Probiotic Properties of Lactic Acid Bacteria Isolated from the Spontaneously Fermented Soybean Foods of the Eastern Himalayas
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Sample
2.2. Microbiological Analysis
2.3. Preliminary Screening of Probiotic Isolates
2.3.1. Acid Tolerance Test
2.3.2. Bile Salt Tolerance Test
2.3.3. Assessment of Cell Surface Hydrophobicity
2.4. Genotypic Identification
2.4.1. Genomic DNA Extraction
2.4.2. PCR Amplification
2.4.3. Purification of PCR
2.4.4. 16S rRNA Gene Sequencing
2.5. In Vitro Screening of Probiotic Properties
2.5.1. Survival to Acid and Bile Salt
2.5.2. Auto-Aggregation and Co-Aggregation Assays
2.5.3. Resistance to Lysozyme
2.5.4. Bile Salt Hydrolase (BSH) Activity
2.5.5. Antagonistic Activity
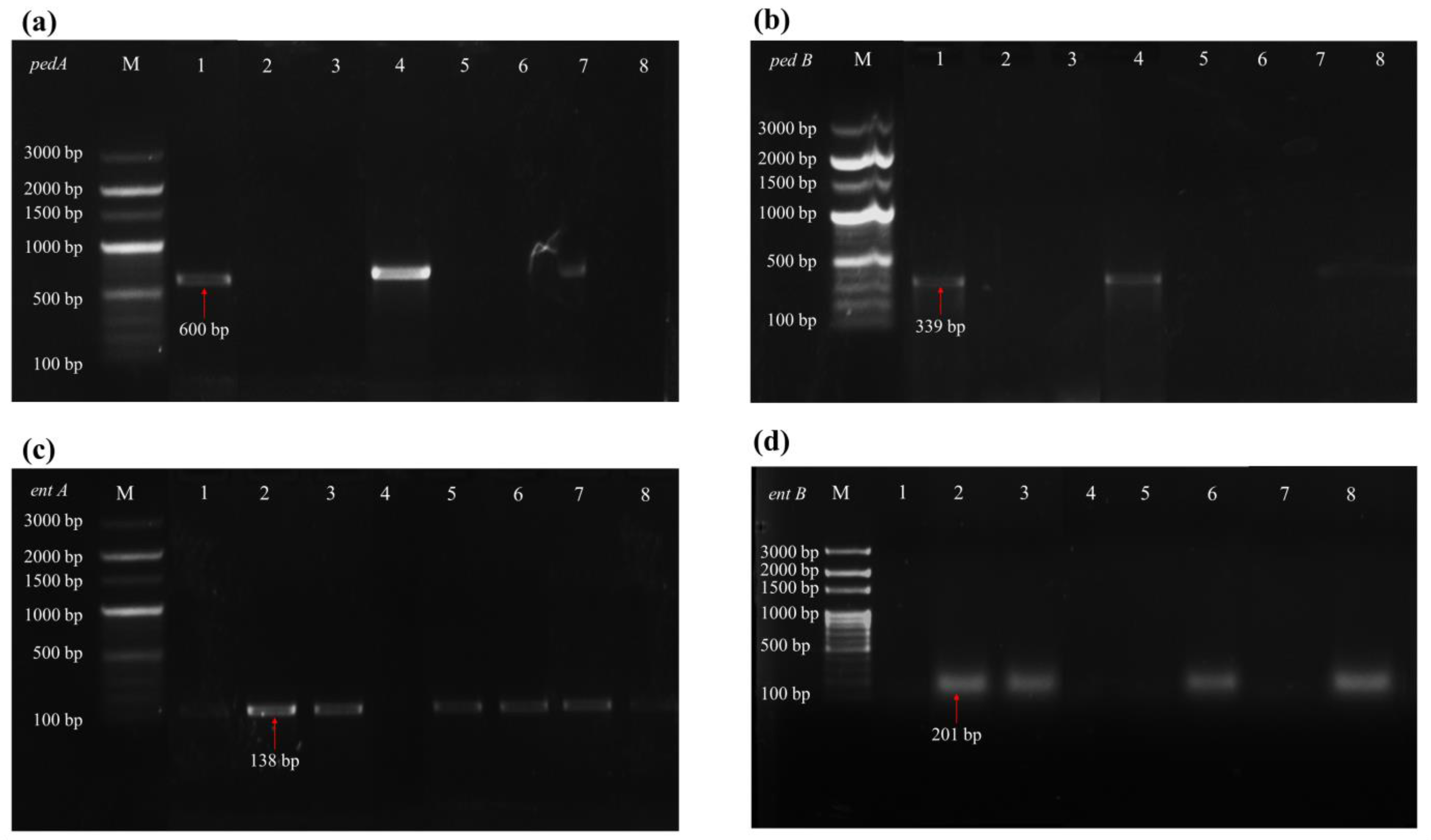
2.5.6. Genetic Screening for Probiotic Functions
2.6. Bioinformatics Analysis
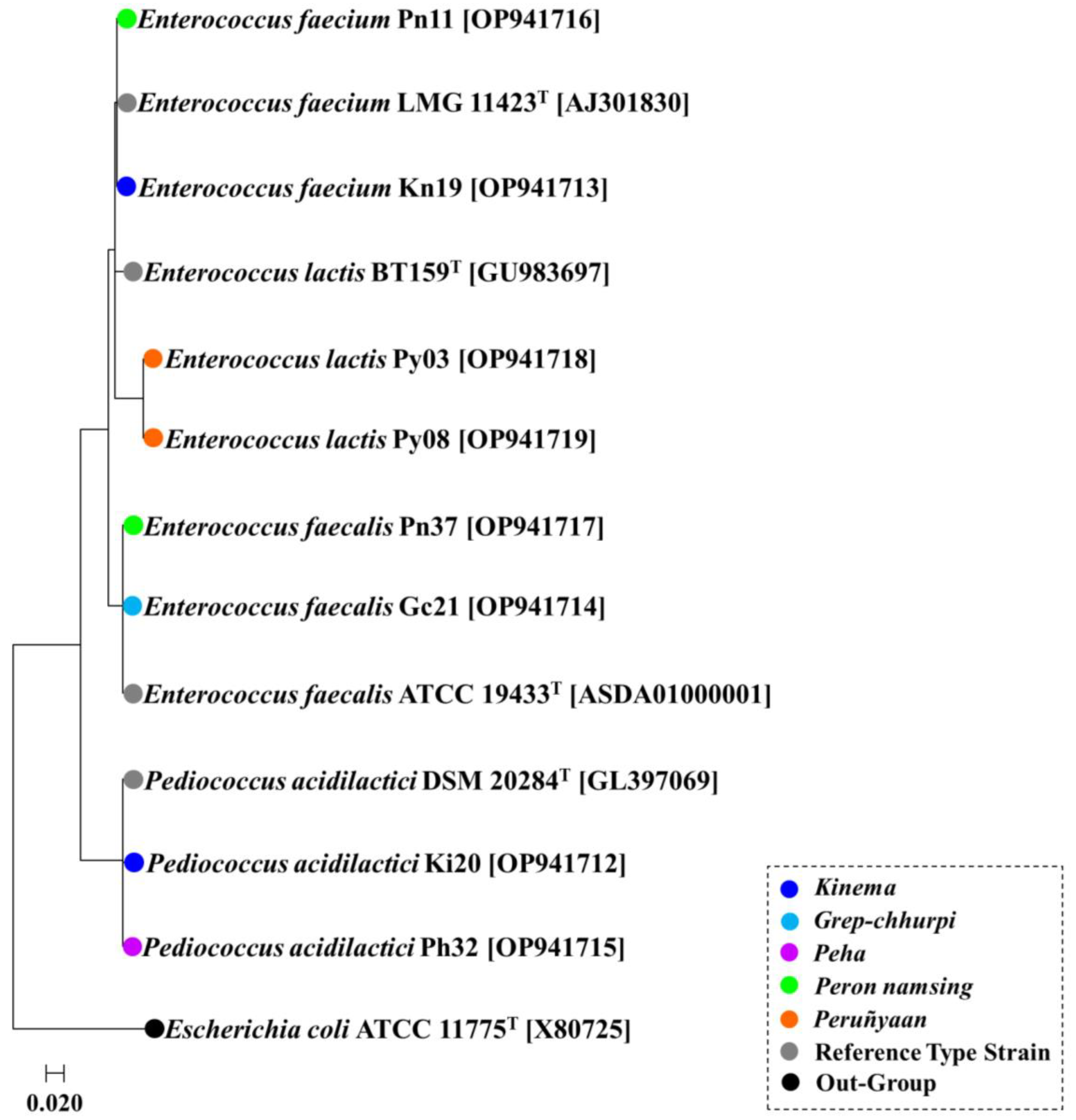
3. Results
3.1. Preliminary Screening
3.2. In-Vitro Probiotic Properties
3.3. Gene Detection of Probiotic Functions
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durazzo, A.; Carocho, M.; Heleno, S.A.; Pedrosa, M.C.; Ueda, J.M.; Barros, L.; Souto, E.B.; Santini, A.; Lucarini, M. Fermented food/beverage and health: Current perspectives. Rend. Lincei Sci. Fis. Nat. 2022, 33, 729–738. [Google Scholar] [CrossRef]
- Nazhand, A.; Souto, E.B.; Lucarini, M.; Souto, S.B.; Durazzo, A.; Santini, A. Ready to use therapeutical beverages: Focus on functional beverages containing probiotics, prebiotics and synbiotics. Beverages 2020, 6, 26. [Google Scholar] [CrossRef]
- Tamang, J.P. Dietary culture and antiquity of the Himalayan fermented foods and alcoholic fermented beverages. J. Ethn. Foods 2022, 9, 30. [Google Scholar] [CrossRef]
- Singh, T.A.; Devi, K.R.; Ahmed, G.; Jeyaram, K. Microbial and endogenous origin of fibrinolytic activity in traditional fermented foods of Northeast India. Food Res. Int. 2014, 55, 356–362. [Google Scholar] [CrossRef]
- Tamang, J.P. Naturally fermented ethnic soybean foods of India. J. Ethn. Foods 2015, 2, 8–17. [Google Scholar] [CrossRef]
- Tamang, J.P.; Jeyaram, K.; Rai, A.K.; Mukherjee, P.K. Diversity of beneficial microorganisms and their functionalities in community-specific ethnic fermented foods of the Eastern Himalayas. Food Res. Int. 2021, 148, 110633. [Google Scholar] [CrossRef]
- Kharnaior, P.; Das, M.; Tamang, J.P. Therapeutic and anti-thrombotic properties of some naturally fermented soybean foods of the Eastern Himalayas. Fermentation 2023, 9, 91. [Google Scholar] [CrossRef]
- Tamang, J.P. Native microorganisms in the fermentation of kinema. Indian J. Microbiol. 2003, 43, 127–130. [Google Scholar]
- Chettri, R.; Tamang, J.P. Bacillus species isolated from Tungrymbai and Bekang, naturally fermented soybean foods of India. Int. J. Food Microbiol. 2015, 197, 72–76. [Google Scholar] [CrossRef]
- Kharnaior, P.; Tamang, J.P. Bacterial and fungal communities and their predictive functional profiles in kinema, a naturally fermented soybean food of India, Nepal and Bhutan. Food Res. Int. 2021, 140, 110055. [Google Scholar] [CrossRef]
- Kharnaior, P.; Tamang, J.P. Metagenomic-Metabolomic Mining of Kinema, a naturally fermented soybean food of the Eastern Himalayas. Front. Microbiol. 2022, 13, 868383. [Google Scholar] [CrossRef] [PubMed]
- Pariyar, P.; Yaduvanshi, P.S.; Raghu, P.; Tamang, J.P. Screening of Poly-Glutamic Acid (PGA)-Producing Bacillus Species from Indian Fermented Soybean Foods and Characterization of PGA. Fermentation 2022, 8, 495. [Google Scholar] [CrossRef]
- Tamang, J.P.; Kharnaior, P.; Pariyar, P.; Thapa, N.; Lar, N.; Win, K.S.; Mar, A.; Nyo, N. Shotgun sequence-based metataxonomic and predictive functional profiles of Pe poke, a naturally fermented soybean food of Myanmar. PLoS ONE 2021, 16, e0260777. [Google Scholar] [CrossRef]
- Chettri, R.; Bhutia, M.; Tamang, J.P. Poly-γ-glutamic acid (PGA)-producing Bacillus species isolated from Kinema, Indian fermented soybean food. Front. Microbiol. 2016, 7, 971. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.K.; Sanjukta, S.; Chourasia, R.; Bhat, I.; Bhardwaj, P.K.; Sahoo, D. Production of bioactive hydrolysate using protease, β-glucosidase and α-amylase of Bacillus spp. isolated from kinema. Bioresour. Technol. 2017, 235, 358–365. [Google Scholar] [CrossRef]
- Chettri, R.; Tamang, J.P. Organoleptic evaluation of Tungrymbai and Bekang, naturally fermented soybean foods, produced by using selected species of Bacillus. J. Sci. Ind. Res. 2016, 75, 416–419. [Google Scholar]
- Sanjukta, S.; Rai, A.K.; Muhammed, A.; Jeyaram, K.; Talukdar, N.C. Enhancement of antioxidant properties of two soybean varieties of Sikkim Himalayan region by proteolytic Bacillus subtilis fermentation. J. Funct. Foods 2015, 14, 650–658. [Google Scholar] [CrossRef]
- Sarkar, P.K.; Tamang, J.P.; Cook, P.E.; Owens, J.D. Kinema—A traditional soybean fermented food: Proximate composition and microflora. Food Microbiol. 1994, 11, 47–55. [Google Scholar] [CrossRef]
- Kumar, J.; Sharma, N.; Kaushal, G.; Samurailatpam, S.; Sahoo, D.; Rai, A.K.; Singh, S.P. Metagenomic insights into the taxonomic and functional features of kinema, a traditional fermented soybean product of Sikkim Himalaya. Front. Microbiol. 2019, 10, 1744. [Google Scholar] [CrossRef]
- Goel, A.; Halami, P.M.; Tamang, J.P. Genome analysis of Lactobacillus plantarum isolated from some Indian fermented foods for bacteriocin production and probiotic marker genes. Front. Microbiol. 2020, 11, 40. [Google Scholar] [CrossRef]
- Lim, E.S. Antibacterial activity of lactic acid bacteria against biogenic amine-producing Bacillus spp. isolated from traditional fermented soybean paste. Korean J. Microbiol. 2018, 54, 398–409. [Google Scholar] [CrossRef]
- Ma, H.; Wang, L.; Yu, H.; Wang, W.; Wu, G.; Qin, G.; Tan, Z.W.Y.; Pang, H. Protease-producing lactic acid bacteria with antibacterial properties and their potential use in soybean meal fermentation. Chem. Biol. Technol. Agric. 2022, 9, 40. [Google Scholar] [CrossRef]
- Sirilun, S.; Sivamaruthi, B.S.; Kesika, P.; Peerajan, S.; Chaiyasut, C. Lactic acid bacteria mediated fermented soybean as a potent nutraceutical candidate. Asian Pac. J. Trop. Biomed. 2017, 7, 930–936. [Google Scholar] [CrossRef]
- Jang, C.H.; Oh, J.; Lim, J.S.; Kim, H.J.; Kim, J.S. Fermented soy products: Beneficial potential in neurodegenerative diseases. Foods 2021, 10, 636. [Google Scholar] [CrossRef]
- Jeong, J.K.; Chang, H.K.; Park, K.Y. Doenjang prepared with mixed starter cultures attenuates azoxymethane and dextran sulfate sodium-induced colitis-associated colon carcinogenesis in mice. J. Carcinog. 2014, 13, 9. [Google Scholar] [CrossRef]
- Fong, F.L.Y.; Lam, K.Y.; San Lau, C.; Ho, K.H.; Kan, Y.H.; Poon, M.Y.; El-Nezami, H.; Sze, E.T.P. Reduction in biogenic amines in douchi fermented by probiotic bacteria. PLoS ONE 2020, 15, e0230916. [Google Scholar] [CrossRef] [PubMed]
- Son, S.H.; Jeon, H.L.; Yang, S.J.; Sim, M.H.; Kim, Y.J.; Lee, N.K.; Paik, H.D. Probiotic lactic acid bacteria isolated from traditional Korean fermented foods based on β-glucosidase activity. Food Sci. Biotechnol. 2018, 27, 123–129. [Google Scholar] [CrossRef]
- Shangpliang, H.N.J.; Tamang, J.P. Phenotypic and genotypic characterisation of lactic acid bacteria isolated from exotic naturally fermented milk (cow and yak) products of Arunachal Pradesh, India. Int. Dairy J. 2021, 118, 105038. [Google Scholar] [CrossRef]
- Nithya, V.; Halami, P.M. Evaluation of probiotic characteristics of Bacillus species isolated from different food sources. Ann. Microbiol. 2013, 63, 129–137. [Google Scholar] [CrossRef]
- Bao, Y.; Zhang, Y.; Zhang, Y.; Liu, Y.; Wang, S.; Dong, X.; Wang, Y.; Zhang, H. Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 2010, 21, 695–701. [Google Scholar] [CrossRef]
- Nath, S.; Sikidar, J.; Roy, M.; Deb, B. In vitro screening of probiotic properties of Lactobacillus plantarum isolated from fermented milk product. Food Qual. Saf. 2020, 4, 213–223. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Mallappa, R.H.; Singh, D.K.; Rokana, N.; Pradhan, D.; Batish, V.K.; Grover, S. Screening and selection of probiotic Lactobacillus strains of Indian gut origin based on assessment of desired probiotic attributes combined with principal component and heatmap analysis. LWT Food Sci. Technol. 2019, 105, 272–281. [Google Scholar] [CrossRef]
- Li, M.; Wang, Y.; Cui, H.; Li, Y.; Sun, Y.; Qiu, H.J. Characterization of lactic acid bacteria isolated from the gastrointestinal tract of a wild boar as potential probiotics. Front. Vet. Sci. 2020, 7, 49. [Google Scholar] [CrossRef] [PubMed]
- Vera-Pingitore, E.; Jimenez, M.E.; Dallagnol, A.; Belfiore, C.; Fontana, C.; Fontana, P.; von Wright, A.; Vignolo, G.; Plumed-Ferrer, C. Screening and characterization of potential probiotic and starter bacteria for plant fermentations. LWT Food Sci. Technol. 2016, 71, 288–294. [Google Scholar] [CrossRef]
- Pradhan, P.; Tamang, J.P. Probiotic properties of lactic acid bacteria isolated from traditionally prepared dry starters of the Eastern Himalayas. World J. Microbiol. Biotechnol. 2021, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Rai, R.; Tamang, J.P. In vitro and genetic screening of probiotic properties of lactic acid bacteria isolated from naturally fermented cow-milk and yak-milk products of Sikkim, India. World J. Microbiol. Biotechnol. 2022, 38, 1–20. [Google Scholar] [CrossRef]
- Turpin, W.; Humblot, C.; Guyot, J.P. Genetic screening of functional properties of lactic acid bacteria in a fermented pearl millet slurry and in the metagenome of fermented starchy foods. Appl. Environ. Microbiol. 2011, 77, 8722–8734. [Google Scholar] [CrossRef]
- Ramiah, K.; Van Reenen, C.A.; Dicks, L.M.T. Expression of the mucus adhesion genes Mub and MapA, adhesion-like factor EF-Tu and bacteriocin gene plaA of Lactobacillus plantarum 423, monitored with real-time PCR. Int. J. Food Microbiol. 2007, 116, 405–409. [Google Scholar] [CrossRef]
- Archer, A.C.; Halami, P.M. Probiotic attributes of Lactobacillus fermentum isolated from human feces and dairy products. Appl. Environ. Microbiol. 2015, 99, 8113–8123. [Google Scholar] [CrossRef]
- Özdemir, G.B.; Oryaşın, E.; Bıyık, H.H.; Özteber, M.; Bozdoğan, B. Phenotypic and genotypic characterization of bacteriocins in enterococcal isolates of different sources. Indian J. Microbiol. 2011, 51, 182–187. [Google Scholar] [CrossRef]
- De Vuyst, L.; Moreno, M.F.; Revets, H. Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int. J. Food Microbiol. 2003, 84, 299–318. [Google Scholar] [CrossRef] [PubMed]
- Rodrıguez, J.M.; Cintas, L.M.; Casaus, P.; Martınez, M.I.; Suárez, A.; Hernández, P.E. Detection of pediocin PA-1-producing pediococci by rapid molecular biology techniques. Food Microbiol. 1997, 14, 363–371. [Google Scholar] [CrossRef]
- El-Arabi, N.I.; Salim, R.G.; Abosereh, N.A.; Abdelhadi, A.A. Molecular characterization of some antilisterial bacteriocin genes from Enterococcus faecium and Pediococcus pentosaceus. Microbiol. Biotechnol. Lett. 2018, 46, 288–299. [Google Scholar] [CrossRef]
- Creti, R.; Imperi, M.; Bertuccini, L.; Fabretti, F.; Orefici, G.; Di Rosa, R.; Baldassarri, L. Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 2004, 53, 13–20. [Google Scholar] [CrossRef]
- Ashelford, K.E.; Chuzhanova, N.A.; Fry, J.C.; Jones, A.J.; Weightman, A.J. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 2006, 72, 5734–5741. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Gopikrishna, T.; Suresh Kumar, H.K.; Perumal, K.; Elangovan, E. Impact of Bacillus in fermented soybean foods on human health. Ann. Microbiol. 2021, 71, 30. [Google Scholar] [CrossRef]
- Tamang, J.P.; Das, S.; Kharnaior, P.; Pariyar, P.; Thapa, N.; Jo, S.W.; Yim, E.J.; Shin, D.H. Shotgun metagenomics of Cheonggukjang, a fermented soybean food of Korea: Community structure, predictive functionalities and amino acids profile. Food Res. Int. 2022, 151, 110904. [Google Scholar] [CrossRef] [PubMed]
- Yongsawas, R.; In-on, A.; Inta, A.; Kampuansai, J.; Pandith, H.; Suwannarach, N.; Lumyong, S.; Chitov, T.; Disayathanoowat, T. Bacterial communities in Lanna fermented soybeans from three different ethnolinguistic groups in Northern Thailand. Microorganisms 2023, 11, 649. [Google Scholar] [CrossRef]
- do Prado, F.G.; Pagnoncelli, M.G.B.; de Melo Pereira, G.V.; Karp, S.G.; Soccol, C.R. Fermented soy products and their potential health benefits: A review. Microorganisms 2022, 10, 1606. [Google Scholar] [CrossRef]
- Liu, Y.; Han, Y.; Cao, L.; Wang, X.; Dou, S. Analysis of main components and prospects of natto. Adv. Enzym Res. 2021, 9, 1–9. [Google Scholar] [CrossRef]
- Elhalis, H.; Chin, X.H.; Chow, Y. Soybean fermentation: Microbial ecology and starter culture technology. Crit. Rev. Food Sci. Nutr. 2023, 14, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Yehuala, G.A.; Bang, W.Y.; Yang, J.; Jung, Y.H.; Park, M.K. Safety evaluation of Bacillus subtilis IDCC1101, newly isolated from cheonggukjang, for industrial applications. Microorganisms 2022, 10, 2494. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Li, M.; Liu, Y.; Zhang, X.; Zheng, Y. Microbial diversity and function of soybean paste in East Asia: What we know and what we don’t. Curr. Opin. Food Sci. 2021, 37, 145–152. [Google Scholar] [CrossRef]
- Urdaneta, V.; Casadesús, J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef]
- Han, S.; Lu, Y.; Xie, J.; Fei, Y.; Zheng, G.; Wang, Z.; Liu, J.; Lv, L.; Ling, Z.; Berglund, B.; et al. Probiotic gastrointestinal transit and colonization after oral administration: A long journey. Front. Cell. Infect. Microbiol. 2021, 11, 609722. [Google Scholar] [CrossRef]
- Garcia-Gonzalez, N.; Prete, R.; Battista, N.; Corsetti, A. Adhesion properties of food-associated lactobacillus plantarum strains on human intestinal epithelial cells and modulation of IL-8 release. Front. Microbiol. 2018, 9, 2392. [Google Scholar] [CrossRef]
- Petrova, P.; Tsvetanova, F.; Petrov, K. Low cell surface hydrophobicity is one of the key factors for high butanol tolerance of Lactic acid bacteria. Eng. Life Sci. 2019, 19, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Krawczyk, B.; Wityk, P.; Gałęcka, M.; Michalik, M. The many faces of Enterococcus spp.-commensal, probiotic and opportunistic pathogen. Microorganisms 2021, 9, 1900. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Huang, L.; Zeng, Y.; Li, W.; Zhou, D.; Xie, J.; Xie, J.; Tu, Q.; Deng, D.; Yin, J. Pediococcus pentosaceus: Screening and application as probiotics in food processing. Front. Microbiol. 2021, 12, 762467. [Google Scholar] [CrossRef]
- Chen, C.; Yu, L.; Tian, F.; Zhao, J.; Zhai, Q. Identification of novel bile salt-tolerant genes in Lactobacillus using comparative genomics and its application in the rapid screening of tolerant strains. Microorganisms 2022, 10, 2371. [Google Scholar] [CrossRef]
- Hernández-Gómez, J.G.; López-Bonilla, A.; Trejo-Tapia, G.; Ávila-Reyes, S.V.; Jiménez-Aparicio, A.R.; Hernández-Sánchez, H. In vitro bile salt hydrolase (BSH) activity screening of different probiotic microorganisms. Foods 2021, 10, 674. [Google Scholar] [CrossRef]
- Mladenović, K.G.; Grujović, M.Ž.; Nikodijević, D.D.; Čomić, L.R. The hydrophobicity of enterobacteria and their co-aggregation with Enterococcus faecalis isolated from Serbian cheese. Biosci. Microbiota Food Health 2020, 39, 227–233. [Google Scholar] [CrossRef]
- Holst, B.; Glenting, J.; Holmstrøm, K.; Israelsen, H.; Vrang, A.; Antonsson, M.; Ahrné, S.; Madsen, S.M. Molecular switch controlling expression of the mannose-specific adhesin, Msa, in Lactobacillus plantarum. Appl. Environ. Microbiol. 2019, 85, e02954-18. [Google Scholar] [CrossRef]
- Sharma, K.; Attri, S.; Goel, G. Selection and evaluation of probiotic and functional characteristics of autochthonous lactic acid bacteria isolated from fermented wheat flour dough babroo. Probiotics Antimicrob. 2019, 11, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Zawistowska-Rojek, A.; Kośmider, A.; Stępień, K.; Tyski, S. Adhesion and aggregation properties of Lactobacillaceae strains as protection ways against enteropathogenic bacteria. Arch. Microbiol. 2022, 204, 285. [Google Scholar] [CrossRef]
- Alp, D.; Kuleaşan, H. Adhesion mechanisms of lactic acid bacteria: Conventional and novel approaches for testing. World J. Microbiol. Biotechnol. 2019, 35, 1–9. [Google Scholar] [CrossRef]
- Samedi, L.; Charles, A.L. Isolation and characterization of potential probiotic Lactobacilli from leaves of food plants for possible additives in pellet feeding. Ann. Agric. Sci. 2019, 64, 55–62. [Google Scholar] [CrossRef]
- Silva, D.R.; Sardi, J.D.C.O.; de Souza Pitangui, N.; Roque, S.M.; da Silva, A.C.B.; Rosalen, P.L. Probiotics as an alternative antimicrobial therapy: Current reality and future directions. J. Funct. Foods 2020, 73, 104080. [Google Scholar] [CrossRef]
- Abanoz, H.S.; Kunduhoglu, B. Antimicrobial activity of a bacteriocin produced by Enterococcus faecalis KT11 against some pathogens and antibiotic-resistant bacteria. Korean J. Food Sci. Anim. Resour. 2018, 38, 1064. [Google Scholar] [CrossRef]
- Sharma, B.R.; Halami, P.M.; Tamang, J.P. Novel pathways in bacteriocin synthesis by lactic acid bacteria with special reference to ethnic fermented foods. Food Sci. Biotechnol. 2021, 31, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.S.; Lee, J.Y.; Singh, B.; Maharjan, S.; Hong, L.; Lee, S.M.; Cui, L.H.; Lee, K.J.; Kim, G.; Yun, C.H.; et al. A new way of producing pediocin in Pediococcus acidilactici through intracellular stimulation by internalized inulin nanoparticles. Sci. Rep. 2018, 8, 5878. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, L.; Zhao, X.; Zhou, Z. Partial characteristics and antimicrobial mode of pediocin produced by Pediococcus acidilactici PA003. Ann. Microbiol. 2015, 65, 1753–1762. [Google Scholar] [CrossRef]
- Hu, C.B.; Malaphan, W.; Zendo, T.; Nakayama, J.; Sonomoto, K. Enterocin X, a novel two-peptide bacteriocin from Enterococcus faecium KU-B5, has an antibacterial spectrum entirely different from those of its component peptides. Appl. Environ. Microbiol. 2010, 76, 4542–4545. [Google Scholar] [CrossRef]
- Wu, Y.; Pang, X.; Wu, Y.; Liu, X.; Zhang, X. Enterocins: Classification, synthesis, antibacterial mechanisms and food applications. Molecules 2022, 27, 2258. [Google Scholar] [CrossRef]

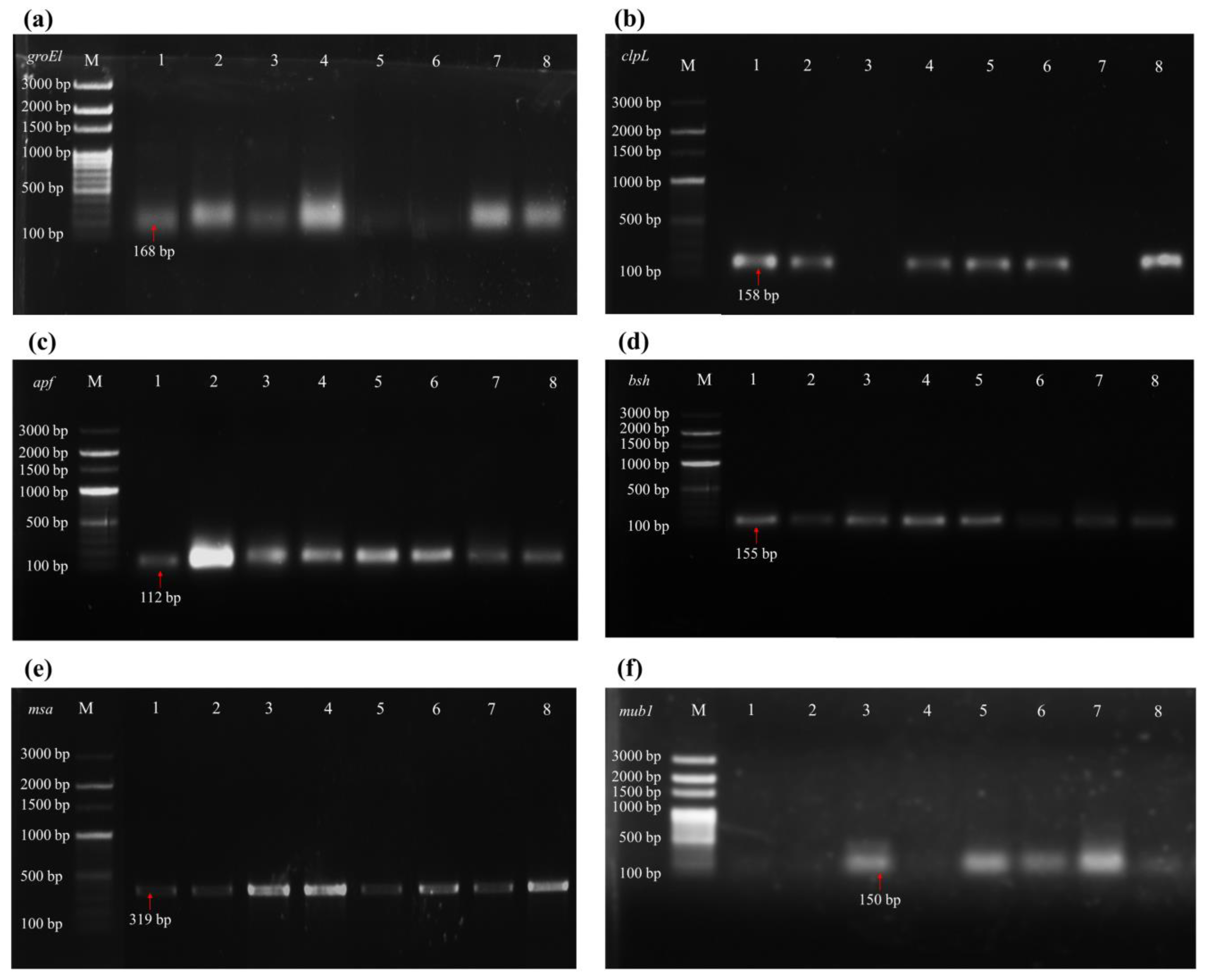
| Genes | Functions | Primer Sequence (5′ → 3′) (F = Forward; R = Reverse) | Annealing Temperature (°C) | Size of Amplicon (bp) | References |
|---|---|---|---|---|---|
| groEl | Survival at low pH | F-TTCCATGGCKTCAGCRATCA R-GCTAAYCCWGTTGGCATTCG | 58 | 168 | [38] |
| clpL | Survival at low pH | F-GCTGCCTTYAAAACATCATCTGG R-AATACAATTTTGAARAACGCAGCTT | 50 | 158 | [38] |
| odc | Survival at low pH | F-TMTWCCAACHGATCGWAATGC R-CRCCCCAWGCACARTCRAA | 52 | 245 | [38] |
| tdc | Survival at low pH | F-CCACTGCTGCATCTGTTTG R-CCRTARTCNGGNATAGCRAARTCNGTRTG | 50 | 370 | [38] |
| Ir0085 | Bile salt | F-RCTTTGACCGRTGGGGCTRT R-NNNATGGCCGCATGGAAA | 57.5 | 150 | [38] |
| Ir1516 | Bile salt | F-TRACCACTYTCWCCATTCAACAA R-CCACTAGCRATGACYAATACKGGT | 56.5 | 143 | [38] |
| apf | Bile salt | F-YAGCAACACGTTCTTGGTTAGCA R-GAATCTGGTGGTTCATAYWCAGC | 53 | 112 | [38] |
| bsh | Bile salt | F-ATTGAAGGCGGAACSGGMTA R-ATWACCGGWCGGAAAGCTG | 58 | 155 | [38] |
| mub1 | Adhesion | F-GTAGTTACTCAGTGACGATCAATG R-TAATTGTAAAGGTATAATCGGAGG | 50 | 150 | [39] |
| msa | Adhesion | F-GCGATTAGGGGTGTGCAAG R-GCAGTTGGTGACGTAGGCA | 55 | 319 | [40] |
| fbp | Adhesion | F-AGTGCTGAAATYATGGGAAGA R-AATTGTCCACCTTGTTGCTG | 60 | 835 | [40] |
| entA | Bacteriocin | F-GGT ACC ACT CAT AGT GGA AA R-CCC TGG AAT TGC TCC ACC TAA | 55 | 138 | [41] |
| entB | Bacteriocin | F-CAA AAT GTA AAA GAA TTA AGT ACG R-AGA GTA TAC ATT TGC TAA CCC | 56 | 201 | [42] |
| pedA | Bacteriocin | F-AAAATATCTAACTAATACTTG R-TAAAAAGATATTTGACCAAAA | 44 | 600 | [43] |
| pedB | Bacteriocin | F-ATGAATAAGACTAAGTCGGAACATATT R-CTATTGGCTAGGCCACGTATTG | 57 | 339 | [44] |
| cylA | Bacteriocin | F-ACTCGGGGATTGATAGGC R-GCTGCTAAAGCTGCGCTT | 54 | 688 | [45] |
| Products | Identity with Sample Code | Type Species (% Similarity) | GenBank Accession Number |
|---|---|---|---|
| Kinema | Pediococcus acidilactici Ki20 | Pediococcus acidilactici DSM 20284 (99.72%) | OP941712 |
| Enterococcus faecium Kn19 | Enterococcus faecium LMG 11423 (99.65%) | OP941713 | |
| Grep chhurpi | Enterococcus faecalis Gc21 | Enterococcus faecalis ATCC 19433 (99.85%) | OP941714 |
| Peha | Pediococcus acidilactici Ph32 | Pediococcus acidilactici DSM 20284 (99.86%) | OP941715 |
| Peron namsing | Enterococcus faecium Pn11 | Enterococcus faecium LMG 11423 (99.52%) | OP941716 |
| Enterococcus faecalis Pn37 | Enterococcus faecalis ATCC 19,433 (99.93%) | OP941717 | |
| Peruñyaan | Enterococcus lactis Py03 | Enterococcus lactis BT159 (99.64%) | OP941718 |
| Enterococcus lactis Py08 | Enterococcus lactis BT159 (99.71%) | OP941719 |
| Samples | Lactic Acid Bacteria | Survival Rate (%) | Cell Surface Hydrophobicity (%) | Auto-Aggregation (%) | Co-Aggregation (%) | Resistance to Lysozyme (%) | BSH Activity | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acid (pH 3) | Bile (0.3% Oxgall) | Bacillus cereus MTCC 1272 | Escherichia coli MTCC 1583 | Salmonella enterica subsp. enterica ser. typhimurium MTCC 3223 | Staphylococcus aureus subsp. aureus MTCC 740 | Sodium Taurodeoxycholate | Sodium Taurocholate | |||||
| Kinema | Pediococcus acidilactici Ki20 | 67.85 ± 0.25 | 78.03 ± 0.21 | 86.67 ± 3.30 | 55.73 ± 0.96 | 79.51 ± 0.35 | 72.20 ± 0.72 | 75.52 ± 0.34 | 75.14 ± 0.52 | 62.38 ± 0.59 | + | + |
| Enterococcus faecium Kn19 | 73.67 ± 1.05 | 74.08 ± 0.38 | 87.84 ± 3.06 | 50.03 ± 1.14 | 65.02 ± 1.04 | 58.48 ± 1.77 | 58.52 ± 0.87 | 58.97 ± 1.13 | 70.80 ± 0.46 | + | + | |
| Grep-chhurpi | Enterococcus faecalis Gc21 | 52.02 ± 1.05 | 66.46 ± 0.88 | 90.50 ± 10.14 | 54.15 ± 0.29 | 59.72 ± 0.93 | 56.31 ± 0.45 | 60.99 ± 0.78 | 59.06 ± 0.25 | 75.09 ± 0.77 | − | − |
| Peha | Pediococcus acidilactici Ph32 | 66.11 ± 2.32 | 79.71 ± 0.13 | 88.97 ± 6.55 | 43.80 ± 0.40 | 64.52 ± 0.25 | 58.58 ± 1.11 | 60.97 ± 0.64 | 60.82 ± 0.41 | 77.76 ± 0.25 | + | + |
| Peron namsing | Enterococcus faecium Pn11 | 55.92 ± 1.85 | 75.32 ± 0.95 | 89.34 ± 6.60 | 48.13 ± 0.33 | 59.13 ± 0.40 | 59.69 ± 0.39 | 66.41 ± 0.77 | 55.93 ± 0.79 | 66.08 ± 0.30 | − | + |
| Enterococcus faecalis Pn37 | 61.43 ± 0.65 | 75.54 ± 0.55 | 85.67 ± 1.89 | 55.05 ± 0.36 | 63.66 ± 0.25 | 56.90 ± 0.42 | 59.58 ± 0.42 | 58.10 ± 1.03 | 67.78 ± 1.05 | − | − | |
| Peruñyaan | Enterococcus lactis Py03 | 53.24 ± 1.17 | 66.41 ± 0.85 | 89.00 ± 11.31 | 55.46 ± 0.48 | 64.01 ± 0.92 | 51.29 ± 0.75 | 60.23 ± 0.50 | 58.54 ± 0.24 | 64.48 ± 1.75 | + | − |
| Enterococcus lactis Py08 | 64.92 ± 1.60 | 71.88 ± 0.14 | 90.34 ± 8.96 | 54.10 ± 0.38 | 58.14 ± 0.27 | 52.69 ± 0.70 | 53.98 ± 0.26 | 54.41 ± 0.52 | 72.53 ± 1.39 | + | − | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kharnaior, P.; Tamang, J.P. Probiotic Properties of Lactic Acid Bacteria Isolated from the Spontaneously Fermented Soybean Foods of the Eastern Himalayas. Fermentation 2023, 9, 461. https://doi.org/10.3390/fermentation9050461
Kharnaior P, Tamang JP. Probiotic Properties of Lactic Acid Bacteria Isolated from the Spontaneously Fermented Soybean Foods of the Eastern Himalayas. Fermentation. 2023; 9(5):461. https://doi.org/10.3390/fermentation9050461
Chicago/Turabian StyleKharnaior, Pynhunlang, and Jyoti Prakash Tamang. 2023. "Probiotic Properties of Lactic Acid Bacteria Isolated from the Spontaneously Fermented Soybean Foods of the Eastern Himalayas" Fermentation 9, no. 5: 461. https://doi.org/10.3390/fermentation9050461
APA StyleKharnaior, P., & Tamang, J. P. (2023). Probiotic Properties of Lactic Acid Bacteria Isolated from the Spontaneously Fermented Soybean Foods of the Eastern Himalayas. Fermentation, 9(5), 461. https://doi.org/10.3390/fermentation9050461







