; Thulesen, E.T.; Mohammadifar, M.A.; Bang-Berthelsen, C.H. Chufa drink: Potential in developing a new plant-based fermented dessert. Foods 2021, 10, 3010. [Google Scholar] [CrossRef]
- Fugaban, J.I.I.; Vazquez Bucheli, J.E.; Kim, B.; Holzapfel, W.H.; Todorov, S.D. Safety and beneficial properties of bacteriocinogenic Pediococcus acidilactici and Pediococcus pentosaceus isolated from silage. Lett. Appl. Microbiol. 2021, 73, 725–734. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- VanOeffelen, M.; Nguyen, M.; Aytan-Aktug, D.; Brettin, T.; Dietrich, E.M.; Kenyon, R.W.; Machi, D.; Mao, C.; Olson, R.; Pusch, G.D.; et al. A genomic data resource for predicting antimicrobial resistance from laboratory-derived antimicrobial susceptibility phenotypes. Brief. Bioinform. 2021, 22, bbab313. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2020, 49, D480–D489. [Google Scholar] [CrossRef]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Bindu, A.; Lakshmidevi, N. Identification and in vitro evaluation of probiotic attributes of lactic acid bacteria isolated from fermented food sources. Arch. Microbiol. 2021, 203, 579–595. [Google Scholar] [CrossRef]
- Estifanos, H. Isolation and identification of probiotic lactic acid bacteria from curd and in vitro evaluation of its growth inhibition activities against pathogenic bacteria. Afr. J. Microbiol. Res. 2014, 8, 1419–1425. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Dubey, S.; Sharma, N.; Thakur, S.; Patel, R.; Reddy, B.M. Bacillus cereus food poisoning in Indian perspective: A review. Pharma Innov. J. 2021, 10, 970–975. [Google Scholar]
- Qian, M.; Liu, D.; Zhang, X.; Yin, Z.; Ismail, B.B.; Ye, X.; Guo, M. A review of active packaging in bakery products: Applications and future trends. Trends Food Sci. Technol. 2021, 114, 459–471. [Google Scholar] [CrossRef]
- Corsetti, A.; Gobbetti, M.; Smacchi, E. Antibacterial activity of sourdough lactic acid bacteria: Isolation of a bacteriocin-like inhibitory substance from Lactobacillus sanfrancisco C57. Food Microbiol. 1996, 13, 447–456. [Google Scholar] [CrossRef]
- Yang, Y.; Tao, W.Y.; Liu, Y.J.; Zhu, F. Inhibition of Bacillus cereus by lactic acid bacteria starter cultures in rice fermentation. Food Control 2008, 19, 159–161. [Google Scholar] [CrossRef]
- Adesulu-Dahunsi, A.T.; Sanni, A.I.; Jeyaram, K. Diversity and technological characterization of Pediococcus pentosaceus strains isolated from Nigerian traditional fermented foods. LWT 2021, 140, 110697. [Google Scholar] [CrossRef]
- Puntillo, M.; Gaggiotti, M.; Oteiza, J.M.; Binetti, A.; Massera, A.; Vinderola, G. Potential of Lactic Acid Bacteria Isolated From Different Forages as Silage Inoculants for Improving Fermentation Quality and Aerobic Stability. Front. Microbiol. 2020, 11, 586716. [Google Scholar] [CrossRef]
- Arena, M.P.; Silvain, A.; Normanno, G.; Grieco, F.; Drider, D.; Spano, G.; Fiocco, D. Use of Lactobacillus plantarum strains as a bio-control strategy against food-borne pathogenic microorganisms. Front. Microbiol. 2016, 7, 464. [Google Scholar] [CrossRef]
- Messens, W.; De Vuyst, L. Inhibitory substances produced by Lactobacilli isolated from sourdoughs—A review. Int. J. Food Microbiol. 2002, 72, 31–43. [Google Scholar] [CrossRef]
- Lindgren, S.E.; Dobrogosz, W.J. Antagonistic activities of lactic acid bacteria in food and feed fermentations. FEMS Microbiol. Lett. 1990, 87, 149–163. [Google Scholar] [CrossRef]
- Thao, T.T.P.; Thoa, L.T.K.; Ngoc, L.M.T.; Lan, T.T.P.; Phuong, T.V.; Truong, H.T.H.; Khoo, K.S.; Manickam, S.; Hoa, T.T.; Tram, N.D.Q.; et al. Characterization halotolerant lactic acid bacteria Pediococcus pentosaceus HN10 and in vivo evaluation for bacterial pathogens inhibition. Chem. Eng. Process. Process Intensif. 2021, 168, 108576. [Google Scholar] [CrossRef]
- Ruiz Rodríguez, L.G.; Mohamed, F.; Bleckwedel, J.; Medina, R.; De Vuyst, L.; Hebert, E.M.; Mozzi, F. Diversity and functional properties of lactic acid bacteria isolated from wild fruits and flowers present in northern Argentina. Front. Microbiol. 2019, 10, 1091. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z. Evaluation of different kinds of organic acids and their antibacterial activity in Japanese Apricot fruits. Afr. J. Agric. Res. 2012, 7, 4911–4918. [Google Scholar] [CrossRef]
- Rishi, L.; Mittal, G.; Agarwal, R.K.; Sharma, T. Melioration in Anti-staphylococcal Activity of Conventional Antibiotic(s) by Organic Acids Present in the Cell Free Supernatant of Lactobacillus paraplantarum. Indian J. Microbiol. 2017, 57, 359–364. [Google Scholar] [CrossRef]
- Parada, J.L.; Caron, C.R.; Medeiros, A.B.P.; Soccol, C.R. Bacteriocins from lactic acid bacteria: Purification, properties and use as biopreservatives. Braz. Arch. Biol. Technol. 2007, 50, 521–542. [Google Scholar] [CrossRef]
- Zhang, T.; Zhang, Y.; Li, L.; Jiang, X.; Chen, Z.; Zhao, F.; Yi, Y. Biosynthesis and Production of Class II Bacteriocins of Food-Associated Lactic Acid Bacteria. Fermentation 2022, 8, 217. [Google Scholar] [CrossRef]
- Pacher, N.; Burtscher, J.; Johler, S.; Etter, D.; Bender, D.; Fieseler, L.; Domig, K.J. Ropiness in Bread—A Re-Emerging Spoilage Phenomenon. Foods 2022, 11, 3021. [Google Scholar] [CrossRef]
- Cho, W.-I.; Chung, M.-S. Antimicrobial effect of a combination of herb extract and organic acid against Bacillus subtilis spores. Food Sci. Biotechnol. 2017, 26, 1423–1428. [Google Scholar] [CrossRef]
- Amit, S.K.; Uddin, M.M.; Rahman, R.; Islam, S.M.R.; Khan, M.S. A review on mechanisms and commercial aspects of food preservation and processing. Agric. Food Secur. 2017, 6, 1–22. [Google Scholar] [CrossRef]
- Gould, G.W. Methods for preservation and extension of shelf life. Int. J. Food Microbiol. 1996, 33, 5–64. [Google Scholar] [CrossRef]
- Su, X.; Wu, F.; Zhang, Y.; Yang, N.; Chen, F.; Jin, Z.; Xu, X. Effect of organic acids on bread quality improvement. Food Chem. 2019, 278, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Ayivi, R.; Gyawali, R.; Krastanov, A.; Aljaloud, S.; Worku, M.; Tahergorabi, R.; Da Silva, R.; Ibrahim, S. Lactic Acid Bacteria: Food Safety and Human Health Applications. Dairy 2020, 1, 202–232. [Google Scholar] [CrossRef]
- Abriouel, H.; Casado Muñoz, M.D.C.; Lavilla Lerma, M.L.; Pérez Montoro, B.; Bockelmann, W.; Pichner, R.; Kabisch, J.; Cho, G.-S.; Franz, C.M.A.P.; Galvez, A.; et al. New insights in antibiotic resistance of Lactobacillus species from fermented foods. Food Res. Int. 2015, 78, 465–481. [Google Scholar] [CrossRef]
- Mathur, S.; Singh, R. Antibiotic resistance in food lactic acid bacteria—A review. Int. J. Food Microbiol. 2005, 105, 281–295. [Google Scholar] [CrossRef] [PubMed]



| Strain Code | Species | Isolation Source |
|---|---|---|
| NFICC10 | Pediococcus pentosaceus | Sourdough |
| NFICC19 | Lactiplantibacillus plantarum | Dill |
| NFICC27 | Lactiplantibacillus plantarum | Sourdough |
| NFICC28 | Leuconostoc citreum | Sourdough |
| NFICC58 | Pediococcus pentosaceus | Sourdough |
| NFICC72 | Lactiplantibacillus plantarum | Gooseberry |
| NFICC87 | Leuconostoc citreum | Beetroot |
| NFICC94 | Leuconostoc citreum | Spinach |
| NFICC103 | Pediococcus pentosaceus | Pumpkin |
| NFICC163 | Lactiplantibacillus platarum | Field pea |
| NFICC207 | Lactiplantibacillus plantarum | Glasswort |
| NFICC293 | Lactiplantibacillus plantarum | Dragsholm plant |
| NFICC341 | Pediococcus pentosaceus | Brewer’s spent grain |
| UMCC 2990 | Fructilactobacillus sanfranciscensis | Sourdough type I |
| UMCC 2996 | Lactiplantibacillus plantarum | Dough for Panettone |
| UMCC 3002 | Furfurilactobacillus rossiae | Dough for Panettone |
| UMCC 3010 | Pediococcus pentosaceus | Gluten-free sourdough |
| UMCC 3011 | Leuconostoc citreum | Dough for Panettone |
| Strain Code | Species | Isolation Source |
|---|---|---|
| DSM 2301 | Bacillus cereus | Food poisoning incident |
| DSM 4222 | Bacillus cereus | - |
| DSM 4312 | Bacillus cereus | Vomit |
| DSM 22905 | Bacillus cytotoxicus | Vegetable puree |
| NFICC119 | Lysinibacillus fusiformis | Beetroot |
| NFICC432 | Paenibacillus polymyxa | Walnut |
| NFICC503 | Bacillus mycoides | Beech leaves |
| NFICC510 | Bacillus altitudinis | Plant |
| NFICC526 | Bacillus mycoides | Red fir |
| NFICC528 | Bacillus subtilis | Sourdough |
| NFICC529 | Lysinibacillus sphaericus | Common Juniper |
| NFICC530 | Bacillus pumilus | Common Juniper |
| NFICC531 | Bacillus simplex | Common Juniper |
| NFICC532 | Lysinibacillus fusiformis | Common Juniper |
| NFICC740 | Bacillus cereus | Plant |
| NFICC781 | Bacillus cereus | Kombucha |
| NFICC816 | Bacillus thuringiensis | Animal feces |
| NFICC855 | Bacillus weihenstephanensis | Potato |
| NFICC869 | Bacillus amyloliquefaciens | Pasteurized BSG |
| NFICC871 | Lysinibacillus sphaericus | Pasteurized BSG |
| NFICC879 | Lysinibacillus boronitolerans | Potato |
| NFICC882 | Lysinibacillus fusiformis | Potato |
| NFICC889 | Lysinibacillus boronitolerans | Potato |
| NFICC906 | Bacillus simplex | Potato |
| NFICC1127 | Bacillus amyloliquefaciens | Pasteurized BSG * |
| NFICC1130 | Bacillus amyloliquefaciens | Pasteurized BSG |
| NFICC1525 | Bacillus subtilis | Herring Garum |
| NFICC1534 | Bacillus subtilis | Miso |
| NFICC1549 | Bacillus licheniformis | Apple pulp |
| Sample/time | MRS | F. sanfranciscensis UMCC 2990 | L. plantarum UMCC2 996 | P. pentosaceus UMCC 3010 | P. pentosaceus NFICC10 | L. plantarum NFICC19 | Lc. citreum NFICC28 | P. pentosaceus NFICC58 | L. plantarum NFICC72 | L. plantarum NFICC163 | L. plantarum NFICC207 | L. plantarum NFICC293 | P. pentosaceus NFICC341 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OXALATE | 14H | 0.31 | 0.37 | 0.36 | 0.38 | 0.39 | 0.95 * | 0.34 | 0.32 | 0.39 | 0.57 * | 0.39 | 0.30 | 0.34 |
| 18H | 0.37 | 0.33 | 0.34 | 0.40 * | 0.93 * | 0.39 * | 0.38 | 0.38 | 0.29 | 0.36 | 0.63 * | 0.39 * | ||
| 24H | 0.37 | 0.44 | 0.44 | 0.48 * | 1.38 * | 0.57 * | 0.41 | 0.70 * | 0.69 * | 0.32 | 0.55 * | 0.37 | ||
| CITRIC ACID | 14H | 1.86 | 0.00 * | 0.00 * | 0.00 * | 0.00 * | 1.77 | 1.04 | 1.18 | 0.00 * | 0.00 * | 0.00 * | 0.00 * | 0.38 * |
| 18H | 1.13 | 0.00 * | 1.06 | 1.12 | 1.05 | 1.05 | 1.86 | 0.92 * | 0.09 * | 1.04 | 0.00 * | 0.00 * | ||
| 24H | 0.99 | 0.90 * | 1.04 | 0.96 | 1.60 | 0.40 * | 2.15 | 0.00 * | 0.00 * | 0.81 * | 0.00 * | 1.10 | ||
| TARTARIC ACID | 14H | 0.00 | 1.12 * | 1.08 | 1.13 * | 1.12 * | 1.17 | 1.47 * | 1.00 | 1.10 * | 1.07 | 1.19 * | 0.97 | 1.08 |
| 18H | 1.12 * | 1.00 | 1.03 | 1.08 * | 0.81 | 1.32 * | 0.00 | 1.02 | 0.61 | 1.17 * | 0.00 | 0.00 | ||
| 24H | 0.00 | 0.00 | 0.00 | 0.00 | 1.22 * | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| GLUCOSE | 14H | 14.62 | 14.33 | 9.34 * | 12.25 | 11.56 | 6.32 * | 6.97 * | 8.00 * | 13.39 | 7.02 * | 13.62 | 6.04 * | 10.13 |
| 18H | 14.99 | 7.06 * | 11.19 * | 11.13 * | 0.95 * | 5.37 * | 5.67 * | 11.28 | 4.87 * | 11.74 | 5.23 * | 8.59 * | ||
| 24H | 14.02 | 11.86 | 9.04 * | 8.98 * | 0.98 * | 2.78 * | 4.03 * | 0.00 * | 0.00 * | 9.52 | 4.07 * | 7.61 * | ||
| SUCROSE | 14H | 1.49 | 1.79 | 0.76 | 1.33 | 1.57 | 0.00 * | 0.00 * | 0.00 * | 1.57 | 0.93 | 1.03 | 0.51 | 1.14 |
| 18H | 1.68 | 0.75 | 1.26 | 1.25 | 0.71 | 0.00 * | 0.00 * | 0.49 * | 0.51 * | 0.92 | 0.49 * | 0.95 | ||
| 24H | 1.72 | 1.33 | 1.22 | 1.34 | 0.70 * | 0.00 * | 0.00 | 0.87 * | 0.83 * | 1.06 | 1.11 | 1.08 | ||
| GLUTAMIC ACID | 14H | 0.23 | 0.08 * | 0.25 | 0.06 * | 0.07 * | 0.00 * | 0.23 | 0.15 | 0.08 * | 0.10 | 0.00 * | 0.17 | 0.18 |
| 18H | 0.13 | 0.13 | 0.11 * | 0.12 * | 0.21 | 0.10 * | 0.12 * | 0.13 | 0.11 * | 0.09 * | 0.13 | 0.18 | ||
| 24H | 0.09 | 0.00 * | 0.00 * | 0.00 * | 0.23 | 0.00 * | 0.00 * | 0.20 | 0.20 | 0.00 * | 0.00 * | 0.00 * | ||
| SUCCINIC | 14H | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.18 | 2.07 * | 0.00 | 0.00 |
| 18H | 0.75 * | 0.59 | 0.70 * | 0.74 * | 0.00 | 0.00 | 0.07 | 0.44 | 0.48 | 1.10 * | 0.44 | 0.00 | ||
| 24H | 0.00 | 0.00 | 0.00 | 0.00 | 1.22 * | 0.00 | 0.08 | 0.00 | 0.00 | 1.15 * | 0.00 | 0.00 | ||
| FORMIC ACID | 14H | 0.00 | 0.03 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.07 * | 0.00 | 0.00 | 0.09 * | 0.07 | 0.15 * |
| 18H | 3.76 * | 0.01 | 0.13 * | 0.15 * | 0.00 | 0.00 | 0.11 * | 0.03 | 0.00 | 0.03 | 0.00 | 0.08 | ||
| 24H | 1.44 * | 0.01 | 0.23 * | 0.18 * | 0.00 | 0.00 | 0.12 | 2.02 * | 0.00 | 0.01 | 0.00 | 0.14 | ||
| ACETIC ACID | 14H | 0.00 | 4.36 | 4.77 * | 4.73 * | 4.48 | 4.90 * | 5.44 * | 0.16 | 4.34 | 4.73 * | 5.14 * | 4.41 | 4.56 |
| 18H | 4.56 * | 4.45 * | 4.38 * | 4.50 * | 4.86 * | 5.52 * | 2.23 | 4.35 | 4.49 * | 4.89 * | 4.49 * | 4.47 * | ||
| 24H | 4.39 | 4.41 | 4.40 | 4.55 * | 4.79 * | 4.77 * | 4.29 | 4.60 * | 4.65 * | 4.84 * | 4.38 | 4.51 * | ||
| 1,3 PROPANDIOL | 14H | 0.00 | 0.71 | 0.85 * | 0.83 * | 0.82 * | 0.86 * | 0.92 * | 0.71 | 0.75 | 0.86 * | 0.89 * | 0.84 * | 0.81 |
| 18H | 0.79 | 0.78 | 0.81 | 0.82 | 0.89 * | 0.86 * | 0.81 | 0.53 | 0.85 * | 0.83 * | 0.88 * | 0.75 | ||
| 24H | 0.67 | 0.74 | 0.79 * | 0.80 * | 0.84 * | 0.67 | 0.84 * | 0.70 | 0.84 * | 0.83 * | 0.86 * | 0.82 * | ||
| 2,3 BUTANDIOL | 14H | 0.00 | 0.05 | 0.06 | 0.21 * | 0.20 * | 0.00 | 0.00 | 0.10 | 0.18 * | 0.00 | 0.29 * | 0.00 | 0.18 * |
| 18H | 0.18 * | 0.00 | 0.16 * | 0.18 * | 0.00 | 0.00 | 0.13 | 0.14 | 0.00 | 0.26 * | 0.00 | 0.06 | ||
| 24H | 0.00 | 0.06 | 0.19 * | 0.17 * | 0.00 | 0.00 | 0.15 | 0.00 | 0.00 | 0.26 * | 0.00 | 0.08 | ||
| LACTIC ACID | 14H | 0.00 | 0.59 | 12.05 * | 6.14 | 5.50 | 14.73 * | 5.47 | 4.97 | 2.88 | 14.25 * | 2.95 | 13.33 * | 7.98 * |
| 18H | 0.86 | 13.08 * | 5.59 | 6.34 | 19.78 * | 6.16 | 7.91 * | 6.56 | 16.14 * | 3.13 | 14.77 * | 9.18 * | ||
| 24H | 0.78 | 5.21 | 9.38 | 10.08 | 19.90 * | 16.91 * | 12.27 * | 20.32 * | 20.78 * | 4.69 | 16.11 * | 11.43 | ||
| ETOH | 14H | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.32 * | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| 18H | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.58 * | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | ||
| 24H | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.37 * | 0.00 | 0.00 | 0.00 | 1.74 * | 0.00 | 0.00 |
| LAB | D(+)-Glucose | D(−)-Fructose | D(+)-Cellobiose | Maltose | Lactose | D(+)-Melibiose | Saccharose | D(+)-Raffinose | Starch | D(+)-Xylose |
|---|---|---|---|---|---|---|---|---|---|---|
| F. sanfranciscensis UMCC 2990 | + | + | + | + | − | − | − | − | − | − |
| L. plantarum UMCC 2996 | + | + | + | + | + | + | + | + | − | − |
| F. rossiae UMCC 3002 | + | +/− | − | + | − | +/− | − | − | − | + |
| P. pentosaceus UMCC 3010 | + | + | + | + | − | − | − | − | − | − |
| Lc. citreum UMCC 3011 | + | +/− | − | + | − | − | − | − | − | + |
| P. pentosaceus NFICC10 | + | + | + | + | + | + | + | + | − | − |
| L. plantarum NFICC19 | + | + | + | + | + | + | + | + | − | − |
| L. plantarum NFICC27 | + | + | + | + | + | + | + | + | − | − |
| Lc. citreum NFICC28 | + | + | + | + | + | + | + | + | − | + |
| L. plantarum NFICC58 | + | + | + | + | + | + | + | + | − | + |
| L. plantarum NFICC72 | + | + | + | + | + | + | + | + | − | − |
| P. pentosaceus NFICC87 | + | + | + | + | − | − | + | − | − | + |
| Lc. citreum NFICC94 | + | + | + | + | − | − | + | − | − | + |
| P. pentosaceus NFICC103 | + | + | + | + | + | + | + | +/− | − | − |
| P. pentosaceus NFICC163 | + | + | + | + | + | + | + | + | − | +/− |
| L. plantarum NFICC207 | + | + | + | + | + | + | + | + | − | + |
| L. plantarum NFICC293 | + | + | + | + | + | + | + | + | − | − |
| P. pentosaceus NFICC341 | + | + | + | + | + | + | + | + | − | +/− |
| Tested Strains | γ-Hemolysis | α-Hemolysis | β-Hemolysis |
|---|---|---|---|
| F. sanfranciscensis UMCC 2990 | x | ||
| L. plantarum UMCC2996 | x | ||
| F. rossiae UMCC 3002 | x | ||
| P. pentosaceus UMCC 3010 | x | ||
| Lc. citreum UMCC 3011 | x | ||
| P. pentosaceus NFICC10 | x | ||
| L. plantarum NFICC19 | x | ||
| L. plantarum NFICC27 | x | ||
| Lc. citreum NFICC28 | x | ||
| L. plantarum NFICC58 | x | ||
| L. plantarum NFICC72 | x | ||
| P. pentosaceus NFICC87 | x | ||
| Lc. citreum NFICC94 | x | ||
| P. pentosaceus NFICC103 | x | ||
| P. pentosaceus NFICC163 | x | ||
| L. plantarum NFICC207 | x | ||
| L. plantarum NFICC293 | x | ||
| P. pentosaceus NFICC341 | x | ||
| S. aureus NFICC1477 | x | ||
| Lc. citreum NFICC88 | x | ||
| B. cereus DMS 2301 | x |
| Tested LAB | Tested Antibiotics | |||||
|---|---|---|---|---|---|---|
| Streptomycin | Ampicillin | Kanamycin | Vancomycin | Chloramphenicol | Gentamycin | |
| F. sanfranciscensis UMCC 2990 | ≤128 | ≤16 | ≤32 | 512 | ≤4 | ≤4 |
| L. plantarum UMCC 2996 | ≤128 | ≤32 | ≤64 | 512 | Resistant | ≤256 |
| F. rossiae UMCC 3002 | ≤128 | ≤4 | ≤32 | 512 | ≤4 | ≤4 |
| P. pentosaceus UMCC 3010 | ≤128 | ≤2 | ≤16 | 512 | ≤4 | ≤4 |
| Lc. citreum UMCC 3011 | ≤128 | ≤4 | ≤32 | 512 | ≤8 | ≤8 |
| P. pentosaceus NFICC10 | ≤128 | ≤1 | ≤16 | 512 | ≤4 | ≤4 |
| L. plantarum NFICC19 | ≤64 | ≤1 | ≤16 | 512 | ≤4 | ≤4 |
| L. plantarum NFICC27 | ≤64 | ≤1 | ≤16 | 512 | ≤4 | ≤4 |
| Lc. citreum NFICC28 | ≤128 | ≤1 | ≤32 | 512 | ≤4 | ≤4 |
| L. plantarum NFICC58 | ≤128 | ≤1 | ≤32 | 512 | ≤4 | ≤4 |
| L. plantarum NFICC72 | ≤64 | ≤1 | ≤16 | 512 | ≤4 | ≤4 |
| Lc. citreum NFICC87 | ≤128 | ≤1 | ≤32 | 512 | ≤4 | ≤4 |
| Lc. citreum NFICC94 | ≤128 | ≤1 | ≤32 | 512 | ≤4 | ≤8 |
| P. pentosaceus NFICC103 | ≤128 | ≤2 | ≤32 | 512 | ≤4 | ≤4 |
| P. pentosaceus NFICC163 | ≤64 | ≤1 | ≤16 | 512 | ≤4 | ≤4 |
| L. plantarum NFICC207 | ≤64 | ≤1 | ≤16 | 512 | ≤4 | ≤4 |
| L. plantarum NFICC293 | ≤128 | ≤1 | ≤8 | ≤8 | ≤4 | ≤4 |
| P. pentosaceus NFICC341 | ≤64 | ≤2 | ≤32 | 512 | ≤4 | ≤4 |
| Strain | Species | Bacteriocins Predicted by BAGEL4 |
|---|---|---|
| NFICC28 | Lc. citreum | None |
| NFICC10 | P. pentosaceus | Penocin A |
| NFICC58 | P. pentosaceus | None |
| NFICC19 | L. plantarum | Plantaricin E, Plantaricin F Plantaricin K, putative class IIc bacteriocin, putative class IIb bacteriocin |
| NFICC72 | L. plantarum | Plantaricin E, Plantaricin F Plantaricin K, putative class IIc bacteriocin, putative class IIb bacteriocin |
| NFICC163 | L. plantarum | Plantaricin E, Plantaricin F Plantaricin K, putative class IIc bacteriocin, putative class IIb bacteriocin |
| NFICC293 | L. plantarum | Plantaricin A, Plantaricin E, Plantaricin F, Plantaricin J, Plantaricin K, Plantaricin N |
| UMCC 2996 | L. plantarum | Plantaricin A, Plantaricin E, Plantaricin F, Plantaricin J, Plantaricin K, Plantaricin N |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iosca, G.; Fugaban, J.I.I.; Özmerih, S.; Wätjen, A.P.; Kaas, R.S.; Hà, Q.; Shetty, R.; Pulvirenti, A.; De Vero, L.; Bang-Berthelsen, C.H. Exploring the Inhibitory Activity of Selected Lactic Acid Bacteria against Bread Rope Spoilage Agents. Fermentation 2023, 9, 290. https://doi.org/10.3390/fermentation9030290
Iosca G, Fugaban JII, Özmerih S, Wätjen AP, Kaas RS, Hà Q, Shetty R, Pulvirenti A, De Vero L, Bang-Berthelsen CH. Exploring the Inhibitory Activity of Selected Lactic Acid Bacteria against Bread Rope Spoilage Agents. Fermentation. 2023; 9(3):290. https://doi.org/10.3390/fermentation9030290
Chicago/Turabian StyleIosca, Giovanna, Joanna Ivy Irorita Fugaban, Süleyman Özmerih, Anders Peter Wätjen, Rolf Sommer Kaas, Quốc Hà, Radhakrishna Shetty, Andrea Pulvirenti, Luciana De Vero, and Claus Heiner Bang-Berthelsen. 2023. "Exploring the Inhibitory Activity of Selected Lactic Acid Bacteria against Bread Rope Spoilage Agents" Fermentation 9, no. 3: 290. https://doi.org/10.3390/fermentation9030290
APA StyleIosca, G., Fugaban, J. I. I., Özmerih, S., Wätjen, A. P., Kaas, R. S., Hà, Q., Shetty, R., Pulvirenti, A., De Vero, L., & Bang-Berthelsen, C. H. (2023). Exploring the Inhibitory Activity of Selected Lactic Acid Bacteria against Bread Rope Spoilage Agents. Fermentation, 9(3), 290. https://doi.org/10.3390/fermentation9030290









