Isolation and Characterization of Enterococcus faecium from Fermented Korean Soybean Paste with Antibacterial Effects
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, and Materials
2.2. Isolation of Probiotics from Fermented Food
2.3. 16s rRNA Sequencing for Identification of Probiotics
2.4. pH and Bile Tolerance of Probiotics
2.5. Co-Aggregation and Auto-Aggregation of Probiotics
2.6. Hydrophobicity of Probiotics
2.7. Antibiotic Susceptibility Assay
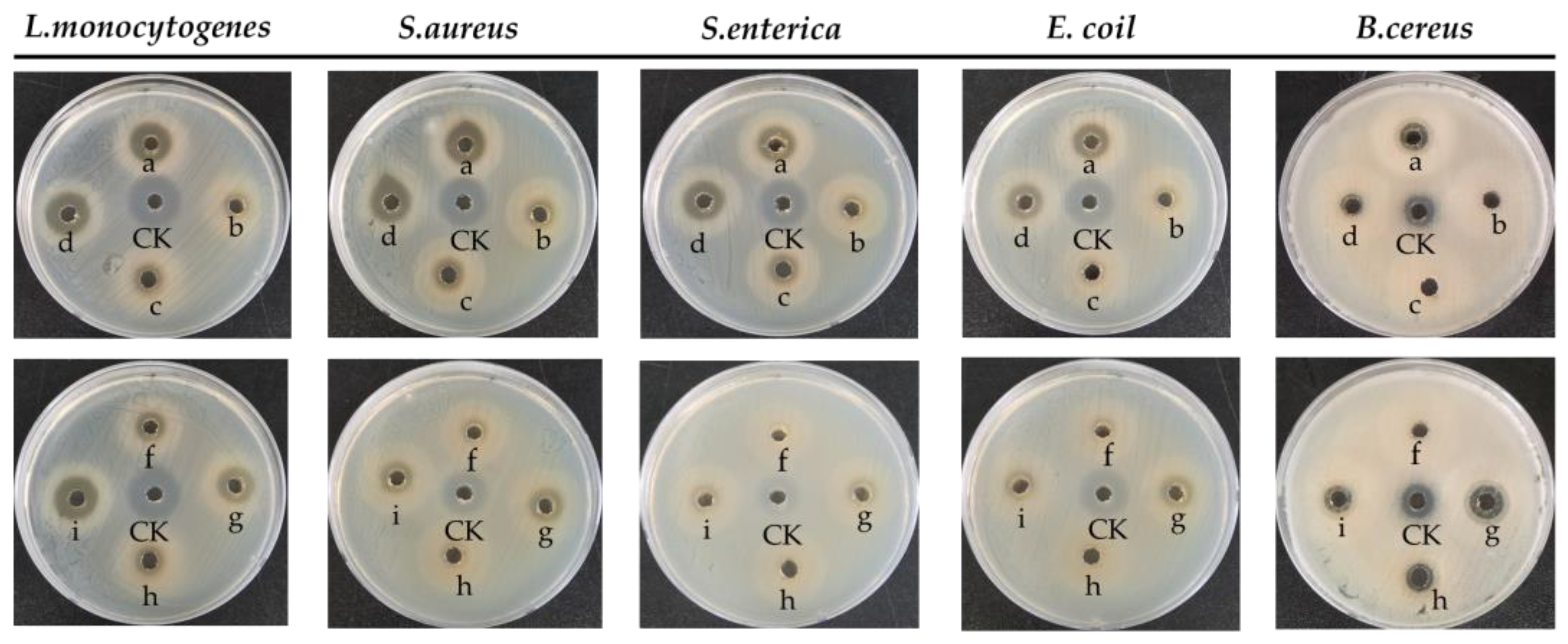
2.8. Antibacterial Activity
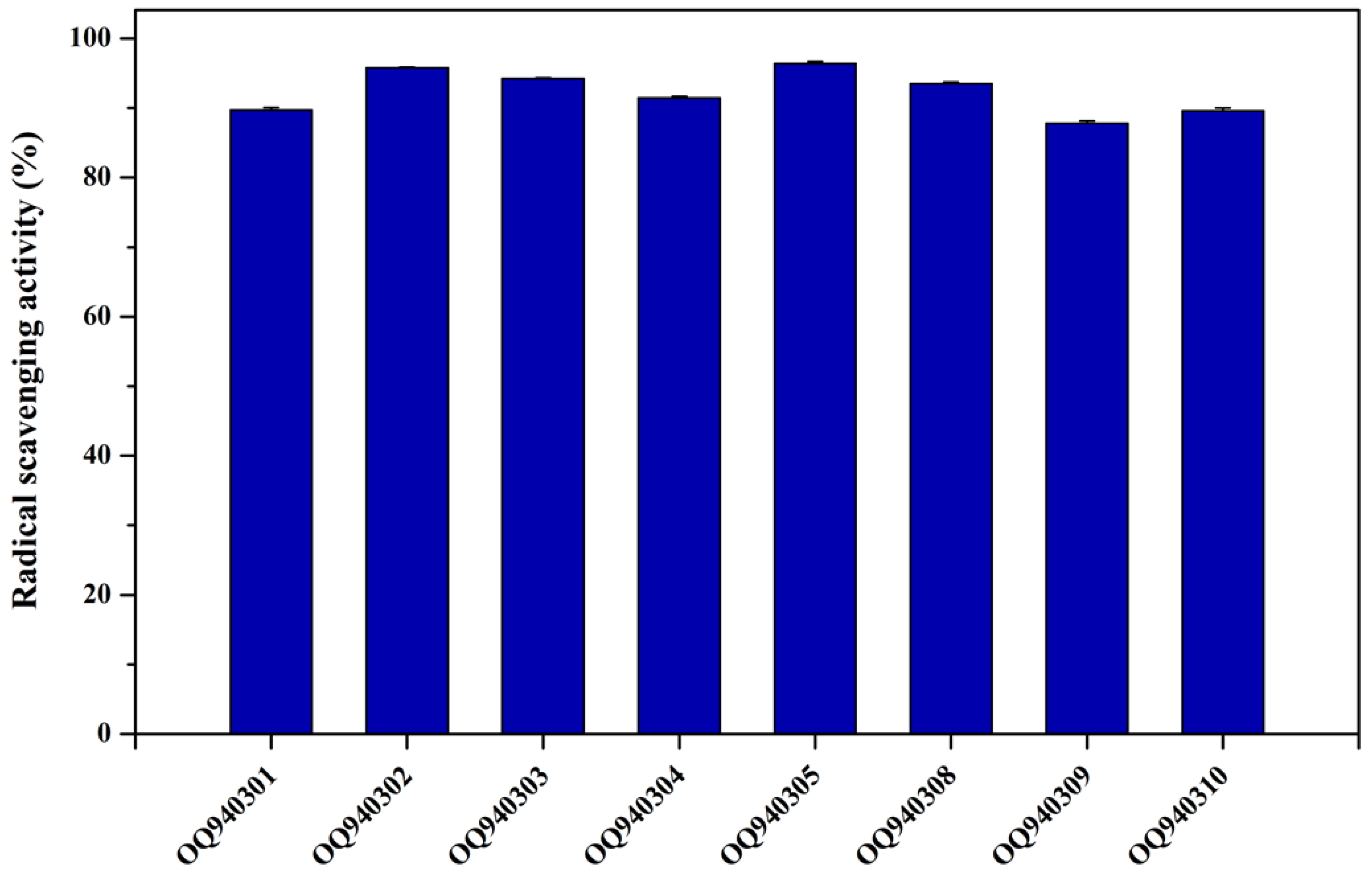
2.9. Antioxidant Activity
2.10. Hemolysis Assay
2.11. Statistical Analysis
3. Result and Discussion
3.1. Isolation and Identification of Probiotics
3.2. Resistance to Low pH and Bile Salt
3.3. Autoaggregation, Coaggregation, and Hydrophobicity Properties
3.4. Hemolytic Property
3.5. Antibiotic Susceptibility
3.6. Antibacterial Activity
3.7. Antioxidant Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gomes, A.C.; Bueno, A.A.; de Souza, R.G.M.; Mota, J.F. Gut microbiota, probiotics and diabetes. Nutr. J. 2014, 13, 60. [Google Scholar] [CrossRef] [Green Version]
- Dubois, A. Spiral bacteria in the human stomach: The gastric helicobacters. Emerg. Infect. Dis. 1995, 1, 79–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salminen, S.; Bouley, C.; Boutron, M.-C.; Cummings, J.H.; Franck, A.; Gibson, G.R.; Isolauri, E.; Moreau, M.-C.; Roberfroid, M.; Rowland, I. Functional food science and gastrointestinal physiology and function. Br. J. Nutr. 1998, 80 (Suppl. S1), S147–S171. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, M.K.; Giri, S.K. Probiotic functional foods: Survival of probiotics during processing and storage. J. Funct. Foods 2014, 9, 225–241. [Google Scholar] [CrossRef]
- Kawai, K.; Kamochi, R.; Oiki, S.; Murata, K.; Hashimoto, W. Probiotics in human gut microbiota can degrade host glycosaminoglycans. Sci. Rep. 2018, 8, 10674. [Google Scholar] [CrossRef] [Green Version]
- Beena Divya, J.; Varsha, K.K.; Nampoothiri, K.M.; Ismail, B.; Pandey, A. Probiotic fermented foods for health benefits. Eng. Life Sci. 2012, 12, 377–390. [Google Scholar] [CrossRef]
- Patangia, D.V.; Ryan, C.A.; Dempsey, E.; Ross, R.P.; Stanton, C. Impact of antibiotics on the human microbiome and consequences for host health. MicrobiologyOpen 2022, 11, e1260. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.L.; Chen, T.; El-Nezami, H.; Savidge, T.C. Food ingredients in human health: Ecological and metabolic perspectives implicating gut microbiota function. Trends Food Sci. Technol. 2020, 100, 103–117. [Google Scholar] [CrossRef]
- Wolfe, W.; Xiang, Z.; Yu, X.; Li, P.; Chen, H.; Yao, M.; Fei, Y.; Huang, Y.; Yin, Y.; Xiao, H. The Challenge of Applications of Probiotics in Gastrointestinal Diseases. Adv. Gut Microbiome Res. 2023, 2023, 1–10. [Google Scholar] [CrossRef]
- Liang, D.; Wu, F.; Zhou, D.; Tan, B.; Chen, T. Commercial probiotic products in public health: Current status and potential limitations. Crit. Rev. Food Sci. Nutr. 2023, 1–22. [Google Scholar] [CrossRef]
- Harahap, I.A.; Suliburska, J. Can probiotics decrease the risk of postmenopausal osteoporosis in women? PharmaNutrition 2023, 24, 100336. [Google Scholar] [CrossRef]
- Sathiyaseelan, A.; Saravanakumar, K.; Han, K.; Naveen, K.V.; Wang, M.-H. Antioxidant and Antibacterial Effects of Potential Probiotics Isolated from Korean Fermented Foods. Int. J. Mol. Sci. 2022, 23, 10062. [Google Scholar] [CrossRef]
- Shin, D.; Chang, S.Y.; Bogere, P.; Won, K.; Choi, J.-Y.; Choi, Y.-J.; Lee, H.K.; Hur, J.; Park, B.-Y.; Kim, Y.; et al. Beneficial roles of probiotics on the modulation of gut microbiota and immune response in pigs. PLoS ONE 2019, 14, e0220843. [Google Scholar] [CrossRef] [Green Version]
- Won, G.; Choi, S.-I.; Park, N.; Kim, J.-E.; Kang, C.-H.; Kim, G.-H. In Vitro Antidiabetic, Antioxidant Activity, and Probiotic Activities of Lactiplantibacillus plantarum and Lacticaseibacillus paracasei Strains. Curr. Microbiol. 2021, 78, 3181–3191. [Google Scholar] [CrossRef]
- Grimoud, J.; Durand, H.; de Souza, S.; Monsan, P.; Ouarné, F.; Theodorou, V.; Roques, C. In vitro screening of probiotics and synbiotics according to anti-inflammatory and anti-proliferative effects. Int. J. Food Microbiol. 2010, 144, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Jafari-Nasab, T.; Khaleghi, M.; Farsinejad, A.; Khorrami, S. Probiotic potential and anticancer properties of Pediococcus sp. isolated from traditional dairy products. Biotechnol. Rep. 2021, 29, e00593. [Google Scholar] [CrossRef]
- Lee, N.K.; Kim, S.-Y.; Han, K.J.; Eom, S.J.; Paik, H.-D. Probiotic potential of Lactobacillus strains with anti-allergic effects from kimchi for yogurt starters. LWT-Food Sci. Technol. 2014, 58, 130–134. [Google Scholar] [CrossRef]
- Courvalin, P. Antibiotic resistance: The pros and cons of probiotics. Dig. Liver Dis. 2006, 38 (Suppl. S2), S261–S265. [Google Scholar] [CrossRef]
- Gueimonde, M.; Sánchez, B.; de Los Reyes-Gavilán, C.G.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef] [Green Version]
- Sharma, P.; Tomar, S.K.; Goswami, P.; Sangwan, V.; Singh, R. Antibiotic resistance among commercially available probiotics. Food Res. Int. 2014, 57, 176–195. [Google Scholar] [CrossRef]
- Kareem, K.Y.; Ling, F.H.; Chwen, L.T.; Foong, O.M.; Asmara, S.A. Inhibitory activity of postbiotic produced by strains of Lactobacillus plantarum using reconstituted media supplemented with inulin. Gut. Pathog. 2014, 6, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraffa, G.; Carminati, D.; Neviani, E. Enterococci Isolated from Dairy Products: A Review of Risks and Potential Technological Use. J. Food Prot. 1997, 60, 732–738. [Google Scholar] [CrossRef] [PubMed]
- Giraffa, G. Enterococci from foods. FEMS Microbiol. Rev. 2002, 26, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.M.; Huch, M.; Abriouel, H.; Holzapfel, W.; Gálvez, A. Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 2011, 151, 125–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, M.F.; Sarantinopoulos, P.; Tsakalidou, E.; De Vuyst, L. The role and application of enterococci in food and health. Int. J. Food Microbiol. 2006, 106, 1–24. [Google Scholar] [CrossRef]
- Park, S.; Saravanakumar, K.; Sathiyaseelan, A.; Park, S.; Hu, X.; Wang, M.-H. Cellular antioxidant properties of nontoxic exopolysaccharide extracted from Lactobacillales (Weissella cibaria) isolated from Korean kimchi. LWT 2022, 154, 112727. [Google Scholar] [CrossRef]
- Ashraf, R.; Shah, N.P. Selective and differential enumerations of Lactobacillus delbrueckii subsp. bulgaricus, Streptococcus thermophilus, Lactobacillus acidophilus, Lactobacillus casei and Bifidobacterium spp. in yoghurt—A review. Int. J. Food Microbiol. 2011, 149, 194–208. [Google Scholar] [CrossRef]
- Mohammed, S.; Con, A.H. Isolation and characterization of potential probiotic lactic acid bacteria from traditional cheese. LWT-Food Sci. Technol. 2021, 152, 112319. [Google Scholar] [CrossRef]
- Vlkova, E.; Rada, V.; Šmehilová, M.; Killer, J. Auto-aggregation and co-aggregation ability in bifidobacteria and clostridia. Folia Microbiol. 2008, 53, 263–269. [Google Scholar] [CrossRef]
- Krausova, G.; Hyrslova, I.; Hynstova, I. In Vitro Evaluation of Adhesion Capacity, Hydrophobicity, and Auto-Aggregation of Newly Isolated Potential Probiotic Strains. Fermentation 2019, 5, 100. [Google Scholar] [CrossRef] [Green Version]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K.; on behalf of the CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing. CLSI Methods Development and Standardization Working Group Best Practices for Evaluation of Antimicrobial Susceptibility Tests. J. Clin. Microbiol. 2018, 56, e01934-17. [Google Scholar] [CrossRef] [Green Version]
- Plessas, S.; Nouska, C.; Karapetsas, A.; Kazakos, S.; Alexopoulos, A.; Mantzourani, I.; Chondrou, P.; Fournomiti, M.; Galanis, A.; Bezirtzoglou, E. Isolation, characterization and evaluation of the probiotic potential of a novel Lactobacillus strain isolated from Feta-type cheese. Food Chem. 2017, 226, 102–108. [Google Scholar] [CrossRef]
- Rojo-Bezares, B.; Sáenz, Y.; Poeta, P.; Zarazaga, M.; Ruiz-Larrea, F.; Torres, C. Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. Int. J. Food Microbiol. 2006, 111, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Soundharrajan, I.; Yoon, Y.H.; Muthusamy, K.; Jung, J.-S.; Lee, H.J.; Han, O.-K.; Choi, K.C. Isolation of Lactococcus lactis from Whole Crop Rice and Determining Its Probiotic and Antimicrobial Properties towards Gastrointestinal Associated Bacteria. Microorganisms 2021, 9, 2513. [Google Scholar] [CrossRef]
- Divyashree, S.; Anjali, P.; Somashekaraiah, R.; Sreenivasa, M. Probiotic properties of Lactobacillus casei—MYSRD 108 and Lactobacillus plantarum-MYSRD 71 with potential antimicrobial activity against Salmonella paratyphi. Biotechnol. Rep. 2021, 32, e00672. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Y.; Jin, M.-M.; Meng, J.; Gao, S.-M.; Lu, R.-R. Exopolysaccharide from Lactobacillus planterum LP6: Antioxidation and the effect on oxidative stress. Carbohydr. Polym. 2013, 98, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Sathiyaseelan, A.; Park, S.; Saravanakumar, K.; Mariadoss, A.V.A.; Wang, M.-H. Evaluation of phytochemicals, antioxidants, and antidiabetic efficacy of various solvent fractions of Gynura procumbens (Lour.) Merr. Process Biochem. 2021, 111, 51–62. [Google Scholar] [CrossRef]
- Mangia, N.P.; Saliba, L.; Deiana, P. Functional and safety characterization of autochthonous Lactobacillus paracasei FS103 isolated from sheep cheese and its survival in sheep and cow fermented milks during cold storage. Ann. Microbiol. 2019, 69, 161–170. [Google Scholar] [CrossRef]
- Al Atya, A.K.; Drider-Hadiouche, K.; Ravallec, R.; Silvain, A.; Vachee, A.; Drider, D. Probiotic potential of Enterococcus faecalis strains isolated from meconium. Front. Microbiol. 2015, 6, 227. [Google Scholar] [CrossRef]
- Gaaloul, N.; Ben Braiek, O.; Berjeaud, J.M.; Arthur, T.; Cavera, V.L.; Chikindas, M.L.; Hani, K.; Ghrairi, T. Evaluation of Antimicrobial Activity and Safety Aspect of E nterococcus Italicus GGN 10 Strain Isolated from T unisian Bovine Raw Milk. J. Food Saf. 2014, 34, 300–311. [Google Scholar] [CrossRef]
- Pieniz, S.; Andreazza, R.; Okeke, B.C.; Camargo, F.A.O.; Brandelli, A. Antimicrobial and antioxidant activities of Enterococcus species isolated from meat and dairy products. Braz. J. Biol. 2015, 75, 923–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamang, J.P.; Watanabe, K.; Holzapfel, W. Review: Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraffa, G. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 2003, 88, 215–222. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Malik, R.K.; Chauhan, P. Functional and safety aspects of enterococci in dairy foods. Indian J. Microbiol. 2008, 48, 317–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Villaluenga, C.; Torino, M.I.; Martín, V.; Arroyo, R.; Garcia-Mora, P.; Estrella Pedrola, I.; Vidal-Valverde, C.; Rodriguez, J.M.; Frias, J. Multifunctional Properties of Soy Milk Fermented by Enterococcus faecium Strains Isolated from Raw Soy Milk. J. Agric. Food Chem. 2012, 60, 10235–10244. [Google Scholar] [CrossRef]
- Beasley, D.E.; Koltz, A.M.; Lambert, J.E.; Fierer, N.; Dunn, R.R. The Evolution of Stomach Acidity and Its Relevance to the Human Microbiome. PLoS ONE 2015, 10, e0134116. [Google Scholar] [CrossRef]
- Hofmann, A.F.; Mysels, K.J. Bile acid solubility and precipitation in vitro and in vivo: The role of conjugation, pH, and Ca2+ ions. J. Lipid Res. 1992, 33, 617–626. [Google Scholar] [CrossRef]
- Strompfova, V.; Laukova, A. In vitro study on bacteriocin production of Enterococci associated with chickens. Anaerobe 2007, 13, 228–237. [Google Scholar] [CrossRef]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140–164. [Google Scholar] [CrossRef]
- Collado, M.C.; Meriluoto, J.; Salminen, S. Measurement of aggregation properties between probiotics and pathogens: In vitro evaluation of different methods. J. Microbiol. Methods 2007, 71, 71–74. [Google Scholar] [CrossRef]
- Van Loosdrecht, M.C.; Lyklema, J.; Norde, W.; Schraa, G.; Zehnder, A.J. The role of bacterial cell wall hydrophobicity in adhesion. Appl. Environ. Microbiol. 1987, 53, 1893–1897. [Google Scholar] [CrossRef] [PubMed]
- Maragkoudakis, P.A.; Zoumpopoulou, G.; Miaris, C.; Kalantzopoulos, G.; Pot, B.; Tsakalidou, E. Probiotic potential of Lactobacillus strains isolated from dairy products. Int. Dairy J. 2006, 16, 189–199. [Google Scholar] [CrossRef]
- Curragh, H.J.; Collins, M. High levels of spontaneous drug resistance in Lactobacillus. J. Appl. Bacteriol. 1992, 73, 31–36. [Google Scholar] [CrossRef]
- Nunziata, L.; Brasca, M.; Morandi, S.; Silvetti, T. Antibiotic resistance in wild and commercial non-enterococcal Lactic Acid Bacteria and Bifidobacteria strains of dairy origin: An update. Food Microbiol. 2022, 104, 103999. [Google Scholar] [CrossRef]
- Temmerman, R.; Pot, B.; Huys, G.; Swings, J. Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int. J. Food Microbiol. 2003, 81, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kunyeit, L.; Rao, R.P.; Anu-Appaiah, K.A. Yeasts originating from fermented foods, their potential as probiotics and therapeutic implication for human health and disease. Crit. Rev. Food Sci. Nutr. 2023, 1–12. [Google Scholar] [CrossRef]
- Sarantinopoulos, P.; Leroy, F.; Leontopoulou, E.; Georgalaki, M.D.; Kalantzopoulos, G.; Tsakalidou, E.; De Vuyst, L. Bacteriocin production by Enterococcus faecium FAIR-E 198 in view of its application as adjunct starter in Greek Feta cheese making. Int. J. Food Microbiol. 2002, 72, 125–136. [Google Scholar] [CrossRef]
- De Kwaadsteniet, M.; Todorov, S.; Knoetze, H.; Dicks, L. Characterization of a 3944 Da bacteriocin, produced by Enterococcus mundtii ST15, with activity against Gram-positive and Gram-negative bacteria. Int. J. Food Microbiol. 2005, 105, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Giraffa, G. Enterococcal bacteriocins: Their potential as anti-Listeria factors in dairy technology. Food Microbiol. 1995, 12, 291–299. [Google Scholar] [CrossRef]
- De Vuyst, L.; Moreno, M.F.; Revets, H. Screening for enterocins and detection of hemolysin and vancomycin resistance in enterococci of different origins. Int. J. Food Microbiol. 2003, 84, 299–318. [Google Scholar] [CrossRef]
- Rakhmanova, A.; Khan, Z.A.; Shah, K. A mini review fermentation and preservation: Role of lactic acid bacteria. MOJ Food Process Technol. 2018, 6, 414–417. [Google Scholar] [CrossRef]
- Rezaee, P.; Kermanshahi, R.K.; Falsafi, T. Antibacterial activity of lactobacilli probiotics on clinical strains of Helicobacter pylori. Iran J. Basic Med. Sci. 2019, 22, 1118–1124. [Google Scholar] [CrossRef] [PubMed]
- Fijan, S. Probiotics and Their Antimicrobial Effect. Microorganisms 2023, 11, 528. [Google Scholar] [CrossRef]
- Aruoma, O.I. Free radicals, oxidative stress, and antioxidants in human health and disease. J. Am. Oil Chem. Soc. 1998, 75, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wu, H.; Gao, L.; Jia, H.; Zhang, Y.; Cui, Z.; Li, Y. Effects of Lactobacillus curvatus and Leuconostoc mesenteroides on Suan Cai Fermentation in Northeast China. J. Microbiol. Biotechnol. 2016, 26, 2148–2158. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Zhang, B.; Liu, C.; Zhou, H.; Wang, X.; Mai, K.; He, G. Effects of dietary raw or Enterococcus faecium fermented soybean meal on growth, antioxidant status, intestinal microbiota, morphology, and inflammatory responses in turbot (Scophthalmus maximus L.). Fish Shellfish. Immunol. 2020, 100, 261–271. [Google Scholar] [CrossRef]
- Abdhul, K.; Ganesh, M.; Shanmughapriya, S.; Kanagavel, M.; Anbarasu, K.; Natarajaseenivasan, K. Antioxidant activity of exopolysaccharide from probiotic strain Enterococcus faecium (BDU7) from Ngari. Int. J. Biol. Macromol. 2014, 70, 450–454. [Google Scholar] [CrossRef]
- Zahrani, A.J.A.; Shori, A.B. Viability of probiotics and antioxidant activity of soy and almond milk fermented with selected strains of probiotic Lactobacillus spp. LWT-Food Sci. Technol. 2023, 176, 114531. [Google Scholar] [CrossRef]


| Isolate Code | Similarity-Based on 16S rRNA | Sequence Length (bp) | GenBank Accession No. |
|---|---|---|---|
| So001 | E. faecium | 826 | OQ940301 |
| So002 | E. faecium | 827 | OQ940302 |
| So003 | E. faecium | 834 | OQ940303 |
| So004 | E. faecium | 838 | OQ940304 |
| So005 | E. faecium | 836 | OQ940305 |
| So008 | E. faecium | 838 | OQ940308 |
| So009 | E. faecium | 838 | OQ940309 |
| So010 | E. faecium | 838 | OQ940310 |
| Cell Viability (log10 CFU/mL) | ||||
|---|---|---|---|---|
| Isolates | Tolerance to Bile Salt | Tolerance to Low pH | ||
| Control | Bile Salt | 0 h | 3 h | |
| OQ940301 | 5.34 ± 0.20 | 5.19 ± 0.02 | 5.66 ± 0.05 | 5.49 ± 0.04 |
| OQ940302 | 5.41 ± 0.18 | 5.24 ± 0.08 | 5.68 ± 0.02 | 5.57 ± 0.09 |
| OQ940303 | 5.43 ± 0.12 | 5.32 ± 0.03 | 5.63 ± 0.01 | 5.54 ± 0.05 |
| OQ940304 | 5.44 ± 0.31 | 5.18 ± 0.05 | 5.62 ± 0.05 | 5.52 ± 0.1 |
| OQ940305 | 5.2 ± 0.28 | 5.06 ± 0.09 | 5.58 ± 0.04 | 5.58 ± 0.03 |
| OQ940308 | 5.34 ± 0.16 | 5.22 ± 0.02 | 5.50 ± 0.05 | 5.59 ± 0.03 |
| OQ940309 | 5.06 ± 0.39 | 4.81 ± 0.03 | 5.62 ± 0.06 | 5.51 ± 0.04 |
| OQ940310 | 4.7 ± 0.51 | 4.15 ± 0.27 | 5.51 ± 0.05 | 5.56 ± 0.05 |
| Strain | Auto-Aggregation (%) | Co-Aggregation (%) | Co-Aggregation (%) | Hydrophobicity (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Salmonella enterica | Staphylococcus aureus | |||||||||
| 1 h | 3 h | 5 h | 1 h | 3 h | 5 h | 1 h | 3 h | 5 h | ||
| OQ940301 | 5.07 ± 2.23 | 15.72 ± 2.87 | 16.16 ± 0.06 | 14.68 ± 4.06 | 23.62 ± 5.89 | 28.10 ± 4.02 | 10.39 ± 2.96 | 23.29 ± 5.1 | 24.19 ± 0.84 | 93.99 ± 0.09 |
| OQ940302 | 12.91 ± 4.65 | 18.18 ± 7.89 | 25.39 ± 3.58 | 9.32 ± 1.17 | 12.77 ± 0.79 | 24.84 ± 1.77 | 7.46 ± 2.84 | 24.09 ± 5.55 | 20.87 ± 5.32 | 94.31 ± 0.04 |
| OQ940303 | 8.9 ± 4.02 | 13.44 ± 3.85 | 18.73 ± 1.44 | 6.07 ± 1.04 | 14.73 ± 0.25 | 21.29 ± 0.66 | 10.57 ± 4.07 | 16.51 ± 1.49 | 24.93 ± 1.08 | 94.49 ± 0.0 |
| OQ940304 | 9.84 ± 5.41 | 16.14 ± 7.22 | 18.28 ± 8.9 | 5.22 ± 1.23 | 9.22 ± 1.66 | 20.38 ± 5.97 | 4.29 ± 3.0 | 12.71 ± 0.59 | 22.16 ± 0.91 | 94.83 ± 0.09 |
| OQ940305 | 6.32 ± 3.19 | 17.08 ± 8.23 | 20.78 ± 1.57 | 5.72 ± 1.69 | 9.22 ± 5.71 | 17.93 ± 3.39 | 7.3 ± 1.15 | 12.28 ± 0.4 | 23.23 ± 1.9 | 95.32 ± 0.03 |
| OQ940308 | 7.7 ± 2.52 | 18.84 ± 2.98 | 20.16 ± 2.95 | 4.15 ± 1.69 | 9.82 ± 4.69 | 14.32 ± 4.07 | 11.65 ± 1.96 | 6.95 ± 9.48 | 12.66 ± 6.63 | 94.99 ± 0.06 |
| OQ940309 | 13.67 ± 7.44 | 25.74 ± 5.54 | 24.52 ± 6.21 | 18.01 ± 7.79 | 18.33 ± 0.1 | 27.62 ± 3.46 | 13.06 ± 5.62 | 15.67 ± 0.85 | 25.62 ± 1.62 | 94.84 ± 0.04 |
| OQ940310 | 10.54 ± 4.65 | 15.42 ± 5.43 | 24.43 ± 9.9 | 9.78 ± 1.7 | 10.58 ± 1.16 | 23.57 ± 3.41 | 7.68 ± 2.35 | 8.6 ± 0.46 | 18.04 ± 2.43 | 94.66 ± 0.04 |
| Zone of Inhibition (mm) | ||||||||
|---|---|---|---|---|---|---|---|---|
| OQ940301 | OQ940302 | OQ940303 | OQ940304 | OQ940305 | OQ940308 | OQ940309 | OQ940310 | |
| Tetracycline (30 μg) | 3.73 ± 0.25 | 2.9 ± 1.42 | 3.87 ± 1.15 | 3.7 ± 0.14 | 3.1 ± 0.26 | 3.87 ± 0.51 | 2.97 ± 0.55 | 2.83 ± 1.26 |
| Vancomycin (30 μg) | 2.20 | 2.00 | 1.7 ± 0.4 | 2.20 | 1.8 ± 0.17 | 2.1 ± 0.14 | 1.83 ± 0.29 | 0.00 |
| Erythromycin (15 μg) | 2.45 ± 1.48 | 0.00 | 1.50 | 2.27 ± 1.33 | 0.00 | 1.50 | 1.30 | 2.4 ± 1.56 |
| Gentamycin (10 μg) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Ampicillin (10 μg) | 3.17 ± 0.58 | 2.9 ± 0.95 | 3.2 ± 0.36 | 3.03 ± 0.06 | 2.97 ± 0.47 | 3.07 ± 0.31 | 2.87 ± 0.4 | 3.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, K.; Park, S.; Sathiyaseelan, A.; Wang, M.-H. Isolation and Characterization of Enterococcus faecium from Fermented Korean Soybean Paste with Antibacterial Effects. Fermentation 2023, 9, 760. https://doi.org/10.3390/fermentation9080760
Han K, Park S, Sathiyaseelan A, Wang M-H. Isolation and Characterization of Enterococcus faecium from Fermented Korean Soybean Paste with Antibacterial Effects. Fermentation. 2023; 9(8):760. https://doi.org/10.3390/fermentation9080760
Chicago/Turabian StyleHan, Kiseok, Soyoung Park, Anbazhagan Sathiyaseelan, and Myeong-Hyeon Wang. 2023. "Isolation and Characterization of Enterococcus faecium from Fermented Korean Soybean Paste with Antibacterial Effects" Fermentation 9, no. 8: 760. https://doi.org/10.3390/fermentation9080760
APA StyleHan, K., Park, S., Sathiyaseelan, A., & Wang, M.-H. (2023). Isolation and Characterization of Enterococcus faecium from Fermented Korean Soybean Paste with Antibacterial Effects. Fermentation, 9(8), 760. https://doi.org/10.3390/fermentation9080760







