Biofilm Inhibition, Antibacterial and Antiadhesive Properties of a Novel Biosurfactant from Lactobacillus paracasei N2 against Multi-Antibiotics-Resistant Pathogens Isolated from Braised Fish
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. Antibiotics’ Resistance Profile of the Pathogens
2.3. Biofilm Formation Ability of the Pathogens
2.4. Production of Biosurfactant
2.5. Characterization of the Biosurfactant
2.6. Antimicrobial Activity of Biosurfactant
2.7. Antiadhesive Activity of Biosurfactant
2.8. Antibiofilm Activity of Biosurfactant
2.8.1. Inhibition of Biofilm Formation
2.8.2. Eradication of Mature Biofilms
2.9. Statistical Analysis
3. Results and Discussion
3.1. Antibiotics’ Susceptibility Profile of the Pathogens
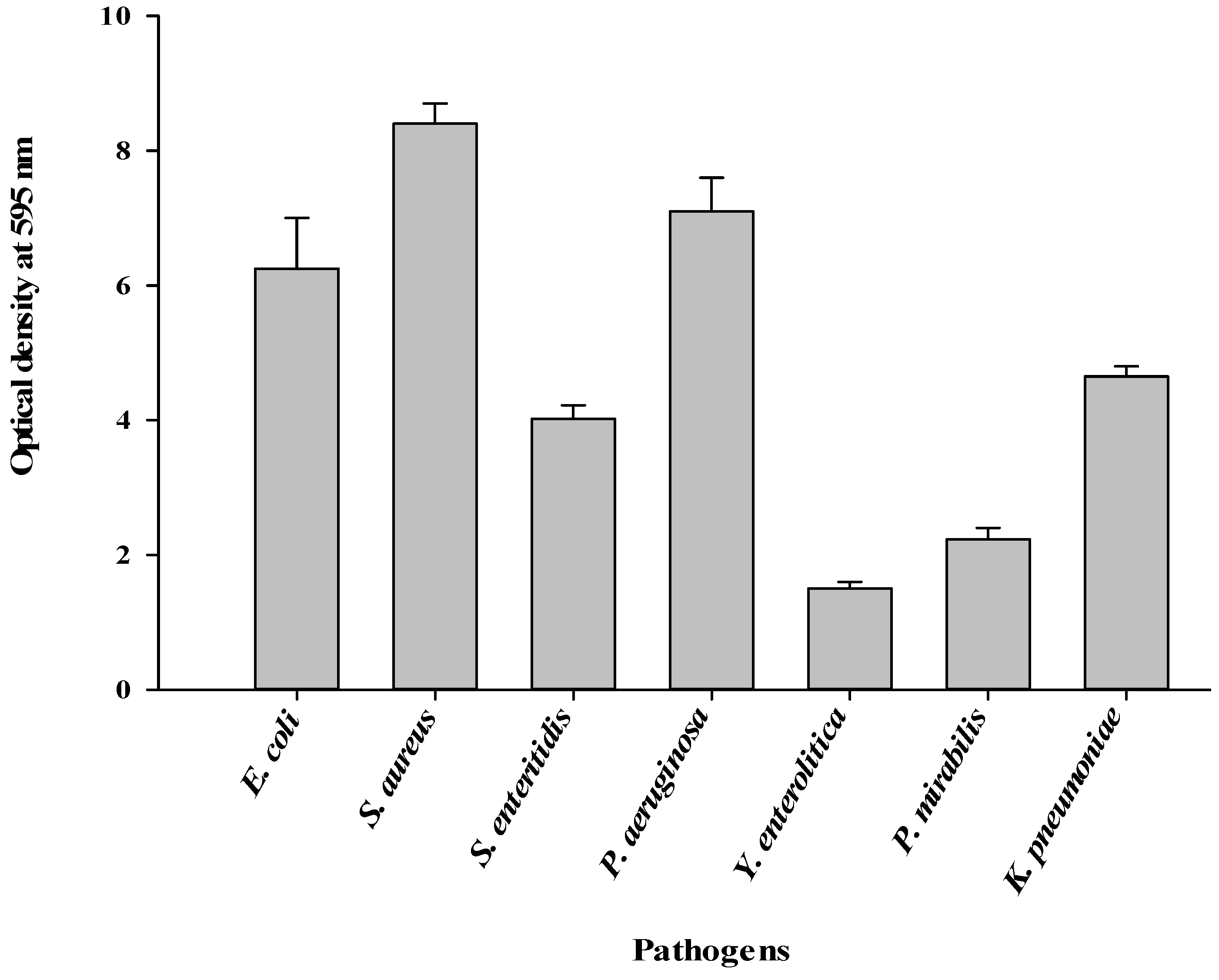
3.2. Formation of Biofilms by the Pathogens
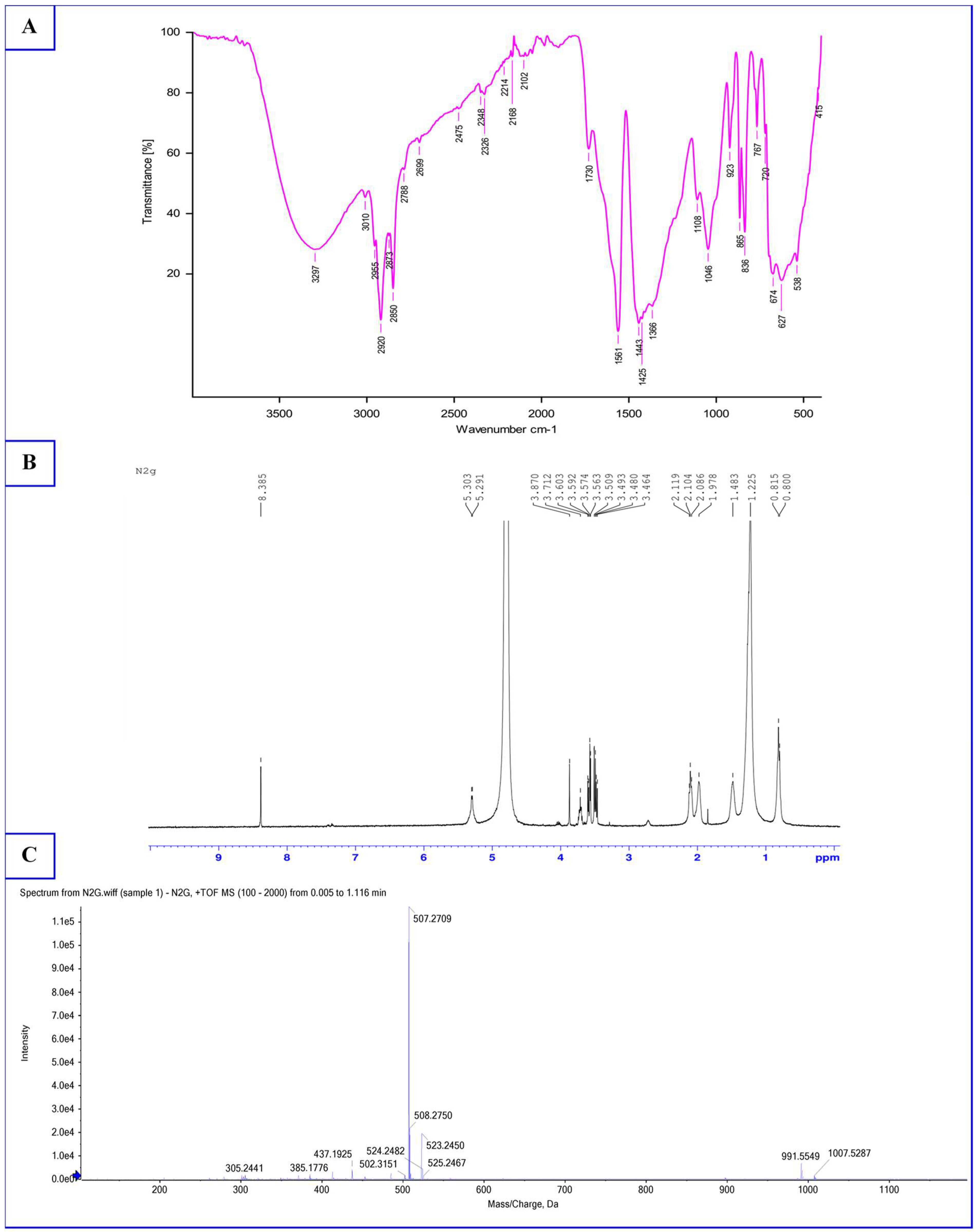
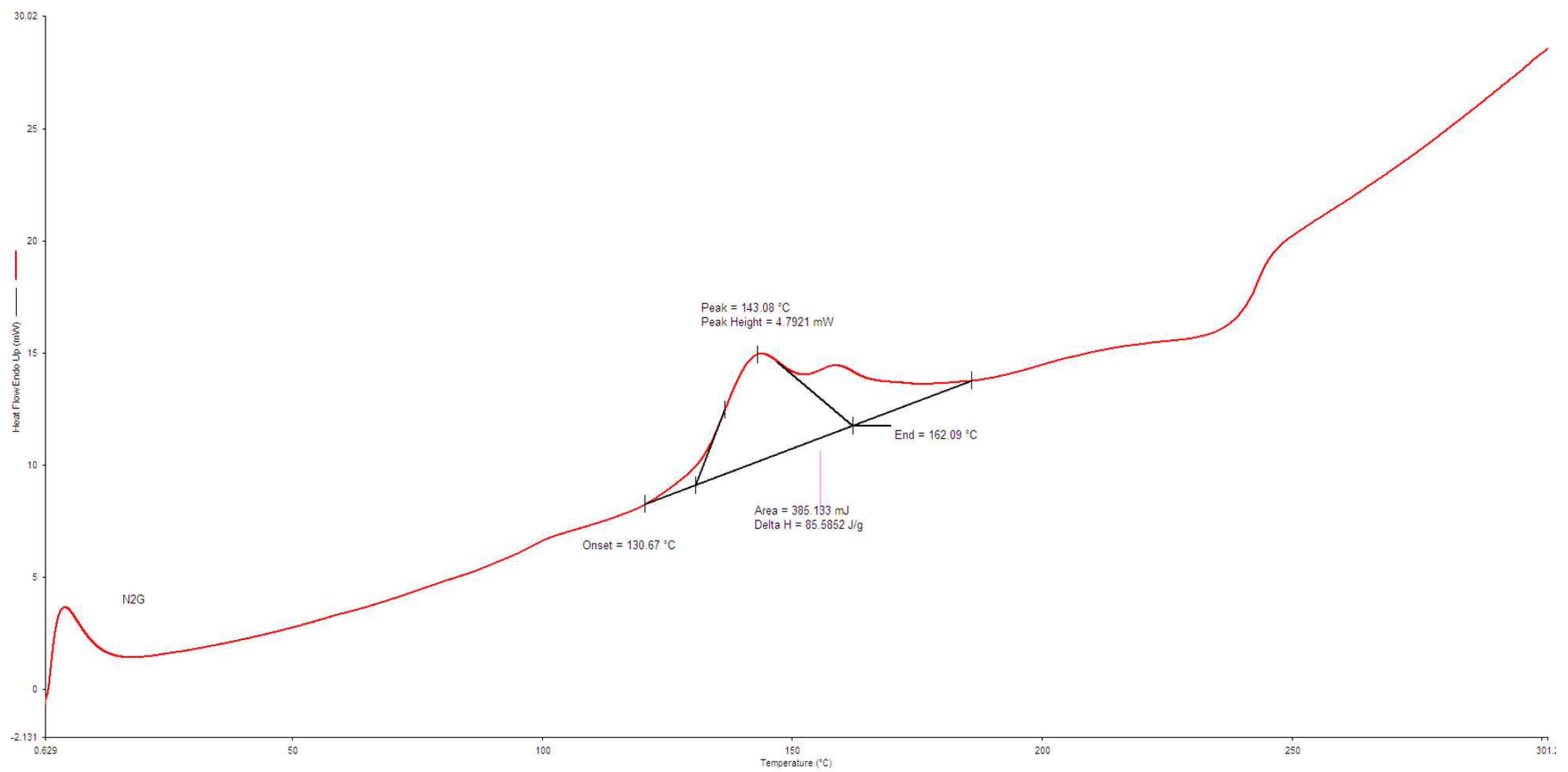
3.3. Characterization of Biosurfactant
3.4. Antimicrobial Activity of Biosurfactant
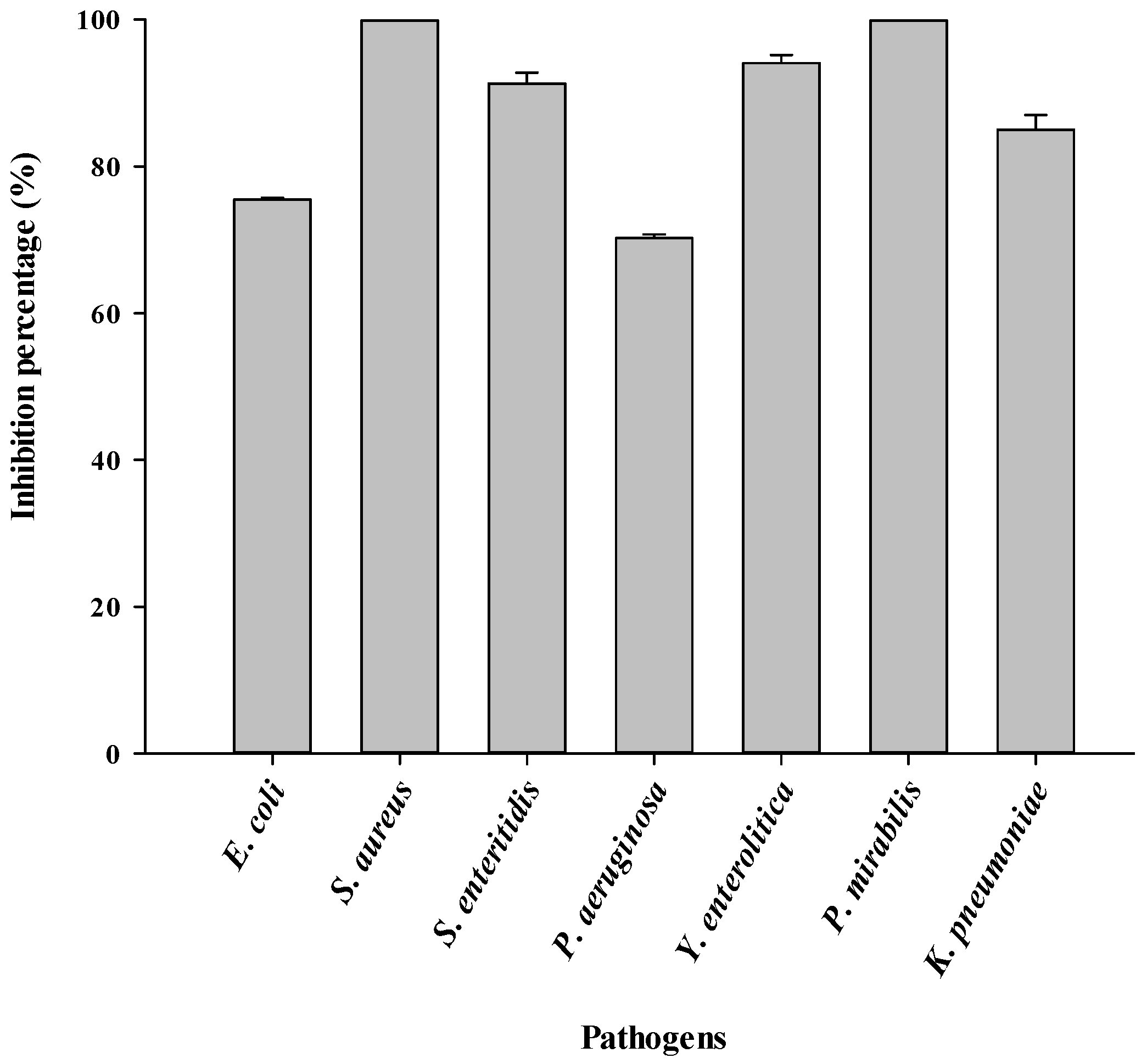
3.5. Antiadhesive Activity of Biosurfactant
3.6. Inhibition of Biofilm Formation by Biosurfactant
3.7. Eradication of Mature Biofilms by Biosurfactant
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mbassi, J.E.G.; Tsafack, A.L.S.; Maboune, A.S.; Eyenga, E.F.; Sophie, N.E.; Bongse, K.P. Quality evaluation of local Cameroonian mackerel (Scomber scombrus) processed by different methods. Int. J. Food Sci. Nutr. 2019, 4, 162–167. [Google Scholar]
- Ntsama, B.I.S.; Tambe, B.A.; Tsafack, T.J.J.; Medoua, G.N.; Kansci, G. Characteristics of fish farming practices and agrochemicals usage therein in four regions of Cameroon. Egypt. J. Aquat. Res. 2018, 44, 145–153. [Google Scholar] [CrossRef]
- Tsafack, T.J.J.; Gondam, K.M.; Yangoua, M.H.; Medjo, K.E.; Ntsama, B.S.I.; Tatfo, K.F.P.; Sasanya, J.; Medoua, N.G. Abridged validation of charm II screening tests for the detection of veterinary drug residues in fish farmed in Cameroon. Food Addit. Contam. Part A 2022, 39, 1691–1704. [Google Scholar] [CrossRef] [PubMed]
- Tsafack, T.J.J.; Tchuenchieu, A.D.K.; Mouafo, T.H.; Baomog, A.M.B.; Adjele, J.J.B.; Medjo, E.K.; Djuikoo, I.L.N.; Ndakoh, B.T.; Matchawe, C.; Sasanya, J.; et al. Microbial assessment and antibiotic susceptibility profile of bacterial fish isolates in an aquaculture production site in Mefou Afamba division of Cameroon. J. Environ. Sci. Eng. 2021, B10, 20–30. [Google Scholar] [CrossRef]
- Maffouo, S.T.M.; Mouafo, T.H.; Mouokeu, R.S.; Manet, L.; Tchuenchieu, K.A.D.; Simo, B.N.; Djeuachi, H.T.; Medoua, G.N.; Tchoumbougnang, F. Evaluation of sanitary risks associated with the consumption of street food in the city of Yaoundé (Cameroon): Case of braised fish from Mvog-Ada, Ngoa Ekélé, Simbock, Ahala and Olézoa. Heliyon 2021, 7, e07780. [Google Scholar] [CrossRef]
- Mouafo, T.H.; Adjele, B.J.J.; Hell, R.H.; Baomog, B.A.M.; Tchuenchieu, K.A.D.; Kamgnia, J.A.; Manet, L.; Bonny, P.; Baleba, M.R.M.; Medoua, G.N. Popular cleaning systems of bottles reused for traditional food packaging in the city of Yaoundé (Cameroon) and study of their prospective effectiveness on biofilms. Front. Food Sci. Technol. 2022, 2, 1060880. [Google Scholar] [CrossRef]
- Guan, C.; Zhang, W.; Su, J.; Li, F.; Chen, D.; Chen, X.; Huang, Y.; Gu, R.; Zhang, C. Antibacterial and antibiofilm potential of Lacticaseibacillus rhamnosus YT and its cell-surface extract. BMC Microbiol. 2023, 12, 12. [Google Scholar] [CrossRef]
- Carrascosa, C.; Raheem, D.; Ramos, F.; Saraiva, A.; Raposo, A. Microbial biofilms in the food industry—A comprehensive review. Int. J. Environ. Res. Public Health 2021, 18, 2014. [Google Scholar] [CrossRef]
- Mouafo, T.H.; Sokamte, T.A.; Mbawala, A.; Ndjouenkeu, R.; Somashekar, D. Biosurfactants from lactic acid bacteria: A critical review on production, extraction, structural characterization and food application. Food Biosci. 2022, 46, 101598. [Google Scholar] [CrossRef]
- Mouafo, T.H.; Mbawala, A.; Somashekar, D.; Tchougang, H.M.; Harohally, N.V.; Ndjouenkeu, R. Biological properties and structural characterization of a novel rhamnolipid like-biosurfactants produced by Lactobacillus casei subsp. casei TM1B. Biotechnol. Appl. Biochem. 2020, 68, 585–596. [Google Scholar] [CrossRef]
- Mouafo, T.H.; Baomog, A.M.B.; Adjele, J.J.B.; Sokamte, T.A.; Mbawala, A.; Ndjouenkeu, R. Microbial profile of fresh beef sold in the markets of Ngaoundéré, Cameroon, and antiadhesive activity of biosurfactants against selected bacterial pathogens. J. Food Qual. 2020, 2020, 5989428. [Google Scholar] [CrossRef]
- Patel, M.; Siddiqui, A.J.; Hamadou, W.S.; Surti, M.; Awadelkareem, A.M.; Ashraf, S.A.; Alreshidi, M.; Snoussi, M.; Rizvi, S.M.D.; Bardakci, F.; et al. Inhibition of Bacterial Adhesion and Antibiofilm Activities of a Glycolipid Biosurfactant from Lactobacillus rhamnosus with Its Physicochemical and Functional Properties. Antibiotics 2021, 10, 1546. [Google Scholar] [CrossRef]
- Santos, D.K.; Rufino, R.D.; Luna, J.M.; Santos, V.A.; Sarubbo, L.A. Biosurfactants: Multifunctional Biomolecules of the 21st Century. Int. J. Mol. Sci. 2016, 17, 401. [Google Scholar] [CrossRef] [Green Version]
- Mouafo, T.H.; Mbawala, A.; Tchougang, M.H.; Ndjouenkeu, R.; Somashekar, D. Application of response surface methodology to improve the production of antimicrobial biosurfactants by Lactobacillus paracasei subsp. tolerans N2 using sugar cane molasses as substrate. Bioresour. Bioprocess. 2018, 5, 48. [Google Scholar] [CrossRef] [Green Version]
- Cavalieri, S.J.; Rankin, I.D.; Harbeck, R.J.; Sautter, R.L.; McCarter, Y.S.; Sharp, S.E.; Ortez, J.H.; Spiegel, C.A. Manual of Antimicrobial Susceptibility Testing; American Society for Microbiology: Washington, DC, USA, 2005. [Google Scholar]
- CLSI (Clinical and Laboratory Standards Institute). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2020. [Google Scholar]
- Krumperman, P.H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983, 46, 165–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambanthamoorthy, K.; Feng, X.; Patel, R.; Patel, S.; Paranavitana, C. Antimicrobial and antibiofilm potential of biosurfactants isolated from lactobacilli against multi-drug-resistant pathogens. BMC Microbiol. 2014, 14, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamali, E.; Jamali, A.; Ardebili, A.; Ezadi, F.; Mohebbi, A. Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC Res. Notes 2020, 13, 27. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.; Hamilton, J.; Rebers, P.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Lobna, A.M.; Ahmed, A.A.A. Identification and characterization of biosurfactants produced by Rodococcus equi and Bacillus Methylotrophicus. J. Biol. Chem. Environ. Sci. 2013, 8, 341–358. [Google Scholar]
- AACC (American Association of Cereal Chemists). Crude Protein-Kjeldahl Method; AACC International Method 46-12.01; American Association of Cereal Chemists: Eagan, MN, USA, 1999; pp. 1–3. [Google Scholar]
- Gudiña, E.J.; Fernandes, E.C.; Teixeira, J.A.; Rodrigues, L.R. Antimicrobial and anti-adhesive activities of cell-bound biosurfactant from Lactobacillus agilis CCUG31450. RSC Adv. 2015, 5, 90960–90968. [Google Scholar] [CrossRef] [Green Version]
- Kim, G.; Xu, Y.J.; Farha, A.K.; Sui, Z.Q.; Corke, H. Bactericidal and antibiofilm properties of Rumex japonicus Houtt. on multidrug-resistant Staphylococcus aureus isolated from milk. J. Dairy Sci. 2022, 105, 2011–2024. [Google Scholar] [CrossRef] [PubMed]
- Pahane, M.M.; Mouafo, T.H.; Djuikoue, I.C.; Nyondo, C.; Tchoumbougnang, F.; Tatsadjieu, L.N.; Njintang, N.Y. Assessment of the microbiological quality of braised products from Bangangté city and antibiotic susceptibility of selected pathogens isolated therein. Asian J. Biotechnol. Bioresour. Technol. 2021, 7, 30–50. [Google Scholar] [CrossRef]
- Jain, N.; Jansone, I.; Obidenova, T.; Simanis, R.; Meisters, J.; Straupmane, D.; Reinis, A. Antimicrobial resistance in nosocomial isolates of Gram-negative bacteria: Public health implications in the Latvian context. Antibiotics 2021, 10, 791. [Google Scholar] [CrossRef]
- Rehman, A.; Patrick, W.M.; Lamont, I.L. Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: New approaches to an old problem. J. Med. Microbiol. 2019, 68, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Beganovic, M.; Luther, M.K.; Rice, L.B.; Arias, C.A.; Rybak, M.J.; LaPlante, K.L. A review of combination antimicrobial therapy for Enterococcus faecalis bloodstream infections and infective endocarditis. Clin. Infect. Dis. 2018, 67, 303–309. [Google Scholar] [CrossRef] [Green Version]
- Champney, W.S. Antibiotics targeting bacterial ribosomal subunit biogenesis. J. Antimicrob. Chemother. 2020, 75, 787–806. [Google Scholar] [CrossRef] [PubMed]
- Gaibani, P.; Giani, T.; Bovo, F.; Lombardo, D.; Amadesi, S.; Lazzarotto, T.; Coppi, M.; Rossolini, G.M.; Ambretti, S. Resistance to ceftazidime/avibactam, meropenem/vaborbactam and imipenem/relebactam in Gram-negative MDR bacilli: Molecular mechanisms and susceptibility testing. Antibiotics 2022, 11, 628. [Google Scholar] [CrossRef] [PubMed]
- Urban-Chmiel, R.; Marek, A.; Stepien-Pysniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria—A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Salem, S.S.; Badawy, M.S.E.M.; Al-Askar, A.A.; Arishi, A.A.; Elkady, F.M.; Hashem, A.H. Green biosynthesis of selenium nanoparticles using orange peel waste: Characterization, antibacterial and antibiofilm activities against multidrug-resistant bacteria. Life 2022, 12, 893. [Google Scholar] [CrossRef]
- Avila-Novoa, M.G.; Iñíguez-Moreno, M.; Solís-Velázquez, O.A.; González-Gomez, J.P.; Guerrero-Medina, P.J.; Gutiérrez-Lomelí, M.J. Biofilm formation by Staphylococcus aureus isolated from food contact surfaces in the dairy industry of Jalisco, Mexico. J. Food Qual. 2018, 2018, 1746139. [Google Scholar] [CrossRef] [Green Version]
- Idrees, M.; Sawant, S.; Karodia, N.; Rahman, A. Staphylococcus aureus Biofilm: Morphology, genetics, pathogenesis and treatment strategies. Int. J. Environ. Res. Public Health 2021, 18, 7602. [Google Scholar] [CrossRef]
- Bai, J.R.; Zhong, K.; Wu, Y.P.; Elena, G.; Gao, H. Antibiofilm activity of shikimic acid against Staphylococcus Aureus. Food Control 2019, 95, 327–333. [Google Scholar] [CrossRef]
- Ferreira, A.; Vecino, X.; Ferreira, D.; Cruz, J.M.; Moldes, A.B.; Rodrigues, L.R. Novel cosmetic formulations containing a biosurfactant from Lactobacillus paracasei. Colloids Surf. B Biointerfaces 2017, 155, 522–529. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Saharan, B.S.; Chauhan, N.; Procha, S.; Lal, S. Isolation and functional characterization of novel biosurfactant produced by Enterococcus faecium. SpringerPlus 2015, 4, 4. [Google Scholar] [CrossRef] [Green Version]
- Vecino, X.; Rodríguez-López, L.; Gudiña, E.J.; Cruz, J.M.; Moldes, A.B.; Rodrigues, L.R. Vineyard pruning waste as an alternative carbon source to produce novel biosurfactants by Lact. Paracasei. J. Ind. Eng. Chem. 2017, 55, 40–49. [Google Scholar] [CrossRef] [Green Version]
- Saravanakumari, P.; Mani, K. Structural characterization of a novel xylolipid biosurfactant from Lactococcus lactis and analysis of antibacterial activity against multi-drug resistant pathogens. Bioresour. Technol. 2010, 101, 8851–8854. [Google Scholar] [CrossRef]
- Hopkinson, R.J.; Leung, I.K.; Smart, T.J.; Rose, N.R.; Henry, L.W.; Claridge, T.D.; Schofield, C.J. Studies on the glutathione-dependent formaldehyde-activating enzyme from Paracoccus denitrificans. PLoS ONE 2015, 10, e0145085. [Google Scholar] [CrossRef] [Green Version]
- White, M.D.; Klecker, M.; Hopkinson, R.J.; Weits, D.A.; Mueller, C.; Naumann, C.; Neill, R.O.; Wickens, J.; Yang, J.; Brooks-bartlett, J.C.; et al. Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat. Commun. 2017, 8, 14690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, S.J.; Al-wahaibi, Y.M.; Al-bahry, S.N.; Elshafie, A.E.; Ennis, C.J. Production, characterization, and application of Bacillus licheniformis W16 biosurfactant in enhancing oil recovery. Front. Microbiol. 2016, 7, 1853. [Google Scholar] [CrossRef] [PubMed]
- Thies, S.; Rausch, S.C.; Kovacic, F.; Wilhelm, S.; Rosenau, F.; Daniel, R.; Streit, W.; Pietruszka, J.; Jaeger, K. Metagenomic discovery of novel enzymes and biosurfactants in a slaughterhouse biofilm microbial community. Sci. Rep. 2016, 6, 27035. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, R.; Ma, J.H.; Jiana, Y.K.; Che, J.N. Extracting keratin from wool by using l-cysteine. Green Chem. 2016, 18, 476–481. [Google Scholar] [CrossRef]
- Garcia-Vello, P.; Sharma, G.; Speciale, I.; Molinaro, A.; Im, S.H.; De Castro, C. Structural features and immunological perception of the cell surface glycans of Lactobacillus plantarum: A novel rhamnose-rich polysaccharide and teichoic acids. Carbohydr. Polym. 2020, 233, 115857. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Saharan, B.S.; Chauhan, N.; Bansal, A.; Procha, S. Production and structural characterization of Lactobacillus helveticus derived biosurfactant. Sci. World J. 2014, 2014, 493548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil, C.V.; Rebocho, A.T.; Esmail, A.; Sevrin, C.; Grandfils, C.; Torres, C.A.V.; Reis, M.A.M.; Freitas, F. Characterization of the thermostable biosurfactant produced by Burkholderia thailandensis DSM 13276. Polymers 2022, 14, 2088. [Google Scholar] [CrossRef]
- Bhadra, S.; Chettri, D.; Verma, A.K. Biosurfactants: Secondary metabolites involved in the process of bioremediation and biofilm removal. Appl. Biochem. Biotechnol. 2022, 1–27. [Google Scholar] [CrossRef]
- Mouafo, T.H.; Mbawala, A.; Ndjouenkeu, R. Effect of different carbon sources on biosurfactants’ production by three strains of Lactobacillus spp. Biomed Res. Int. 2018, 2018, 5034783. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Li, P.; Chen, X.; Zou, Y.; Li, H.; Yuan, G.; Hu, H. Activity of sodium lauryl sulfate, rhamnolipids, and n-acetylcysteine against biofilms of five common pathogens. Microb. Drug Resist. 2019, 26, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Neopane, P.; Nepal, H.P.; Shrestha, R.; Uehara, O.; Abiko, Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int. J. Gen. Med. 2018, 11, 25–32. [Google Scholar] [CrossRef]
- Cochis, A.; Fracchia, L.; Martinotti, M.G.; Rimondini, L. Biosurfactants prevent in vitro Candida albicans biofilm formation on resins and silicon materials for prosthetic devices. Oral Med. 2012, 113, 755–761. [Google Scholar] [CrossRef]
- Sharma, D.; Saharan, B.S. Functional characterization of biomedical potential of biosurfactant produced by Lactobacillus helveticus. Biotechnol. Rep. 2016, 11, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Biswa, P.; Doble, M. Production of acylated homoserine lactone by gram-positive bacteria isolated from marine water. FEMS Microbiol. Lett. 2013, 343, 34–41. [Google Scholar] [CrossRef] [Green Version]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.; Re, S.D.; Henry, N.; Fontaine, T.; Balestrino, D.; Latour-Lambert, P.; Ghigo, J.M. Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. USA 2006, 103, 12558–12563. [Google Scholar] [CrossRef] [PubMed]
- Bissong, M.E.A.; Ateba, C.N. Genotypic and phenotypic evaluation of biofilm production and antimicrobial resistance in Staphylococcus aureus isolated from milk, north west province, South Africa. Antibiotics 2020, 9, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Yasir, M.; Willcox, M.D.P.; Dutta, D. Action of antimicrobial peptides against bacterial biofilms. Materials 2018, 11, 2468. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Gu, S.; Cui, X.; Shi, Y.; Wen, S.; Chen, H.; Ge, J. Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microb. Pathog. 2019, 127, 12–20. [Google Scholar] [CrossRef]
- Oppenheimer-Shaanan, Y.; Steinberg, N.; Kolodkin-Gal, I. Small molecules are natural triggers for the disassembly of biofilms. Trends Microbiol. 2013, 21, 594–601. [Google Scholar] [CrossRef]
- Singh, A.K.; Sharma, P. Disinfectant-like activity of lipopeptide biosurfactant produced by Bacillus tequilensis strain SDS21. Colloids Surf. B Biointerfaces 2020, 185, 110514. [Google Scholar] [CrossRef]


| Pathogens | Antimicrobial Resistance Profile * | MAR Index |
|---|---|---|
| E. coli EM2 | AMP, AX, C, CXM, F, FX, GEN, IMP, NA, NOR, SXT, TE | 0.75 |
| S. enteritidis PE1 | AMC, AMP, C, CAZ, F, NOR, SXT, TE | 0.50 |
| S. aureus SA1 | AMC, AMP, AX, CAZ, CXM, F, FX, NA, NOR, TE | 0.62 |
| P. aeruginosa CT3 | AX, AMP, CIP, CXM, F, SXT, FX, IMP, NA, NOR, TE | 0.68 |
| Y. enterolitica MH5 | AMP, CAZ, ERY, NA, NOR, SXT, TE | 0.43 |
| P. mirabilis MR2 | AMP, AMC, C, CIP, NA, SXT | 0.37 |
| K. pneumoniae AG5 | AMC, AMP, AX, CAZ, NA, TE | 0.37 |
| Parameters | Composition | Retention Time (min) | Proportions (%) |
|---|---|---|---|
| Fatty acids | |||
| Lauric acid | 4.51 | 19.71 | |
| Myristic acid | 6.75 | 5.38 | |
| Palmitic acid | 9.38 | 27.90 | |
| 15-Methyl palmitic acid | 12.28 | 2.77 | |
| Oleic acid | 12.81 | 29.26 | |
| Linoleic acid | 14.01 | 4.26 | |
| γ-Homo linolenic acid | 18.39 | 2.89 | |
| Monosaccharides | |||
| Maltol | 4.34 | 2.50 | |
| 2-Deoxy-D-ribose, | 6.44 | 14.18 | |
| DL-Arabinose, | 8.85 | 2.05 | |
| D-Glucitol, 1,4-anhydro- | 9.22 | 47.70 | |
| 1,4-Anhydro-d-galactitol | 9.66 | 1.26 | |
| Methyl 2,4-di-O-acetyl-3,6-dideoxy-D-glucopyranoside | 9.95 | 12.51 |
| Pathogens | MIC (mg/mL) | MBC (mg/mL) | MBC/MIC |
|---|---|---|---|
| Escherichia coli EM2 | 7.50 | 10.00 | 1.33 |
| Staphylococcus aureus SA1 | 10.00 | 15.00 | 1.5 |
| Salmonella enteritidis PE1 | 2.50 | 5.00 | 2 |
| Pseudomonas aeruginosa CT3 | 7.50 | 10.00 | 1.33 |
| Yersinia enterolitica MH5 | 0.50 | 1.00 | 2 |
| Proteus mirabilis MR2 | 1.00 | 2.50 | 2.5 |
| Klebsiella pneumoniae AG5 | 5.00 | 7.50 | 1.5 |
| Pathogens | Biosurfactant Concentrations (mg/mL) | |||||
|---|---|---|---|---|---|---|
| 0 | 2.5 | 5.0 | 10.0 | 15.0 | 20.0 | |
| E. coli EM2 | 0.00 ± 0.00 aA | 10.25 ± 0.10 cB | 22.75 ± 1.50 eC | 41.20 ± 1.30 dD | 48.50 ± 0.77 dE | 62.10 ± 1.80 eF |
| S. aureus SA1 | 0.00 ± 0.00 aA | 15.25 ± 0.25 dB | 30.70 ± 1.00 fC | 45.63 ± 1.50 eD | 61.30 ± 2.50 eE | 81.90 ± 3.60 fF |
| S. enteritidis PE1 | 0.00 ± 0.00 aA | 1.50 ± 0.00 bB | 14.30 ± 0.00 dC | 25.80 ± 0.62 cD | 35.80 ± 1.09 cE | 55.90 ± 2.00 dF |
| P. aeruginosa CT3 | 0.00 ± 0.00 aA | 0.00 ± 0.00 aA | 2.90 ± 0.10 aB | 12.60 ± 0.26 aC | 26.70 ± 0.20 aD | 40.20 ± 1.60 aE |
| Y. enterolitica MH5 | 0.00 ± 0.00 aA | 18.30 ± 0.50 eB | 28.90 ± 2.10 fC | 47.30 ± 0.95 eD | 62.75 ± 0.74 eE | 80.25 ± 1.01 fF |
| P. mirabilis MR2 | 0.00 ± 0.00 aA | 0.00 ± 0.00 aA | 4.90 ± 0.40 bB | 15.50 ± 1.10 bC | 29.30 ± 0.00 bD | 42.80 ± 1.70 bE |
| K. pneumoniae AG5 | 0.00 ± 0.00 aA | 0.00 ± 0.00 aA | 7.50 ± 0.90 cB | 17.20 ± 0.85 bC | 30.90 ± 1.70 bD | 47.90 ± 0.10 cE |
| Pathogens | Biosurfactant Concentrations (mg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 5.0 | 10.0 | 20.0 | 30.0 | 40.0 | 50.0 | |
| E. coli EM2 | 0.00 ± 0.00 aA | 0.25 ± 0.01 cB | 3.15 ± 0.10 cC | 13.90 ± 0.10 bD | 28.50 ± 0.77 dE | 42.10 ± 0.10 cF | 60.50 ± 0.80 dG |
| S. aureus SA1 | 0.00 ± 0.00 aA | 2.25 ± 0.15 dB | 6.13 ± 0.40 dC | 25.93 ± 1.90 dD | 42.10 ± 2.50 eE | 58.90 ± 1.60 eF | 75.40 ± 2.10 eG |
| S. enteritidis PE1 | 0.00 ± 0.00 aA | 0.10 ± 0.01 bB | 3.10 ± 0.00 cC | 15.80 ± 0.12 cD | 25.70 ± 0.39 cE | 45.80 ± 1.00 dF | 59.90 ± 2.00 dG |
| P. aeruginosa CT3 | 0.00 ± 0.00 aA | 0.00 ± 0.00 aA | 0.90 ± 0.02 aB | 9.70 ± 0.36 aC | 20.40 ± 0.50 aD | 35.30 ± 0.60 aE | 50.60 ± 2.60 aF |
| Y. enterolitica MH5 | 0.00 ± 0.00 aA | 2.30 ± 0.10 dB | 8.20 ± 0.10 eC | 27.30 ± 2.25 dD | 42.25 ± 0.14 eE | 60.25 ± 2.01 eF | 78.15 ± 0.81 eG |
| P. mirabilis MR2 | 0.00 ± 0.00 aA | 0.00 ± 0.00 aA | 2.90 ± 0.20 bB | 13.80 ± 2.10 bcC | 22.30 ± 0.03 bD | 40.70 ± 0.30 bE | 54.50 ± 0.70 bF |
| K. pneumoniae AG5 | 0.00 ± 0.00 aA | 0.00 ± 0.00 aA | 3.10 ± 0.10 cB | 14.40 ± 1.84 bcC | 23.90 ± 0.10 bD | 39.10 ± 0.90 bE | 57.20 ± 0.50 cF |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mouafo, H.T.; Sokamte, A.T.; Manet, L.; Mbarga, A.J.M.; Nadezdha, S.; Devappa, S.; Mbawala, A. Biofilm Inhibition, Antibacterial and Antiadhesive Properties of a Novel Biosurfactant from Lactobacillus paracasei N2 against Multi-Antibiotics-Resistant Pathogens Isolated from Braised Fish. Fermentation 2023, 9, 646. https://doi.org/10.3390/fermentation9070646
Mouafo HT, Sokamte AT, Manet L, Mbarga AJM, Nadezdha S, Devappa S, Mbawala A. Biofilm Inhibition, Antibacterial and Antiadhesive Properties of a Novel Biosurfactant from Lactobacillus paracasei N2 against Multi-Antibiotics-Resistant Pathogens Isolated from Braised Fish. Fermentation. 2023; 9(7):646. https://doi.org/10.3390/fermentation9070646
Chicago/Turabian StyleMouafo, Hippolyte Tene, Alphonse Tegang Sokamte, Linda Manet, Arsene Joseph Manga Mbarga, Sachivkina Nadezdha, Somashekhar Devappa, and Augustin Mbawala. 2023. "Biofilm Inhibition, Antibacterial and Antiadhesive Properties of a Novel Biosurfactant from Lactobacillus paracasei N2 against Multi-Antibiotics-Resistant Pathogens Isolated from Braised Fish" Fermentation 9, no. 7: 646. https://doi.org/10.3390/fermentation9070646
APA StyleMouafo, H. T., Sokamte, A. T., Manet, L., Mbarga, A. J. M., Nadezdha, S., Devappa, S., & Mbawala, A. (2023). Biofilm Inhibition, Antibacterial and Antiadhesive Properties of a Novel Biosurfactant from Lactobacillus paracasei N2 against Multi-Antibiotics-Resistant Pathogens Isolated from Braised Fish. Fermentation, 9(7), 646. https://doi.org/10.3390/fermentation9070646







