Nature’s Antimicrobial Arsenal: Non-Ribosomal Peptides from PGPB for Plant Pathogen Biocontrol
Abstract
:1. Introduction
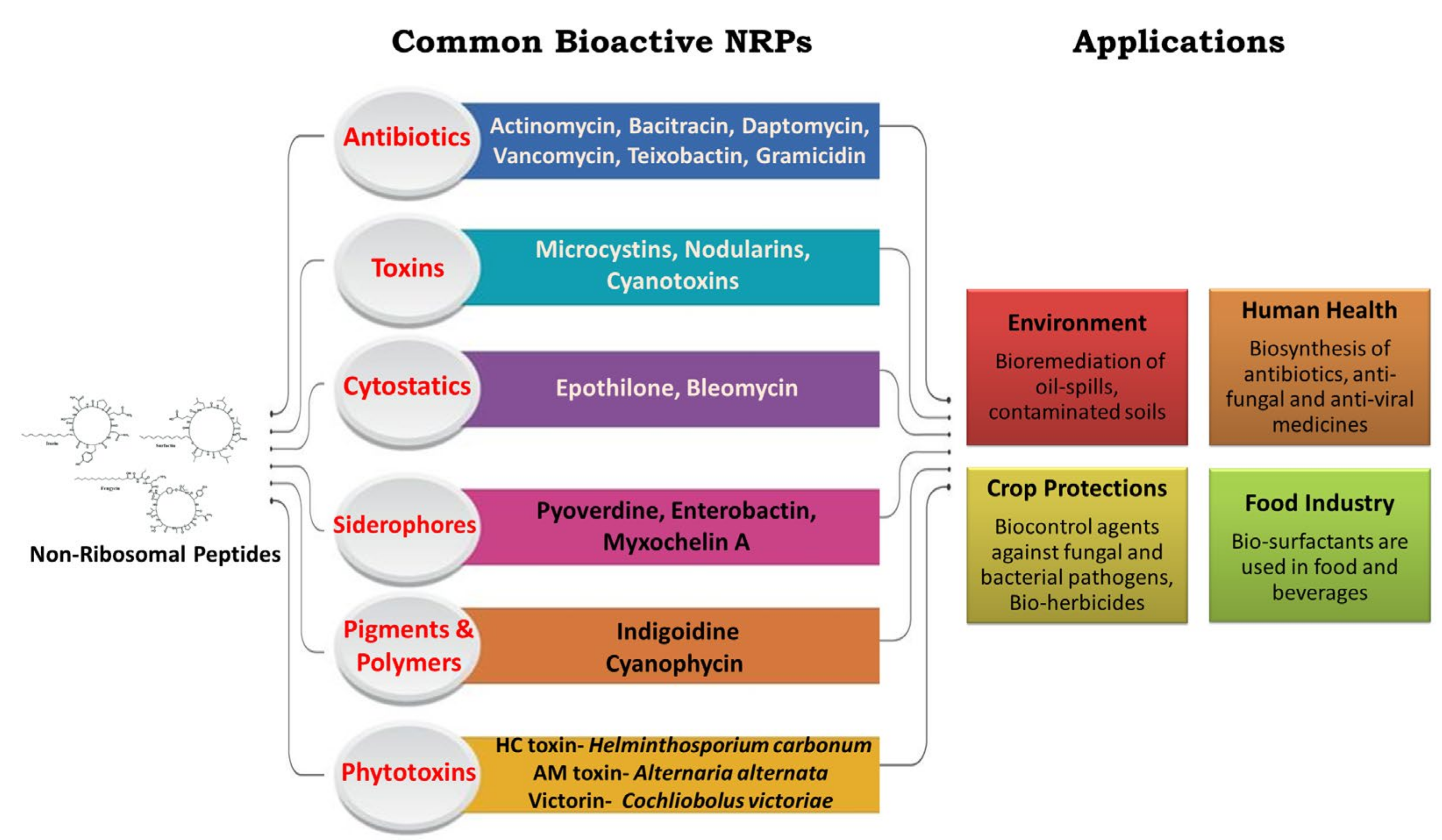
2. Potential Non-Ribosomal Peptides for Biocontrol
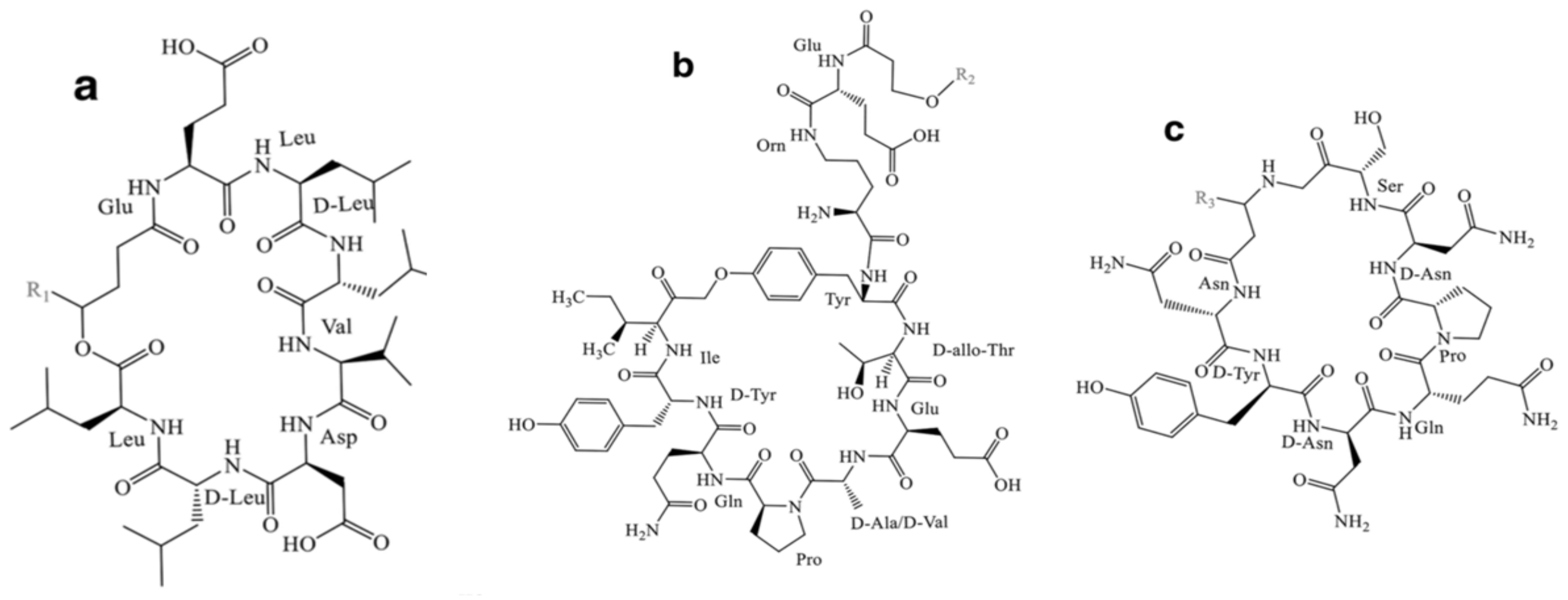
- Iturin
- b.
- Surfactin
- c.
- Fengycin
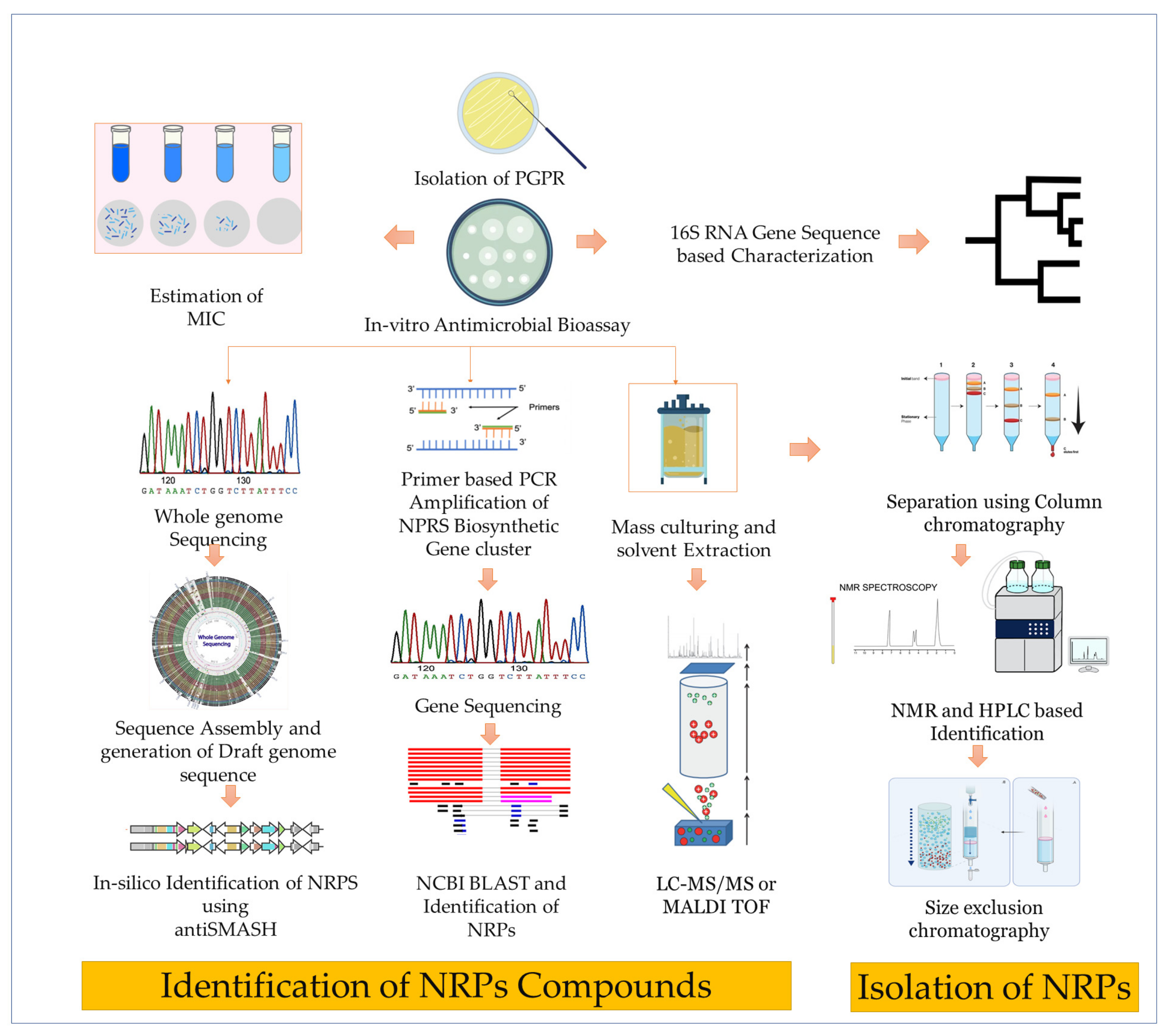
3. Synthesis, Mode of Action, and Characterization
- Synthesis of NRPs
- b.
- Mode of action
- c.
- Characterization
- Mass spectrometry (MS) is a powerful technique for the analysis of NRPs [105]. MS allows for the determination of the mass-to-charge ratio (m/z) of the NRP and its fragments [106]. This information helps identify the exact chemical structure of the NRP, as well as to determine its molecular weight and the presence of various functional groups [107]. Several types of MS techniques can be used for the analysis of NRPs, including matrix-assisted laser desorption/ionization (MALDI-TOF) MS [108], electrospray ionization (ESI) MS [109], and tandem mass spectrometry (MS/MS) [110].
- The use of nuclear magnetic resonance (NMR) spectroscopy has also been reported in NMR characterization; however, it can be tedious, error-prone, and require substantial quantities of purified material [114]. This can be especially problematic since NRPs are often produced by microorganisms that are difficult to cultivate, making it challenging to obtain enough material for NMR-based sequencing [115]. As a solution, a new nanomolar scale approach to NRP sequencing is needed. A recent study has claimed to identify three compounds, keanumycins A-C, from Pseudomonas sp. QS1027 using NMR that is found to have significant potential against phytopathogen Botrytis cinerea [116].
- Genome mining is a relatively new approach to the identification of NRPs. It involves the analysis of the genome sequence of microorganisms to identify BGCs for NRPSs and other biosynthetic enzymes [120]. It can also be used to predict the chemical structures of NRPs based on the presence of specific biosynthetic genes. There are several computational tools available for genome mining, such as antiSMASH and NRPSpredictor [121]. These tools can be used to predict the chemical structures of NRPs, as well as to identify potential new NRPs.
4. Application of NRPs in Sustainable Agriculture
4.1. Impact on Bacterial Pathogens
4.2. Impact on Fungal Pathogens
4.3. Biocontrol in Soilless Agriculture
| S. No | Type of PGPB | Media | Crops | References |
|---|---|---|---|---|
| 1. | Bacillus sp, Halobacillus sp., B.gibsonii, Staphylococcus succinus, Zhihengliuella halotolerans, Oceanobacillus oncorhynchi, Exiguobacterium aurantiacum, B.atrophaeus, Zhihengliuella sp., Halomonas sp., Virgibacillus picturae, Oceanobacillus sp., and Thalassobacillus sp | Hydroponic | Triticum aestivum | [174] |
| 2. | Bacillus amyloliquefaciens, B. brevis, B. circulans, B. coagulans, B. firmus, B. halodenitrificans, B. laterosporus, B. licheniformis, B. megaterium, B. mycoides, B. pasteurii, B. subtilis, and Paenibacillus polymyxa | Hydroponic floating system | Lactuca sativa L. var. Crispa | [175] |
| 3. | Enterobacter hormoechei | Hydroponics | Cucumis sativus | [176] |
| 4. | Acinetobacter calcoaceticus | Hydroponics | Lactuca sativa | [177] |
| 5. | BFD160 Enterobacter asburiae, TFD26 P. koreensis, and BFS112 P. linii | Soilless culture comprising rock wool blocks placed in plastic pots containing perlite and peat (1:1) | Cucumis melo L | [178] |
| 6. | Pseudarthrobactr chlorophenolicus BF2P4-5 | Cocopeat media | S. lycopersicum | [179] |
| 7. | Flavobacterium crocinum HYN0056T | Soilless culture comprising cocopeat (51.5%), peat moss (10%), Vermiculite (13%), perlite (15%), zeolite (10%), humic acid (0.1%), fertilizer (0.4%) | Arabidopsis thaliana | [180] |
| 8. | Pantoea dispersa, Pantoea ananatis, Burkholderia arboris, Burkholderia pyrrocinia, and Burkholderia pyrrocinia | Cocopeat substrate | Solanum melongena | [181] |
| 9. | B. megaterium TV-91C, Pantoea agglomerans RK-92, and B. subtilis TV17C | Peat | Brassica oleracea var. capitata ‘Yalova1’ | [182] |
| 10. | B. amyloliquefaciens, B. subtilis, B. pumilus, and B. sphaericus | Peat moss | Cucurbita pepo and S. lycopersicum | [183] |
| 11. | Rhizobacterium, B. subtilis | Perlite | Lactuca sativa ‘Partavousi’ | [70] |
| 12. | Bacillus sp. | Hydroponics | Zea Mays | [184] |
| 13. | P. pseudoalcaligenes and Bacillus subtilis | Hydroponics | Glycine max L. | [185] |
| 14. | Sinorhizobium meliloti and P. fluorescence | Soilless media containing sand and sterile perlite (v/v, 2:1) | Medicago sativa L. | [186] |
4.4. Future Prospective
4.5. Commercial Aspects of PGPB Formulations for Biocontrol Strategies
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
| ADP | Adenosine-Diphosphate |
| BGC | Biosynthetic gene cluster |
| CID | Collision-induced dissociation |
| CLP | Cyclic lipopeptide |
| DNA | Deoxyribose Nucleic Acid |
| EPA | Environmental Protection Agency |
| ESI | Electron spray ionization |
| ETD | Electron transfer dissociation |
| FAO | Food and Agriculture Organization |
| FDA | Food and Drug Administration |
| HCN | Hydrogen cyanide |
| HPLC | High-performance liquid chromatography |
| LC | Liquid Chromatography |
| MALDI TOF | Matrix-assisted laser desorption/ionization-Time of Flight |
| MAPK | Mitogen-Activated Protein Kinase |
| MIC | Minimum Inhibitory Concentration |
| MMP | Mitochondrial membrane potential |
| MPA | Methylphenyl acetate |
| MS | Mass Spectroscopy |
| NMR | Nuclear Magnetic Resonance |
| NRPs | Non-Ribosomal Peptides |
| NRPS | Non-Ribosomal Peptide synthetase |
| PAA | Phenylacetic acid |
| PARP | Poly (ADP-ribose) polymerase |
| PGPB | Plant-Growth-Promoting Bacteria |
| PKS | Polyketides |
| RNA | Ribose Nucleic Acid |
| ROS | Reactive Oxygen Species |
| TLC | Thin-Layer chromatography |
| UPLC | Ultra-high-performance liquid chromatography |
| VOC | Volatile organic compounds |
References
- WFP; WHO; UNICEF. The State of Food Security and Nutrition in the World 2022. 2022. Available online: https://policycommons.net/artifacts/2483950/the-state-of-food-security-and-nutrition-in-the-world-2022/3506270/ (accessed on 28 May 2023).
- Chavas, J.; Rivieccio, G.; Di Falco, S.; De Luca, G.; Capitanio, F. Agricultural Diversification, Productivity, and Food Security across Time and Space. Agric. Econ. 2022, 53, 41–58. [Google Scholar] [CrossRef]
- Pixley, K.V.; Falck-Zepeda, J.B.; Paarlberg, R.L.; Phillips, P.W.B.; Slamet-Loedin, I.H.; Dhugga, K.S.; Campos, H.; Gutterson, N. Genome-Edited Crops for Improved Food Security of Smallholder Farmers. Nat. Genet. 2022, 54, 364–367. [Google Scholar] [CrossRef]
- FAO 2021. Available online: https://www.fao.org/news/story/en/item/1402920/icode/ (accessed on 21 June 2023).
- Bernardo-Cravo, A.P.; Schmeller, D.S.; Chatzinotas, A.; Vredenburg, V.T.; Loyau, A. Environmental Factors and Host Microbiomes Shape Host–Pathogen Dynamics. Trends Parasitol. 2020, 36, 616–633. [Google Scholar] [CrossRef]
- Von Wintersdorff, C.J.H.; Penders, J.; Van Niekerk, J.M.; Mills, N.D.; Majumder, S.; Van Alphen, L.B.; Savelkoul, P.H.M.; Wolffs, P.F.G. Dissemination of Antimicrobial Resistance in Microbial Ecosystems through Horizontal Gene Transfer. Front. Microbiol. 2016, 7, 173. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Beltrán, J.; DelaFuente, J.; Leon-Sampedro, R.; MacLean, R.C.; San Millan, A. Beyond Horizontal Gene Transfer: The Role of Plasmids in Bacterial Evolution. Nat. Rev. Microbiol. 2021, 19, 347–359. [Google Scholar] [CrossRef]
- Zeilinger, S.; Gupta, V.K.; Dahms, T.E.S.; Silva, R.N.; Singh, H.B.; Upadhyay, R.S.; Gomes, E.V.; Tsui, C.K.M.; Chandra Nayak, S. Friends or Foes? Emerging Insights from Fungal Interactions with Plants. FEMS Microbiol. Rev. 2016, 40, 182–207. [Google Scholar] [CrossRef] [Green Version]
- Brundrett, M.C. Mycorrhizal Associations and Other Means of Nutrition of Vascular Plants: Understanding the Global Diversity of Host Plants by Resolving Conflicting Information and Developing Reliable Means of Diagnosis. Plant. Soil. 2009, 320, 37–77. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Zobel, M. How Mycorrhizal Associations Drive Plant Population and Community Biology. Science 2020, 367, eaba1223. [Google Scholar] [CrossRef]
- Ranjan, A.; Chauhan, A.; Rajput, V.D.; Basniwal, R.K.; Minkina, T.; Sushkova, S.; Jindal, T. Genetic Basis of Fungal Endophytic Bioactive Compounds Synthesis, Modulation, and Their Biotechnological Application. In Bacterial Endophytes for Sustainable Agriculture and Environmental Management; Springer: Berlin/Heidelberg, Germany, 2022; pp. 157–186. [Google Scholar]
- Doehlemann, G.; Ökmen, B.; Zhu, W.; Sharon, A. Plant Pathogenic Fungi. Microbiol. Spectr. 2017, 5, 1–23. [Google Scholar] [CrossRef]
- Lockwood, J.L. Evolution of Concepts Associated with Soilborne Plant Pathogens. Annu. Rev. Phytopathol. 1988, 26, 93–121. [Google Scholar] [CrossRef]
- Ranjan, A.; Jindal, T.; Ranjan, A.; Jindal, T. Overview of Organophosphate Compounds. Toxicol. Organophosphate Poisoning New Insights 2022, 1, 1–25. [Google Scholar]
- Ranjan, A.; Jindal, T. Toxicology of Organophosphate Poisoning in Human. In Toxicology of Organophosphate Poisoning; Springer: Berlin/Heidelberg, German, 2022; pp. 27–43. [Google Scholar]
- Curl, C.L.; Spivak, M.; Phinney, R.; Montrose, L. Synthetic Pesticides and Health in Vulnerable Populations: Agricultural Workers. Curr. Environ. Health Rep. 2020, 7, 13–29. [Google Scholar] [CrossRef]
- John, C.J.; Kumar, S.; Ge, M. Probiotic Prospects of PGPR for Green and Sustainable Agriculture. Arch. Phytopathol. Plant Prot. 2020, 53, 899–914. [Google Scholar] [CrossRef]
- Dimkić, I.; Janakiev, T.; Petrović, M.; Degrassi, G.; Fira, D. Plant-Associated Bacillus and Pseudomonas Antimicrobial Activities in Plant Disease Suppression via Biological Control Mechanisms—A Review. Physiol. Mol. Plant. Pathol. 2022, 117, 101754. [Google Scholar] [CrossRef]
- Migunova, V.D.; Sasanelli, N. Bacteria as Biocontrol Tool against Phytoparasitic Nematodes. Plants 2021, 10, 389. [Google Scholar] [CrossRef]
- Wang, H.; Liu, R.; You, M.P.; Barbetti, M.J.; Chen, Y. Pathogen Biocontrol Using Plant Growth-Promoting Bacteria (PGPR): Role of Bacterial Diversity. Microorganisms 2021, 9, 1988. [Google Scholar] [CrossRef]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability—A Review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef] [Green Version]
- Wani, S.P.; Gopalakrishnan, S. Plant Growth-Promoting Microbes for Sustainable Agriculture. In Plant Growth Promoting Rhizobacteria (PGPR): Prospects for Sustainable Agriculture; Springer: Berlin/Heidelberg, Germany, 2019; pp. 19–45. [Google Scholar]
- Dimkic, I.; Stankovic, S.; Nišavic, M.; Petkovic, M.; Ristivojevic, P.; Fira, D.; Beric, T. The Profile and Antimicrobial Activity of Bacillus Lipopeptide Extracts of Five Potential Biocontrol Strains. Front. Microbiol. 2017, 8, 44–55. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; de Vries, R.H.; Chakraborty, P.; Song, C.; Zhao, X.; Scheffers, D.-J.; Roelfes, G.; Kuipers, O.P. Novel Modifications of Nonribosomal Peptides from Brevibacillus laterosporus MG64 and Investigation of Their Mode of Action. Appl. Environ. Microbiol. 2020, 86, e01981-20. [Google Scholar] [CrossRef]
- Rajput, D.V.; Minkina, T.; Kumari, A.; Shende, S.S.; Ranjan, A.; Faizan, M.; Barakvov, A.; Gromovik, A.; Gorbunova, N.; Rajput, P. A Review on Nanobioremediation Approaches for Restoration of Contaminated Soil. Eurasian J. Soil Sci. 2022, 11, 43–60. [Google Scholar] [CrossRef]
- Gu, Q.; Qiao, J.; Wang, R.; Lu, J.; Wang, Z.; Li, P.; Zhang, L.; Ali, Q.; Khan, A.R.; Gao, X. The Role of Pyoluteorin from Pseudomonas protegens Pf-5 in Suppressing the Growth and Pathogenicity of Pantoea ananatis on Maize. Int. J. Mol. Sci. 2022, 23, 6431. [Google Scholar] [CrossRef]
- Hua, G.K.H.; Wang, L.; Chen, J.; Ji, P. Biological Control of Fusarium wilt on Watermelon by Fluorescent Pseudomonads. Biocontrol Sci. Technol. 2020, 30, 212–227. [Google Scholar] [CrossRef]
- Wang, X.; Mavrodi, D.V.; Ke, L.; Mavrodi, O.V.; Yang, M.; Thomashow, L.S.; Zheng, N.; Weller, D.M.; Zhang, J. Biocontrol and Plant Growth-promoting Activity of Rhizobacteria from C Hinese Fields with Contaminated Soils. Microb. Biotechnol. 2015, 8, 404–418. [Google Scholar] [CrossRef]
- Caulier, S.; Gillis, A.; Colau, G.; Licciardi, F.; Liépin, M.; Desoignies, N.; Modrie, P.; Legrève, A.; Mahillon, J.; Bragard, C. Versatile Antagonistic Activities of Soil-Borne Bacillus spp. and Pseudomonas spp. against Phytophthora infestans and Other Potato Pathogens. Front. Microbiol. 2018, 9, 143. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of Action of Plant Growth Promoting Bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef] [Green Version]
- Hur, G.H.; Vickery, C.R.; Burkart, M.D. Explorations of Catalytic Domains in Non-Ribosomal Peptide Synthetase Enzymology. Nat. Prod. Rep. 2012, 29, 1074–1098. [Google Scholar] [CrossRef] [Green Version]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological Control of Plant Pathogens by Bacillus Species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Sivasakthi, S.; Usharani, G.; Saranraj, P. Biocontrol Potentiality of Plant Growth Promoting Bacteria (PGPR)-Pseudomonas fluorescens and Bacillus subtilis: A Review. Afr. J. Agric. Res. 2014, 9, 1265–1277. [Google Scholar]
- Niu, X.; Thaochan, N.; Hu, Q. Diversity of Linear Non-Ribosomal Peptide in Biocontrol Fungi. J. Fungi 2020, 6, 61. [Google Scholar] [CrossRef]
- Miller, B.R.; Gulick, A.M. Structural Biology of Nonribosomal Peptide Synthetases. Nonribosomal Pept. Polyketide Biosynth. Methods Protos. 2016, 1401, 3–29. [Google Scholar]
- Strieker, M.; Tanović, A.; Marahiel, M.A. Nonribosomal Peptide Synthetases: Structures and Dynamics. Curr. Opin. Struct. Biol. 2010, 20, 234–240. [Google Scholar] [CrossRef]
- Schwarzer, D.; Finking, R.; Marahiel, M.A. Nonribosomal Peptides: From Genes to Products. Nat. Prod. Rep. 2003, 20, 275–287. [Google Scholar] [CrossRef]
- Walsh, C.T.; Chen, H.; Keating, T.A.; Hubbard, B.K.; Losey, H.C.; Luo, L.; Marshall, C.G.; Miller, D.A.; Patel, H.M. Tailoring Enzymes That Modify Nonribosomal Peptides during and after Chain Elongation on NRPS Assembly Lines. Curr. Opin. Chem. Biol. 2001, 5, 525–534. [Google Scholar] [CrossRef]
- Wan, C.; Fan, X.; Lou, Z.; Wang, H.; Olatunde, A.; Rengasamy, K.R.R. Iturin: Cyclic Lipopeptide with Multifunction Biological Potential. Crit. Rev. Food Sci. Nutr. 2022, 62, 7976–7988. [Google Scholar] [CrossRef]
- Mnif, I.; Ghribi, D. Potential of Bacterial Derived Biopesticides in Pest Management. Crop. Prot. 2015, 77, 52–64. [Google Scholar] [CrossRef]
- Delcambe, L.; Peypoux, F.; Besson, F.; Guinand, M.; Michel, G. Structure of Iturin and Iturin-like Substances. Biochem. Soc. Trans. 1977, 5, 1122–1124. [Google Scholar] [CrossRef] [Green Version]
- Maldonado Desena, F.; De la Cruz Ceferino, N.; Gómez Cornelio, S.; Alvarez Villagomez, C.; Herrera Candelario, J.L.; De la Rosa García, S. Bacteria Halotolerant from Karst Sinkholes as a Source of Biosurfactants and Bioemulsifiers. Microorganisms 2022, 10, 1264. [Google Scholar] [CrossRef]
- Besson, F.; Peypoux, F.; Michel, G.; Delcambe, L. Identification of Antibiotics of Iturin Group in Various Strains of Bacillus subtilis. J. Antibiot. 1978, 31, 284–288. [Google Scholar] [CrossRef] [Green Version]
- Stein, T. Bacillus subtilis Antibiotics: Structures, Syntheses and Specific Functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P. Bacillus Lipopeptides: Versatile Weapons for Plant Disease Biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef]
- Thimon, L.; Peypoux, F.; Dana Maget, R.; Roux, B.; Michel, G. Interactions of Bioactive Lipopeptides, Iturin A and Surfactin from Bacillus subtilis. Biotechnol. Appl. Biochem. 1992, 16, 144–151. [Google Scholar]
- Tsuge, K.; Akiyama, T.; Shoda, M. Cloning, Sequencing, and Characterization of the Iturin A Operon. J. Bacteriol. 2001, 183, 6265–6273. [Google Scholar] [CrossRef] [Green Version]
- Das, P.; Mukherjee, S.; Sen, R. Genetic Regulations of the Biosynthesis of Microbial Surfactants: An Overview. Biotechnol. Genet. Eng. Rev. 2008, 25, 165–186. [Google Scholar] [CrossRef]
- Singh, D.; Yadav, D.K.; Sinha, S.; Mondal, K.K.; Singh, G.; Pandey, R.R.; Singh, R. Genetic Diversity of Iturin Producing Strains of Bacillus Species Antagonistic to Ralstonia solanacerarum Causing Bacterial Wilt Disease in Tomato. Afr. J. Microbiol. Res. 2013, 7, 5459–5470. [Google Scholar]
- Rasiya, K.T.; Sebastian, D. Iturin and Surfactin from the Endophyte Bacillus amyloliquefaciens Strain RKEA3 Exhibits Antagonism against Staphylococcus aureus. Biocatal. Agric. Biotechnol. 2021, 36, 102125. [Google Scholar]
- Ntushelo, K.; Ledwaba, L.K.; Rauwane, M.E.; Adebo, O.A.; Njobeh, P.B. The Mode of Action of Bacillus Species against Fusarium graminearum, Tools for Investigation, and Future Prospects. Toxins 2019, 11, 606. [Google Scholar] [CrossRef] [Green Version]
- Balleza, D.; Alessandrini, A.; Beltrán García, M.J. Role of Lipid Composition, Physicochemical Interactions, and Membrane Mechanics in the Molecular Actions of Microbial Cyclic Lipopeptides. J. Membr. Biol. 2019, 252, 131–157. [Google Scholar] [CrossRef]
- Ferrarini, E.; De Roo, V.; Geudens, N.; Martins, J.C.; Höfte, M. Altering in Vivo Membrane Sterol Composition Affects the Activity of the Cyclic Lipopeptides Tolaasin and Sessilin against Pythium. Biochim. Biophys. Acta (BBA)-Biomembr. 2022, 1864, 184008. [Google Scholar] [CrossRef]
- Gong, A.-D.; Li, H.-P.; Yuan, Q.-S.; Song, X.-S.; Yao, W.; He, W.-J.; Zhang, J.-B.; Liao, Y.-C. Antagonistic Mechanism of Iturin A and Plipastatin A from Bacillus amyloliquefaciens S76-3 from Wheat Spikes against Fusarium graminearum. PLoS ONE 2015, 10, e0116871. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zhang, C.; Liang, J.; Wu, L.; Gao, W.; Jiang, J. Iturin A Extracted from Bacillus subtilis WL-2 Affects Phytophthora infestans via Cell Structure Disruption, Oxidative Stress, and Energy Supply Dysfunction. Front. Microbiol. 2020, 11, 536083. [Google Scholar] [CrossRef]
- Jacques, P. Surfactin and Other Lipopeptides from Bacillus spp. Biosurfactants Genes Appl. 2011, 57–91. [Google Scholar]
- Kakinuma, A.; Sugino, H.; Isono, M.; Tamura, G.; Arima, K. Determination of Fatty Acid in Surfactin and Elucidation of the Total Structure of Surfactin. Agric. Biol. Chem. 1969, 33, 973–976. [Google Scholar] [CrossRef]
- Chen, X.; Lu, Y.; Shan, M.; Zhao, H.; Lu, Z.; Lu, Y. A Mini-Review: Mechanism of Antimicrobial Action and Application of Surfactin. World J. Microbiol. Biotechnol. 2022, 38, 143. [Google Scholar] [CrossRef]
- Abbasi Moud, A. Rheology and Microscopy Analysis of Polymer–Surfactant Complexes. Colloid. Polym. Sci. 2022, 300, 733–762. [Google Scholar] [CrossRef]
- Sharma, D.; Singh, S.S.; Baindara, P.; Sharma, S.; Khatri, N.; Grover, V.; Patil, P.B.; Korpole, S. Surfactin like Broad Spectrum Antimicrobial Lipopeptide Co-Produced with Sublancin from Bacillus subtilis Strain A52: Dual Reservoir of Bioactives. Front. Microbiol. 2020, 11, 1167. [Google Scholar] [CrossRef]
- Abdel-Mawgoud, A.M.; Aboulwafa, M.M.; Hassouna, N.A.-H. Characterization of Surfactin Produced by Bacillus subtilis Isolate BS5. Appl. Biochem. Biotechnol. 2008, 150, 289–303. [Google Scholar] [CrossRef]
- Liu, L.; Jin, X.; Lu, X.; Guo, L.; Lu, P.; Yu, H.; Lv, B. Mechanisms of Surfactin from Bacillus subtilis SF1 against Fusarium foetens: A Novel Pathogen Inducing Potato Wilt. J. Fungi 2023, 9, 367. [Google Scholar] [CrossRef]
- Tang, Q.; Bie, X.; Lu, Z.; Lv, F.; Tao, Y.; Qu, X. Effects of Fengycin from Bacillus subtilis FmbJ on Apoptosis and Necrosis in Rhizopus stolonifer. J. Microbiol. 2014, 52, 675–680. [Google Scholar] [CrossRef]
- Yang, H.; Li, X.; Li, X.; Yu, H.; Shen, Z. Identification of Lipopeptide Isoforms by MALDI-TOF-MS/MS Based on the Simultaneous Purification of Iturin, Fengycin, and Surfactin by RP-HPLC. Anal. Bioanal. Chem. 2015, 407, 2529–2542. [Google Scholar] [CrossRef]
- Wu, C.-Y.; Chen, C.-L.; Lee, Y.-H.; Cheng, Y.-C.; Wu, Y.-C.; Shu, H.-Y.; Goötz, F.; Liu, S.-T. Nonribosomal Synthesis of Fengycin on an Enzyme Complex Formed by Fengycin Synthetases. J. Biol. Chem. 2007, 282, 5608–5616. [Google Scholar] [CrossRef] [Green Version]
- Medeot, D.B.; Fernandez, M.; Morales, G.M.; Jofré, E. Fengycins from Bacillus amyloliquefaciens MEP218 Exhibit Antibacterial Activity by Producing Alterations on the Cell Surface of the Pathogens Xanthomonas axonopodis pv. vesicatoria and Pseudomonas aeruginosa PA01. Front. Microbiol. 2020, 10, 3107. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.-H.; Wang, L.-C.; Chen, W.-C.; Chen, S.-Y. Production and Characterization of Fengycin by Indigenous Bacillus subtilis F29-3 Originating from a Potato Farm. Int. J. Mol. Sci. 2010, 11, 4526–4538. [Google Scholar] [CrossRef] [Green Version]
- Karthika, S.; Midhun, S.J.; Jisha, M.S. A Potential Antifungal and Growth-Promoting Bacterium bacillus sp. KTMA4 from Tomato Rhizosphere. Microb. Pathog. 2020, 142, 104049. [Google Scholar] [CrossRef]
- Abd El-Daim, I.A.; Bejai, S.; Fridborg, I.; Meijer, J. Identifying Potential Molecular Factors Involved in Bacillus amyloliquefaciens 5113 Mediated Abiotic Stress Tolerance in Wheat. Plant. Biol. 2018, 20, 271–279. [Google Scholar] [CrossRef]
- Seifi Kalhor, M.; Aliniaeifard, S.; Seif, M.; Javadi, E.; Bernard, F.; Li, T.; Lastochkina, O. Rhizobacterium Bacillus subtilis Reduces Toxic Effects of High Electrical Conductivity in Soilless Culture of Lettuce. In Proceedings of the International Symposium on New Technologies for Environment Control, Energy-Saving and Crop Production in Greenhouse and Plant 1227, Beijing, China, 20–24 August 2017; pp. 471–478. [Google Scholar]
- Romero, D.; De Vicente, A.; Rakotoaly, R.H.; Dufour, S.E.; Veening, J.-W.; Arrebola, E.; Cazorla, F.M.; Kuipers, O.P.; Paquot, M.; Pérez-García, A. The Iturin and Fengycin Families of Lipopeptides Are Key Factors in Antagonism of Bacillus subtilis toward Podosphaera fusca. Mol. Plant-Microbe Interact. 2007, 20, 430–440. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Sun, C. Fengycins, Cyclic Lipopeptides from Marine Bacillus subtilis Strains, Kill the Plant-Pathogenic Fungus Magnaporthe Grisea by Inducing Reactive Oxygen Species Production and Chromatin Condensation. Appl. Environ. Microbiol. 2018, 84, e00445-18. [Google Scholar] [CrossRef] [Green Version]
- Haddoudi, I.; Cabrefiga, J.; Mora, I.; Mhadhbi, H.; Montesinos, E.; Mrabet, M. Biological Control of Fusarium wilt Caused by Fusarium equiseti in Vicia faba with Broad Spectrum Antifungal Plant-Associated Bacillus spp. Biol. Control 2021, 160, 104671. [Google Scholar] [CrossRef]
- Nerling, D.; Castoldi, C.T.; Ehrhardt-Brocardo, N.C.M. The Role of PGPR-Polar Metabolites, Metal-Chelator Compounds and Antibiotics on Plant Growth. Second. Metab. Volatiles PGPR Plant-Growth Promot. 2022, 7, 77–93. [Google Scholar]
- Siddiqui, Z.A. PGPR: Prospective Biocontrol Agents of Plant Pathogens. In PGPR: Biocontrol and Biofertilization; Springer: Berlin/Heidelberg, Germany, 2005; pp. 111–142. [Google Scholar]
- Gimenez, D.; Phelan, A.; Murphy, C.D.; Cobb, S.L. Fengycin A Analogues with Enhanced Chemical Stability and Antifungal Properties. Org. Lett. 2021, 23, 4672–4676. [Google Scholar] [CrossRef]
- Wang, H.; Fewer, D.P.; Holm, L.; Rouhiainen, L.; Sivonen, K. Atlas of Nonribosomal Peptide and Polyketide Biosynthetic Pathways Reveals Common Occurrence of Nonmodular Enzymes. Proc. Natl. Acad. Sci. USA 2014, 111, 9259–9264. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, S.; Acharya, D.; Adholeya, A.; Barrow, C.J.; Deshmukh, S.K. Nonribosomal Peptides from Marine Microbes and Their Antimicrobial and Anticancer Potential. Front. Pharmacol. 2017, 8, 828. [Google Scholar] [CrossRef]
- Weissman, K.J. The Structural Biology of Biosynthetic Megaenzymes. Nat. Chem. Biol. 2015, 11, 660–670. [Google Scholar] [CrossRef]
- Kittilä, T.; Kittel, C.; Tailhades, J.; Butz, D.; Schoppet, M.; Büttner, A.; Goode, R.J.A.; Schittenhelm, R.B.; van Pee, K.-H.; Süssmuth, R.D. Halogenation of Glycopeptide Antibiotics Occurs at the Amino Acid Level during Non-Ribosomal Peptide Synthesis. Chem. Sci. 2017, 8, 5992–6004. [Google Scholar] [CrossRef] [Green Version]
- Abderrahmani, A.; Tapi, A.; Nateche, F.; Chollet, M.; Leclère, V.; Wathelet, B.; Hacene, H.; Jacques, P. Bioinformatics and Molecular Approaches to Detect NRPS Genes Involved in the Biosynthesis of Kurstakin from Bacillus thuringiensis. Appl. Microbiol. Biotechnol. 2011, 92, 571–581. [Google Scholar] [CrossRef]
- Martínez-Núñez, M.A. Nonribosomal Peptides Synthetases and Their Applications in Industry. Sustain. Chem. Process. 2016, 4, 13. [Google Scholar] [CrossRef] [Green Version]
- Bloudoff, K.; Schmeing, T.M. Structural and Functional Aspects of the Nonribosomal Peptide Synthetase Condensation Domain Superfamily: Discovery, Dissection and Diversity. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2017, 1865, 1587–1604. [Google Scholar] [CrossRef]
- Luo, C.; Liu, X.; Zhou, X.; Guo, J.; Truong, J.; Wang, X.; Zhou, H.; Li, X.; Chen, Z. Unusual Biosynthesis and Structure of Locillomycins from Bacillus subtilis 916. Appl. Environ. Microbiol. 2015, 81, 6601–6609. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Zhao, L.; Bode, H.B. Engineering of Specific Single-Module Nonribosomal Peptide Synthetases of the RXP Type for the Production of Defined Peptides. ACS Synth. Biol. 2022, 12, 203–212. [Google Scholar] [CrossRef]
- Oide, S.; Turgeon, B.G. Natural Roles of Nonribosomal Peptide Metabolites in Fungi. Mycoscience 2020, 61, 101–110. [Google Scholar] [CrossRef]
- Mengin-Lecreulx, D.; Allen, N.E.; Hobbs, J.N.; van Heijenoort, J. Inhibition of Peptidoglycan Biosynthesis in Bacillus Megaterium by Daptomycin. FEMS Microbiol. Lett. 1990, 69, 245–248. [Google Scholar] [CrossRef]
- Wang, F.; Zhou, H.; Olademehin, O.P.; Kim, S.J.; Tao, P. Insights into Key Interactions between Vancomycin and Bacterial Cell Wall Structures. ACS Omega 2018, 3, 37–45. [Google Scholar] [CrossRef]
- Liu, J.; Volk, K.J.; Lee, M.S.; Pucci, M.; Handwerger, S. Binding Studies of Vancomycin to the Cytoplasmic Peptidoglycan Precursors by Affinity Capillary Electrophoresis. Anal. Chem. 1994, 66, 2412–2416. [Google Scholar] [CrossRef]
- Seydlová, G.; Sokol, A.; Lišková, P.; Konopásek, I.; Fišer, R. Daptomycin Pore Formation and Stoichiometry Depend on Membrane Potential of Target Membrane. Antimicrob. Agents Chemother. 2019, 63, e01589-18. [Google Scholar] [CrossRef] [Green Version]
- Siewert, G.; Strominger, J.L. Bacitracin: An Inhibitor of the Dephosphorylation of Lipid Pyrophosphate, an Intermediate in the Biosynthesis of the Peptidoglycan of Bacterial Cell Walls. Proc. Natl. Acad. Sci. USA 1967, 57, 767–773. [Google Scholar] [CrossRef] [Green Version]
- Economou, N.J.; Cocklin, S.; Loll, P.J. High-Resolution Crystal Structure Reveals Molecular Details of Target Recognition by Bacitracin. Proc. Natl. Acad. Sci. USA 2013, 110, 14207–14212. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Lu, J.; Sun, J.; Zhu, X.; Zhou, L.; Lu, Z.; Lu, Y. C16-Fengycin A Affect the Growth of Candida albicans by Destroying Its Cell Wall and Accumulating Reactive Oxygen Species. Appl. Microbiol. Biotechnol. 2019, 103, 8963–8975. [Google Scholar] [CrossRef]
- Zakharova, A.A.; Efimova, S.S.; Ostroumova, O.S. Lipid Microenvironment Modulates the Pore-Forming Ability of Polymyxin B. Antibiotics 2022, 11, 1445. [Google Scholar] [CrossRef]
- Zhang, H.; Srinivas, S.; Xu, Y.; Wei, W.; Feng, Y. Genetic and Biochemical Mechanisms for Bacterial Lipid A Modifiers Associated with Polymyxin Resistance. Trends Biochem. Sci. 2019, 44, 973–988. [Google Scholar] [CrossRef]
- Gray, D.A.; Wenzel, M. More than a Pore: A Current Perspective on the in Vivo Mode of Action of the Lipopeptide Antibiotic Daptomycin. Antibiotics 2020, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Killian, J.A. Gramicidin and Gramicidin-Lipid Interactions. Biochim. Biophys. Acta (BBA)-Rev. Biomembr. 1992, 1113, 391–425. [Google Scholar] [CrossRef]
- Fang, S.-T.; Huang, S.-H.; Yang, C.-H.; Liou, J.-W.; Mani, H.; Chen, Y.-C. Effects of Calcium Ions on the Antimicrobial Activity of Gramicidin A. Biomolecules 2022, 12, 1799. [Google Scholar] [CrossRef]
- Qian, S.; Lu, H.; Sun, J.; Zhang, C.; Zhao, H.; Lu, F.; Bie, X.; Lu, Z. Antifungal Activity Mode of Aspergillus ochraceus by Bacillomycin D and Its Inhibition of Ochratoxin A (OTA) Production in Food Samples. Food Control. 2016, 60, 281–288. [Google Scholar] [CrossRef]
- Nain, Z.; Mansur, F.J.; Syed, S.B.; Islam, M.A.; Azakami, H.; Islam, M.R.; Karim, M.M. Inhibition of Biofilm Formation, Quorum Sensing and Other Virulence Factors in Pseudomonas aeruginosa by Polyphenols of Gynura procumbens Leaves. J. Biomol. Struct. Dyn. 2022, 40, 5357–5371. [Google Scholar] [CrossRef]
- Stincone, P.; Fonseca Veras, F.; Micalizzi, G.; Donnarumma, D.; Vitale Celano, G.; Petras, D.; de Angelis, M.; Mondello, L.; Brandelli, A. Listeria Monocytogenes Exposed to Antimicrobial Peptides Displays Differential Regulation of Lipids and Proteins Associated to Stress Response. Cell. Mol. Life Sci. 2022, 79, 263. [Google Scholar] [CrossRef]
- Silva-Stenico, M.E.; Silva, C.S.P.; Lorenzi, A.S.; Shishido, T.K.; Etchegaray, A.; Lira, S.P.; Moraes, L.A.B.; Fiore, M.F. Non-Ribosomal Peptides Produced by Brazilian Cyanobacterial Isolates with Antimicrobial Activity. Microbiol. Res. 2011, 166, 161–175. [Google Scholar] [CrossRef]
- Kaniusaite, M.; Goode, R.J.A.; Tailhades, J.; Schittenhelm, R.B.; Cryle, M.J. Exploring Modular Reengineering Strategies to Redesign the Teicoplanin Non-Ribosomal Peptide Synthetase. Chem. Sci. 2020, 11, 9443–9458. [Google Scholar] [CrossRef]
- Wollinsky, B.; Li, S.-M. Detection and Purification of Non-Ribosomal Peptide Synthetase Products in Neosartorya fischeri. Fungal Second. Metab. Methods Protoc. 2012, 944, 111–119. [Google Scholar]
- Behsaz, B.; Bode, E.; Gurevich, A.; Shi, Y.-N.; Grundmann, F.; Acharya, D.; Caraballo-Rodríguez, A.M.; Bouslimani, A.; Panitchpakdi, M.; Linck, A. Integrating Genomics and Metabolomics for Scalable Non-Ribosomal Peptide Discovery. Nat. Commun. 2021, 12, 3225. [Google Scholar] [CrossRef]
- Novák, J.; Lemr, K.; Schug, K.A.; Havlíček, V. CycloBranch: De Novo Sequencing of Nonribosomal Peptides from Accurate Product Ion Mass Spectra. J. Am. Soc. Mass. Spectrom. 2015, 26, 1780–1786. [Google Scholar] [CrossRef] [Green Version]
- Sarnowski, C.P.; Bikaki, M.; Leitner, A. Cross-Linking and Mass Spectrometry as a Tool for Studying the Structural Biology of Ribonucleoproteins. Structure 2022, 30, 441–461. [Google Scholar] [CrossRef]
- Munakata, Y.; Heuson, E.; Daboudet, T.; Deracinois, B.; Duban, M.; Hehn, A.; Coutte, F.; Slezack-Deschaumes, S. Screening of Antimicrobial Activities and Lipopeptide Production of Endophytic Bacteria Isolated from Vetiver Roots. Microorganisms 2022, 10, 209. [Google Scholar] [CrossRef]
- Fortinez, C.M.; Bloudoff, K.; Harrigan, C.; Sharon, I.; Strauss, M.; Schmeing, T.M. Structures and Function of a Tailoring Oxidase in Complex with a Nonribosomal Peptide Synthetase Module. Nat. Commun. 2022, 13, 548. [Google Scholar] [CrossRef]
- Castro, G.S.; Sousa, T.F.; da Silva, G.F.; Pedroso, R.C.N.; Menezes, K.S.; Soares, M.A.; Dias, G.M.; Santos, A.O.; Yamagishi, M.E.B.; Faria, J.V. Characterization of Peptaibols Produced by a Marine Strain of the Fungus Trichoderma endophyticum via Mass Spectrometry, Genome Mining and Phylogeny-Based Prediction. Metabolites 2023, 13, 221. [Google Scholar] [CrossRef]
- Kersten, R.D.; Yang, Y.-L.; Xu, Y.; Cimermancic, P.; Nam, S.-J.; Fenical, W.; Fischbach, M.A.; Moore, B.S.; Dorrestein, P.C. A Mass Spectrometry–Guided Genome Mining Approach for Natural Product Peptidogenomics. Nat. Chem. Biol. 2011, 7, 794–802. [Google Scholar] [CrossRef] [Green Version]
- Dorrestein, P.C.; Kelleher, N.L. Dissecting Non-Ribosomal and Polyketide Biosynthetic Machineries Using Electrospray Ionization Fourier-Transform Mass Spectrometry. Nat. Prod. Rep. 2006, 23, 893–918. [Google Scholar] [CrossRef]
- Wills, R.H.; O’connor, P.B. Structural Characterization of Actinomycin D Using Multiple Ion Isolation and Electron Induced Dissociation. J. Am. Soc. Mass. Spectrom. 2013, 25, 186–195. [Google Scholar] [CrossRef]
- Molinski, T.F. Microscale Methodology for Structure Elucidation of Natural Products. Curr. Opin. Biotechnol. 2010, 21, 819–826. [Google Scholar] [CrossRef] [Green Version]
- Mohimani, H.; Liu, W.-T.; Kersten, R.D.; Moore, B.S.; Dorrestein, P.C.; Pevzner, P.A. NRPquest: Coupling Mass Spectrometry and Genome Mining for Nonribosomal Peptide Discovery. J. Nat. Prod. 2014, 77, 1902–1909. [Google Scholar] [CrossRef] [Green Version]
- Götze, S.; Vij, R.; Burow, K.; Thome, N.; Urbat, L.; Schlosser, N.; Pflanze, S.; Müller, R.; Hänsch, V.G.; Schlabach, K. Ecological Niche-Inspired Genome Mining Leads to the Discovery of Crop-Protecting Nonribosomal Lipopeptides Featuring a Transient Amino Acid Building Block. J. Am. Chem. Soc. 2023, 145, 2342–2353. [Google Scholar] [CrossRef]
- Reimer, J.M.; Eivaskhani, M.; Harb, I.; Guarné, A.; Weigt, M.; Schmeing, T.M. Structures of a Dimodular Nonribosomal Peptide Synthetase Reveal Conformational Flexibility. Science 2019, 366, eaaw4388. [Google Scholar] [CrossRef]
- Tarry, M.J.; Haque, A.S.; Bui, K.H.; Schmeing, T.M. X-Ray Crystallography and Electron Microscopy of Cross-and Multi-Module Nonribosomal Peptide Synthetase Proteins Reveal a Flexible Architecture. Structure 2017, 25, 783–793. [Google Scholar] [CrossRef]
- Corpuz, J.C.; Sanlley, J.O.; Burkart, M.D. Protein-Protein Interface Analysis of the Non-Ribosomal Peptide Synthetase Peptidyl Carrier Protein and Enzymatic Domains. Synth. Syst. Biotechnol. 2022, 7, 677–688. [Google Scholar] [CrossRef]
- Kresna, I.D.M.; Wuisan, Z.G.; Pohl, J.-M.; Mettal, U.; Otoya, V.L.; Gand, M.; Marner, M.; Otoya, L.L.; Boöhringer, N.; Vilcinskas, A. Genome-Mining-Guided Discovery and Characterization of the PKS-NRPS-Hybrid Polyoxyperuin Produced by a Marine-Derived Streptomycete. J. Nat. Prod. 2022, 85, 888–898. [Google Scholar] [CrossRef]
- Boddy, C.N. Bioinformatics Tools for Genome Mining of Polyketide and Non-Ribosomal Peptides. J. Ind. Microbiol. Biotechnol. 2014, 41, 443–450. [Google Scholar] [CrossRef]
- Martínez-Núñez, M.A.; Rodríguez-Escamilla, Z.; y López, V.L. New Strategies to Discover Non-Ribosomal Peptides as a Source of Antibiotics Molecules. In Pharmaceutical Biocatalysis; Jenny Stanford Publishing: Dubai, United Arab Emirates, 2019; pp. 701–720. ISBN 0429353111. [Google Scholar]
- Wu, G.; Liu, Y.; Xu, Y.; Zhang, G.; Shen, Q.; Zhang, R. Exploring Elicitors of the Beneficial Rhizobacterium Bacillus amyloliquefaciens SQR9 to Induce Plant Systemic Resistance and Their Interactions with Plant Signaling Pathways. Mol. Plant-Microbe Interact. 2018, 31, 560–567. [Google Scholar] [CrossRef] [Green Version]
- Borriss, R.; Wu, H.; Gao, X. Secondary Metabolites of the Plant Growth Promoting Model Rhizobacterium Bacillus velezensis FZB42 Are Involved in Direct Suppression of Plant Pathogens and in Stimulation of Plant-Induced Systemic Resistance. Second. Metab. Plant Growth Promot. Rhizomicroorg. Discov. Appl. 2019, 147–168. [Google Scholar]
- Zhou, G.; Qiu, X.; Wu, X.; Lu, S. Horizontal Gene Transfer Is a Key Determinant of Antibiotic Resistance Genes Profiles during Chicken Manure Composting with the Addition of Biochar and Zeolite. J. Hazard. Mater. 2021, 408, 124883. [Google Scholar] [CrossRef]
- Liu, Y.; Ding, S.; Shen, J.; Zhu, K. Nonribosomal Antibacterial Peptides That Target Multidrug-Resistant Bacteria. Nat. Prod. Rep. 2019, 36, 573–592. [Google Scholar] [CrossRef] [Green Version]
- Sharma, V.; Salwan, R. Antimicrobial Peptides from Biocontrol Agents: Future Wave in Plant Disease Management. In Plant Pathogens; Apple Academic Press: Palm Bay, FL, USA, 2020; pp. 241–267. ISBN 0429057210. [Google Scholar]
- Esmaeel, Q.; Pupin, M.; Jacques, P.; Leclère, V. Nonribosomal Peptides and Polyketides of Burkholderia: New Compounds Potentially Implicated in Biocontrol and Pharmaceuticals. Environ. Sci. Pollut. Res. 2018, 25, 29794–29807. [Google Scholar] [CrossRef]
- Ishikawa, F.; Ohnishi, R.; Uchida, C.; Tanabe, G. Activity-Based Protein Profiling of a Surfactin-Producing Nonribosomal Peptide Synthetase in Bacillus subtilis. STAR Protoc. 2022, 3, 101462. [Google Scholar] [CrossRef]
- Dutta, S.; Yu, S.M.; Lee, Y.H. Assessment of the Contribution of Antagonistic Secondary Metabolites to the Antifungal and Biocontrol Activities of Pseudomonas fluorescens Nbc275. Plant. Pathol. J. 2020, 36, 491–496. [Google Scholar] [CrossRef]
- Jähne, J.; Le Thi, T.T.; Blumenscheit, C.; Schneider, A.; Pham, T.L.; Le Thi, P.T.; Blom, J.; Vater, J.; Schweder, T.; Lasch, P. Novel Plant-Associated Brevibacillus and Lysinibacillus Genomospecies Harbor a Rich Biosynthetic Potential of Antimicrobial Compounds. Microorganisms 2023, 11, 168. [Google Scholar] [CrossRef]
- Tian, Y.; Ji, S.; Zhang, E.; Chen, Y.; Xu, G.; Chen, X.; Fan, J.; Tang, X. Complete Genome Analysis of Bacillus subtilis TY-1 Reveals Its Biocontrol Potential against Tobacco Bacterial Wilt. Mar. Genom. 2023, 68, 101018. [Google Scholar] [CrossRef]
- Clough, S.E.; Jousset, A.; Elphinstone, J.G.; Friman, V. Combining in Vitro and in Vivo Screening to Identify Efficient Pseudomonas Biocontrol Strains against the Phytopathogenic Bacterium Ralstonia solanacearum. Microbiologyopen 2022, 11, e1283. [Google Scholar] [CrossRef]
- Deng, X.; Zhang, N.; Li, Y.; Zhu, C.; Qu, B.; Liu, H.; Li, R.; Bai, Y.; Shen, Q.; Falcao Salles, J. Bio-organic Soil Amendment Promotes the Suppression of Ralstonia solanacearum by Inducing Changes in the Functionality and Composition of Rhizosphere Bacterial Communities. New Phytol. 2022, 235, 1558–1574. [Google Scholar] [CrossRef]
- Qi, P.; Sun, D.; Wu, T.; Li, Y. Stress Proteins, Nonribosomal Peptide Synthetases, and Polyketide Synthases Regulate Carbon Sources-Mediated Bio-Demulsifying Mechanisms of Nitrate-Reducing Bacterium Gordonia sp. TD-4. J. Hazard. Mater. 2022, 422, 126900. [Google Scholar] [CrossRef]
- Zhou, L.; Song, C.; Li, Z.; Kuipers, O.P. Antimicrobial Activity Screening of Rhizosphere Soil Bacteria from Tomato and Genome-Based Analysis of Their Antimicrobial Biosynthetic Potential. BMC Genom. 2021, 22, 29. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, J.; Xia, Z.; Wei, H.-L. Characterization of a Versatile Plant Growth-Promoting Rhizobacterium Pseudomonas mediterranea Strain S58. Microorganisms 2020, 8, 334. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.M.; Yang, H.J.; Huang, J.G.; Yuan, L. Lysobacter enzymogenes LE16 Autolysates Have Potential as Biocontrol Agents—Lysobacter sp. Autolysates as Biofungicide. J. Appl. Microbiol. 2020, 129, 1684–1692. [Google Scholar] [CrossRef]
- Abdelli, F.; Jardak, M.; Elloumi, J.; Stien, D.; Cherif, S.; Mnif, S.; Aifa, S. Antibacterial, Anti-Adherent and Cytotoxic Activities of Surfactin (s) from a Lipolytic Strain Bacillus safensis F4. Biodegradation 2019, 30, 287–300. [Google Scholar] [CrossRef]
- Pusztahelyi, T.; Holb, I.J.; Pócsi, I. Secondary Metabolites in Fungus-Plant Interactions. Front. Plant. Sci. 2015, 6, 573. [Google Scholar] [CrossRef] [Green Version]
- Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk-ściseł, J. Trichoderma: The Current Status of Its Application in Agriculture for the Biocontrol of Fungal Phytopathogens and Stimulation of Plant Growth. Int. J. Mol. Sci. 2022, 23, 29. [Google Scholar] [CrossRef]
- Daoubi, M.; Pinedo-Rivilla, C.; Rubio, M.B.; Hermosa, R.; Monte, E.; Aleu, J.; Collado, I.G. Hemisynthesis and Absolute Configuration of Novel 6-Pentyl-2H-Pyran-2-One Derivatives from Trichoderma spp. Tetrahedron 2009, 65, 4834–4840. [Google Scholar] [CrossRef] [Green Version]
- Vizcaino, J.A.; Sanz, L.; Cardoza, R.E.; Monte, E.; Gutierrez, S. Detection of Putative Peptide Synthetase Genes in Trichoderma Species: Application of This Method to the Cloning of a Gene from T. Harzianum CECT 2413. FEMS Microbiol. Lett. 2005, 244, 139–148. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Selvaraj, J.N.; Xing, F.; Zhou, L.; Wang, Y.; Song, H.; Tan, X.; Sun, L.; Sangare, L.; Folly, Y.M.E.; et al. Antagonistic Action of Bacillus subtilis Strain SG6 on Fusarium graminearum. PLoS ONE 2014, 9, e92486. [Google Scholar] [CrossRef]
- Jiang, J.; Gao, L.; Bie, X.; Lu, Z.; Liu, H.; Zhang, C.; Lu, F.; Zhao, H. Identification of Novel Surfactin Derivatives from NRPS Modification of Bacillus subtilis and Its Antifungal Activity against Fusarium moniliforme. BMC Microbiol. 2016, 16, 31. [Google Scholar] [CrossRef] [Green Version]
- Sarwar, A.; Hassan, M.N.; Imran, M.; Iqbal, M.; Majeed, S.; Brader, G.; Sessitsch, A.; Hafeez, F.Y. Biocontrol Activity of Surfactin A Purified from Bacillus NH-100 and NH-217 against Rice Bakanae Disease. Microbiol. Res. 2018, 209, 1–13. [Google Scholar] [CrossRef]
- Leclère, V.; Béchet, M.; Adam, A.; Guez, J.-S.; Wathelet, B.; Ongena, M.; Thonart, P.; Gancel, F.; Chollet-Imbert, M.; Jacques, P. Mycosubtilin Overproduction by Bacillus subtilis BBG100 Enhances the Organism’s Antagonistic and Biocontrol Activities. Appl. Environ. Microbiol. 2005, 71, 4577–4584. [Google Scholar] [CrossRef] [Green Version]
- Niemhom, N.; Kittiwongwattana, C. Biocontrol Potential, Genome and Nonribosomal Peptide Synthetase Gene Expression of Bacillus velezensis 2211. Curr. Appl. Sci. Technol. 2023, 10, 1–17. [Google Scholar] [CrossRef]
- Kaur, C.; Fidenza, M.; Ervin, E.; Bais, H.P. SpoA-Dependent Antifungal Activity of a Plant Growth Promoting Rhizobacteria Bacillus subtilis Strain UD1022 against the Dollar Spot Pathogen (Clarireedia jacksonii). Available online: https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4394400 (accessed on 28 May 2023).
- Rosier, A.; Pomerleau, M.; Beauregard, P.B.; Samac, D.A.; Bais, H.P. Surfactin and Spo0A-Dependent Antagonism by Bacillus subtilis Strain UD1022 against Medicago sativa Phytopathogens. Plants 2023, 12, 1007. [Google Scholar] [CrossRef]
- Doty, S.L.; Joubert, P.M.; Firrincieli, A.; Sher, A.W.; Tournay, R.; Kill, C.; Parikh, S.S.; Okubara, P. Potential Biocontrol Activities of Populus Endophytes against Several Plant Pathogens Using Different Inhibitory Mechanisms. Pathogens 2023, 12, 13. [Google Scholar] [CrossRef]
- Songwattana, P.; Boonchuen, P.; Piromyou, P.; Wongdee, J.; Greetatorn, T.; Inthaisong, S.; Tantasawat, P.A.; Teamtisong, K.; Tittabutr, P.; Boonkerd, N. Insights into Antifungal Mechanisms of Bacillus velezensis S141 against Cercospora Leaf Spot in Mungbean (V. radiata). Microbes Environ. 2023, 38, ME22079. [Google Scholar] [CrossRef]
- Khatri, S.; Sazinas, P.; Strube, M.L.; Ding, L.; Dubey, S.; Shivay, Y.S.; Sharma, S.; Jelsbak, L. Pseudomonas Is a Key Player in Conferring Disease Suppressiveness in Organic Farming. Plant Soil 2023, 1–20. [Google Scholar] [CrossRef]
- Qiao, J.; Zhang, R.; Liu, Y.; Liu, Y. Evaluation of the Biocontrol Efficiency of Bacillus subtilis Wettable Powder on Pepper Root Rot Caused by Fusarium solani. Pathogens 2023, 12, 225. [Google Scholar] [CrossRef]
- Yabaneri, C.; Sevim, A. Endophytic Fungi from the Common Walnut and Their in Vitro Antagonistic Activity against Ophiognomonia leptostyla. Biologia 2023, 78, 361–371. [Google Scholar] [CrossRef]
- Ahmad, T.; Xing, F.; Nie, C.; Cao, C.; Xiao, Y.; Yu, X.; Moosa, A.; Liu, Y. Biocontrol Potential of Lipopeptides Produced by the Novel Bacillus subtilis Strain Y17B against Postharvest Alternaria Fruit Rot of Cherry. Front. Microbiol. 2023, 14, 689. [Google Scholar] [CrossRef]
- Diabankana, R.G.C.; Shulga, E.U.; Validov, S.Z.; Afordoanyi, D.M. Genetic Characteristics and Enzymatic Activities of Bacillus velezensis KS04AU as a Stable Biocontrol Agent against Phytopathogens. Int. J. Plant Biol. 2022, 13, 201–222. [Google Scholar] [CrossRef]
- Costa, A.; Corallo, B.; Amarelle, V.; Stewart, S.; Pan, D.; Tiscornia, S.; Fabiano, E. Paenibacillus sp. Strain UY79, Isolated from a Root Nodule of Arachis Villosa, Displays a Broad Spectrum of Antifungal Activity. Appl. Environ. Microbiol. 2022, 88, e01645-21. [Google Scholar] [CrossRef]
- Matilla, M.A.; Monson, R.E.; Murphy, A.; Schicketanz, M.; Rawlinson, A.; Duncan, C.; Mata, J.; Leeper, F.; Salmond, G.P.C. Solanimycin: Biosynthesis and Distribution of a New Antifungal Antibiotic Regulated by Two Quorum-Sensing Systems. mBio 2022, 13, 22. [Google Scholar] [CrossRef]
- Pandin, C.; Le Coq, D.; Deschamps, J.; Védie, R.; Rousseau, T.; Aymerich, S.; Briandet, R. Complete Genome Sequence of Bacillus velezensis QST713: A Biocontrol Agent That Protects Agaricus bisporus Crops against the Green Mould Disease. J. Biotechnol. 2018, 278, 10–19. [Google Scholar] [CrossRef]
- Palazzini, J.M.; Dunlap, C.A.; Bowman, M.J.; Chulze, S.N. Bacillus velezensis RC 218 as a Biocontrol Agent to Reduce Fusarium Head Blight and Deoxynivalenol Accumulation: Genome Sequencing and Secondary Metabolite Cluster Profiles. Microbiol. Res. 2016, 192, 30–36. [Google Scholar] [CrossRef]
- Korangi Alleluya, V.; Argüelles Arias, A.; Ribeiro, B.; De Coninck, B.; Helmus, C.; Delaplace, P.; Ongena, M. Bacillus Lipopeptide-Mediated Biocontrol of Peanut Stem Rot Caused by Athelia rolfsii. Front. Plant. Sci. 2023, 14, 1069971. [Google Scholar] [CrossRef]
- Al-Mutar, D.M.K.; Alzawar, N.S.A.; Noman, M.; Azizullah; Li, D.; Song, F. Suppression of Fusarium wilt in Watermelon by Bacillus smyloliquefaciens DHA55 through Extracellular Production of Antifungal Lipopeptides. J. Fungi 2023, 9, 336. [Google Scholar] [CrossRef]
- ALTRÃO, C.S. Characterization of Disease Suppression Activity of Bacillus Cyclic Lipopeptide Depending on the Induced Disease Resistance in Plant. Ph.D. Thesis, Tokyo University of Agriculture, Tokyo, Japan, 2022. [Google Scholar]
- Höfte, M. The Use of Pseudomonas spp. as Bacterial Biocontrol Agents to Control Plant Diseases. Burleigh Dodds Ser. Agric. Sci. 2021, 301–374. [Google Scholar] [CrossRef]
- Hoff, G.; Arias, A.A.; Boubsi, F.; Pršic, J.; Meyer, T.; Ibrahim, H.M.M.; Steels, S.; Luzuriaga, P.; Legras, A.; Franzil, L.; et al. Surfactin Stimulated by Pectin Molecular Patterns and Root Exudates Acts as a Key Driver of the Bacillus-Plant Mutualistic Interaction. mBio 2021, 12, e01774-21. [Google Scholar] [CrossRef]
- Laird, M.; Piccoli, D.; Weselowski, B.; McDowell, T.; Renaud, J.; MacDonald, J.; Yuan, Z.C. Surfactin-Producing Bacillus velezensis 1B-23 and Bacillus sp. 1D-12 Protect Tomato against Bacterial Canker Caused by Clavibacter michiganensis subsp. Michiganensis. J. Plant Pathol. 2020, 102, 451–458. [Google Scholar] [CrossRef]
- Abdallah, D.B.; Krier, F.; Jacques, P.; Tounsi, S.; Frikha-Gargouri, O. Agrobacterium Tumefaciens C58 Presence Affects Bacillus velezensis 32a Ecological Fitness in the Tomato Rhizosphere. Environ. Sci. Pollut. Res. 2020, 27, 28429–28437. [Google Scholar] [CrossRef]
- Wu, J.J.; Huang, J.W.; Deng, W.L. Phenylacetic Acid and Methylphenyl Acetate From the Biocontrol Bacterium Bacillus mycoides BM02 Suppress Spore Germination in Fusarium oxysporum f. sp. lycopersici. Front. Microbiol. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Azizoglu, U.; Yilmaz, N.; Simsek, O.; Ibal, J.C.; Tagele, S.B.; Shin, J.-H. The Fate of Plant Growth-Promoting Rhizobacteria in Soilless Agriculture: Future Perspectives. 3 Biotech. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Singh, G.; Biswas, D.R.; Marwaha, T.S. Mobilization of Potassium from Waste Mica by Plant Growth Promoting Rhizobacteria and Its Assimilation by Maize (Zea Mays) and Wheat (Triticum aestivum L.): A Hydroponics Study under Phytotron Growth Chamber. J. Plant Nutr. 2010, 33, 1236–1251. [Google Scholar] [CrossRef]
- Strigul, N.S.; Kravchenko, L.V. Mathematical Modeling of PGPR Inoculation into the Rhizosphere. Environ. Model. Softw. 2006, 21, 1158–1171. [Google Scholar] [CrossRef]
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.Z.; Reddy, M.S.; El Enshasy, H. Plant Growth Promoting Rhizobacteria (PGPR) as Green Bioinoculants: Recent Developments, Constraints, and Prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Orhan, F. Alleviation of Salt Stress by Halotolerant and Halophilic Plant Growth-Promoting Bacteria in Wheat (Triticum aestivum). Braz. J. Microbiol. 2016, 47, 621–627. [Google Scholar] [CrossRef] [Green Version]
- Moncada, A.; Vetrano, F.; Miceli, A. Alleviation of Salt Stress by Plant Growth-Promoting Bacteria in Hydroponic Leaf Lettuce. Agronomy 2020, 10, 1523. [Google Scholar] [CrossRef]
- Prajapati, K.; Modi, H. Growth Promoting Effect of Potassium Solubilizing Enterobacter Hormaechei (KSB-8) on Cucumber (Cucumis sativus) under Hydroponic Conditions. Int. J. Adv. Res. Biol. Sci. 2016, 3, 168–173. [Google Scholar]
- Suzuki, W.; Sugawara, M.; Miwa, K.; Morikawa, M. Plant Growth-Promoting Bacterium Acinetobacter calcoaceticus P23 Increases the Chlorophyll Content of the Monocot Lemna Minor (Duckweed) and the Dicot Lactuca Sativa (Lettuce). J. Biosci. Bioeng. 2014, 118, 41–44. [Google Scholar] [CrossRef]
- Murgese, P.; Santamaria, P.; Leoni, B.; Crecchio, C. Ameliorative Effects of PGPB on Yield, Physiological Parameters, and Nutrient Transporter Genes Expression in Barattiere (Cucumis melo L.). J. Soil. Sci. Plant. Nutr. 2020, 20, 784–793. [Google Scholar] [CrossRef]
- Issifu, M.; Songoro, E.K.; Onguso, J.; Ateka, E.M.; Ngumi, V.W. Potential of Pseudarthrobacter Chlorophenolicus BF2P4-5 as a Biofertilizer for the Growth Promotion of Tomato Plants. Bacteria 2022, 1, 191–206. [Google Scholar] [CrossRef]
- Kim, J.; Woo, O.-G.; Bae, Y.; Keum, H.L.; Chung, S.; Sul, W.J.; Lee, J.-H. Enhanced Drought and Salt Stress Tolerance in Arabidopsis by Flavobacterium Crocinum HYN0056 T. J. Plant Biol. 2020, 63, 63–71. [Google Scholar] [CrossRef]
- Li, X.; Yan, J.; Li, D.; Jiang, Y.; Zhang, Y.; Wang, H.; Zhang, J.; Ahmed, T.; Li, B. Isolation and Molecular Characterization of Plant-Growth-Promoting Bacteria and Their Effect on Eggplant (Solanum Melongena) Growth. Agriculture 2021, 11, 1258. [Google Scholar] [CrossRef]
- Turan, M.; Ekinci, M.; Yildirim, E.; Güneş, A.; Karagöz, K.; Kotan, R.; Dursun, A. Plant Growth-Promoting Rhizobacteria Improved Growth, Nutrient, and Hormone Content of Cabbage (Brassica oleracea) Seedlings. Turk. J. Agric. For. 2014, 38, 327–333. [Google Scholar] [CrossRef]
- Abdalla, O.A.; Bibi, S.; Zhang, S. Application of Plant Growth-Promoting Rhizobacteria to Control Papaya Ringspot Virus and Tomato Chlorotic Spot Virus. Arch. Phytopathol. Plant Prot. 2017, 50, 584–597. [Google Scholar] [CrossRef]
- de Sousa, S.M.; de Oliveira, C.A.; Andrade, D.L.; de Carvalho, C.G.; Ribeiro, V.P.; Pastina, M.M.; Marriel, I.E.; de Paula Lana, U.G.; Gomes, E.A. Tropical Bacillus Strains Inoculation Enhances Maize Root Surface Area, Dry Weight, Nutrient Uptake and Grain Yield. J. Plant Growth Regul. 2021, 40, 867–877. [Google Scholar] [CrossRef]
- Yasmin, H.; Naeem, S.; Bakhtawar, M.; Jabeen, Z.; Nosheen, A.; Naz, R.; Keyani, R.; Mumtaz, S.; Hassan, M.N. Halotolerant Rhizobacteria Pseudomonas pseudoalcaligenes and Bacillus subtilis Mediate Systemic Tolerance in Hydroponically Grown Soybean (Glycine max L.) against Salinity Stress. PLoS ONE 2020, 15, e0231348. [Google Scholar] [CrossRef]
- Sepehri, M.; Khatabi, B. Combination of Siderophore-Producing Bacteria and Piriformospora indica Provides an Efficient Approach to Improve Cadmium Tolerance in Alfalfa. Microb. Ecol. 2021, 81, 717–730. [Google Scholar] [CrossRef]

| S. No. | Source Organism | Compound | Assessment Method | Plant-Pathogen | Disease Caused | Reference |
|---|---|---|---|---|---|---|
| 1 | B. porteri HB1.4B Brevibacillus RS1.1 Brevibacillus DP1.3A Brevibacillus HB2.2 | Uncharacterized NRPs and NRP-PK hybrids. | Antagonist activity was identified through in vitro antibacterial activity and gene clusters were identified after whole-genome sequencing, analysis, and annotations | Clavibacter michiganensis | Bacterial wilt and canker of tomato | [131] |
| 2 | B. porteri HB1.1 Brevibacillus DP1.3A | Xanthomonas campestris | black rot in cruciferous | [131] | ||
| 3 | B. subtilis TY-1 | surfactin, bacillibactin, and fengycin | Identified using AntiSMASH | Ralstonia solanacearum | Bacterial wilt of tobacco | [132] |
| 4 | Pseudomonas strain CHA0 | orfamide A | Direct contact inhibition, indirectly using supernatant assays Whole-genome sequencing and AntiSMASH | R. solanacearum | bacterial wilt in a range of host plants, e.g., potato, tobacco, tomato | [133] |
| 5 | Sphingomonas, Pseudoxanthomonas and Stenotrophomonas | Not Specified | NPRS BGC enrichment was confirmed using metagenomic studies. Pathogen inhibition was further confirmed in pot experiments | R. solanacearum | bacterial wilt in a range of host plants, e.g., potato, tobacco, tomato | [134] |
| 6 | Gordonia sp. TD-4 | Not Specified | NPRS BGC enrichment was confirmed using metagenomic studies | Not Specified | Not Specified | [135] |
| 7 | Pseudomonas strain CHA0 | orfamide A | Direct contact inhibition, indirectly using supernatant assays. Whole-genome sequencing and AntiSMASH | R. solanacearum | bacterial wilt in a range of host plants, e.g., potato, tobacco, tomato | [133] |
| 8 | B. subtilis EH11 Paenibacillus sp. EDO6 B. endophyticus FH5 | Surfactin, fengycin, bacillibactin, petrobactin, lichenysin, and bacillaene. | In vitro inhibition assay of bacterial pathogens and NRP bio-clusters were confirmed by genome-based analysis | Erwinia carotovora | soft rot disease of potato, carrot, and cabbage | [136] |
| 9 | B. subtilis EH11 Paenibacillus sp. EDO6 B. endophyticus FH5 | P. syringae | bacterial blight and bacterial speck among several plants | [136] | ||
| 10 | Brevibacillus laterosporus MG64 | Bogorols K | MIC = 4 μg/mL LC-MS/MSantiSMASH | X. campestris pv. campestris NCCB92058 | Black rot disease in crucifers | [24] |
| Bogorols L | MIC = 2 μg/mL LC-MS/MSantiSMASH | X. campestris pv. campestris NCCB92058 | ||||
| Brevibacillin | MIC = 2 μg/mL LC-MS/MSantiSMASH | X. campestris pv. campestris NCCB92058 | ||||
| 11 | Brevibacillin | MIC = 2 μg/mL LC-MS/MSantiSMASH | X. translucens pv. graminis LMG587 | leaf streak in wheat and cereal crop. | [24] | |
| Bogorol L | MIC = 8 μg/mL LC-MS/MSantiSMASH | P. syringae pv. tomato DC3000 | ||||
| 12 | P. mediterranea Strain S58 | Fengycin, Crochelin A, Entolysin, Orfamide B, Siderophore, Syringomycin | Antagonistic test and Disease control assay confirmed the biocontrol potential. NRPS BGCs were identified, encoding for NRPs | P. syringae pv. tabaci | Wild fire, Angular leaf spot in Tobacco | [137] |
| 13 | Lysobacter enzymogenes LE16 | surfactin | antiSMASH | P. syringae pv. tabaci | tobacco wildfire disease | [138] |
| 14 | B.safensis F4 | Surfactin | MIC = 1.56 mg mL−1 (crude biosurfactant)Identified using HPLC | P. savastanoi | olive knot disease | [139] |
| 15 | Surfactin | MIC = 3.125 mg mL−1 (crude biosurfactant)Identified using HPLC | Agrobacterium tumefaciens | Crown gall | [139] | |
| 16 | B. amyloliquefaciens SQR9 | Fengycin | In vitro inhibition assay | P. syringae pv. tomato DC3000 (Studied on Arabidopsis) | Model organism for studying plant–bacterial interactions. This strain can infect Arabidopsis too | [123] |
| S. No. | Source Organism | NRP Compound | Assessment Method | Pathogen | Disease Caused | Reference |
|---|---|---|---|---|---|---|
| 1 | B. velezensis 2211 | Bacillomycin D (bmyA), fengycin (fenB) |
| Colletotrichum fructicola | The causal agent of anthracnose and soft rot in avocado fruits | [148] |
| 2 | B. subtilis Strain UD1022 | - |
| Clarireedia jacksonii | Dollar spot of the grass | [149] |
| 3 | B. subtilis Strain UD1022 | Surfactin |
| Ascochyta medicaginicola StC 306-5 | Black stem of Alfalfa and Medicago truncatula. | [150] |
| 4 | Burkholderia vietnamiensis strain WPB | Occidiofungin |
| Gaeumannomyces graminis var. tritici | Take all diseases, roots of grass and cereal plants | [151] |
| 5 | B. velezensis S141 | surfactin, bacilysin, and bacillomycin D |
| Cercospora canescens | leaf spot disease of amaranth | [152] |
| 6 | Pseudomonas spp. SK2, and SK3 | Obafuorin and cupriachelin |
| F. oxysporum | yellowing, stunting, and death of seedlings and yellowing and stunting of older plants | [153] |
| 7 | Pseudomonas spp. SK2, and SK3 | Obafuorin and cupriachelin |
| Verticillium dahliae | verticillium wilt causes the leaves to curl and discolor. | [153] |
| 8 | B. subtilis PTS-394 | Surfactin, Iturin, and Fengcyin |
| F. solani | pepper root rot | [154] |
| 9 | Alternaria sp. CC-3 | NRPS BGCs were identified |
| Ophiognomonia leptostyla | walnut anthracnose or walnut black spot | [155] |
| 10 | B. subtilis strain Y17B | surfactin |
| Alternaria alternata | Leaf spots, rots, blights and affects other plant parts in over 380 host plants | [156] |
| 11 | Bacillus. velezensis KS04AU. | Surfactin |
| F. oxysporum f. sp. radicis-lycopersici ZUM2407 (Forl ZUM2407) | Fusarium wilt disease in Tomatoes causes heavy loss | [157] |
| 12 | Paenibacillus sp. strain (UY79) | fusaricidin B, tridecaptin, |
| F. verticillioides A71. | seedling blight, or stalk or ear rot in maize | [158] |
| 13 | Dickeya solani MK10 | Solanimycin |
| V. dahliae | verticillium wilt in many host plants | [159] |
| 14 | B. subtilis (B. velezensis QST713) | Ericin + NRPS |
| T. aggressivum | green mold disease | [160] |
| 15 | Bacillus subtilis (B. velezensis RC 218) | Iturin, Fengycin |
| F. graminearum | Fusarium head blight | [161] |
| S. No. | NRP Compound | Source of NRP (Only Bacteria) | Pathogen | Plant Name | Reference |
|---|---|---|---|---|---|
| 1 | Purified surfactin | B. velezensis GA1 | Athelia rolfsii | Peanut | [162] |
| 2 | Iturin, bacillomycin D, surfactin, and fengycin | B. amyloliquefaciens DHA55 | F. oxysporum f. sp. niveum (fon) | Watermelon | [163] |
| 3 | Surfactin, Iturin A | Bacillus spp. | P. syringae pv. maculicola MAFF 302783 | Cabbage | [164] |
| 4 | Lokisin | P. koreensis 2.74 (CBS 125413) | Pythium ultimum | Tomato | [165] |
| 5 | Surfactin | B. velezensis | Botrytis cinerea | Tobacco | [166] |
| 6 | Surfactin A, surfactin B, Surfactin C | B. velezenis 1B-23 or Bacillus sp. 1D-12 | Clavibacter michiganensis subsp. michiganensis | Tomato | [167] |
| 7 | Surfactin | B. velezensis 32a | Agrobacterium tumefaciens C58 | Tomato | [168] |
| 8 | Phenylacetic acid (PAA) and methylphenyl acetate (MPA) | B. mycoides BM02 | F. oxysporum f. sp. lycopersici (Fol) | Tomato | [169] |
| Brand Name | PGPB Used | Manufacturer | Country of Origin |
|---|---|---|---|
| Taegro 2 | B. subtilis var. amyloliquefaciens Strain FZB24 | Novozymes | Denmark |
| Rhizolizer | B. amyloliquefaciens along with a fungus Trichoderma harzianum | Locus Agricultural Solutions | United States |
| LALGUARD M52 OD | B. subtilis | Lallemand Plant Care | Canada |
| RHIZOVITAL® 42 | B. amyloliquefaciens | Andermatt | Canada |
| GeumanoControl | Not Disclosed | Bio-lide | Poland |
| CEASE | B. subtilis strain QST 713 | BioWorks Inc. | United States |
| Nutri-Life Platform | Consortium of Bacillus, Pseudomonas, and Trichoderma | Nutri-Tech Solutions | Australia |
| Serenade® SOIL | Bacillus subtilis QST 713 | Bayer | Canada |
| Subtilex® | B. subtilis strain MBI 600 | Bioglobal | Turkey |
| Rizovital 42 | Bacillus amyloliquefaciens | Organic Crop Protectants (OCP) | Australia |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ranjan, A.; Rajput, V.D.; Prazdnova, E.V.; Gurnani, M.; Bhardwaj, P.; Sharma, S.; Sushkova, S.; Mandzhieva, S.S.; Minkina, T.; Sudan, J.; et al. Nature’s Antimicrobial Arsenal: Non-Ribosomal Peptides from PGPB for Plant Pathogen Biocontrol. Fermentation 2023, 9, 597. https://doi.org/10.3390/fermentation9070597
Ranjan A, Rajput VD, Prazdnova EV, Gurnani M, Bhardwaj P, Sharma S, Sushkova S, Mandzhieva SS, Minkina T, Sudan J, et al. Nature’s Antimicrobial Arsenal: Non-Ribosomal Peptides from PGPB for Plant Pathogen Biocontrol. Fermentation. 2023; 9(7):597. https://doi.org/10.3390/fermentation9070597
Chicago/Turabian StyleRanjan, Anuj, Vishnu D. Rajput, Evgeniya Valeryevna Prazdnova, Manisha Gurnani, Pallavi Bhardwaj, Shikha Sharma, Svetlana Sushkova, Saglara S. Mandzhieva, Tatiana Minkina, Jebi Sudan, and et al. 2023. "Nature’s Antimicrobial Arsenal: Non-Ribosomal Peptides from PGPB for Plant Pathogen Biocontrol" Fermentation 9, no. 7: 597. https://doi.org/10.3390/fermentation9070597
APA StyleRanjan, A., Rajput, V. D., Prazdnova, E. V., Gurnani, M., Bhardwaj, P., Sharma, S., Sushkova, S., Mandzhieva, S. S., Minkina, T., Sudan, J., Zargar, S. M., Chauhan, A., & Jindal, T. (2023). Nature’s Antimicrobial Arsenal: Non-Ribosomal Peptides from PGPB for Plant Pathogen Biocontrol. Fermentation, 9(7), 597. https://doi.org/10.3390/fermentation9070597












