Exploring the Potential of Non-Conventional Yeasts in Wine Fermentation with a Focus on Saccharomycopsis fermentans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Media
2.2. Trifluoro-Leucine (TFL)-Resistant Strain Selection
2.3. PCR Amplification and Sequencing of LEU4 Amplicons
2.4. Fermentation Conditions
2.5. Analytical Methods of Must Analysis
3. Results
3.1. Verification of Obtained TFL-Resistant Mutants
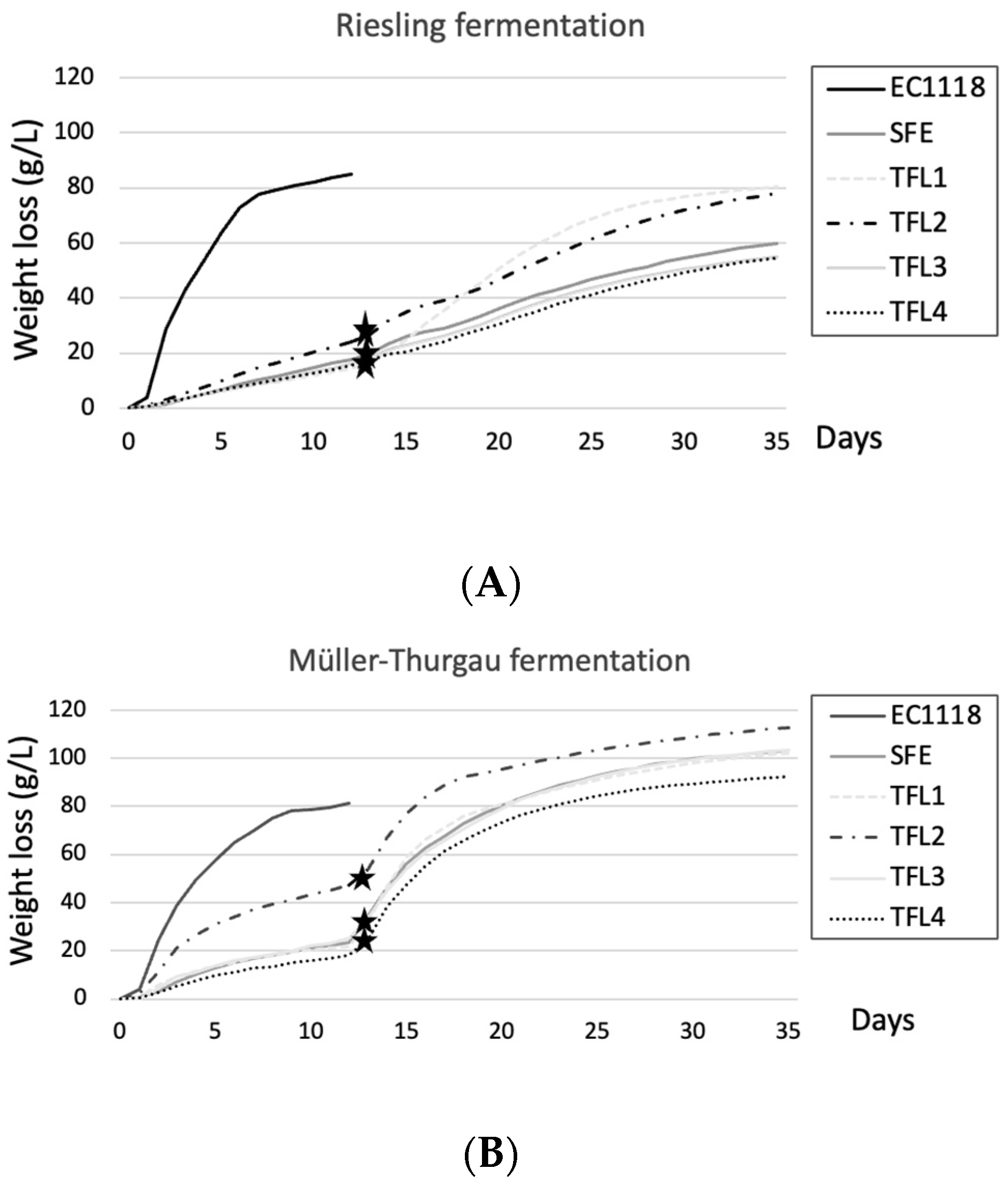
3.2. Fermentation Performance
3.3. Amino Acid Concentrations after Single Fermentation of Riesling Must
3.4. Aroma Analysis of Fermentations
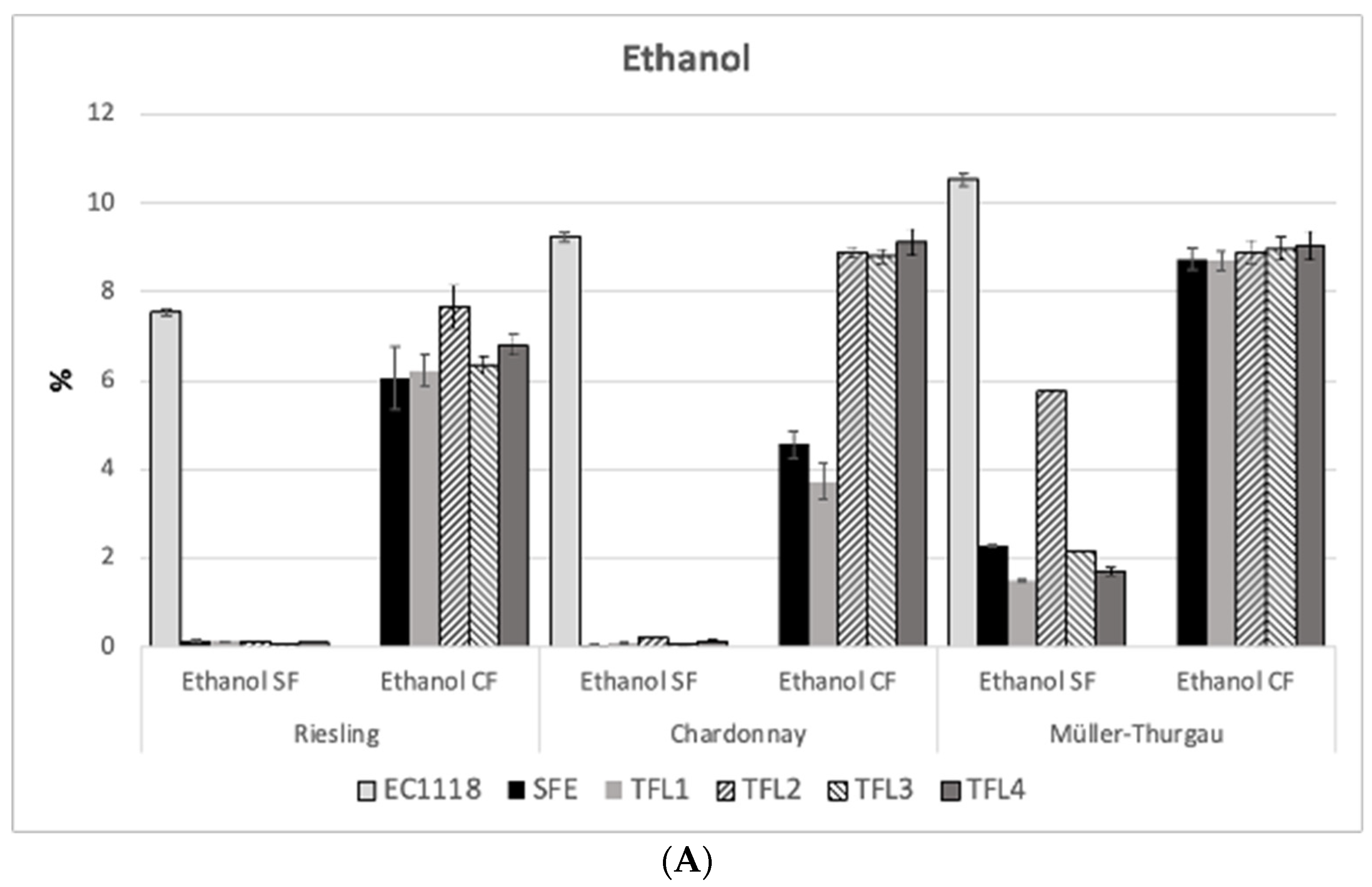
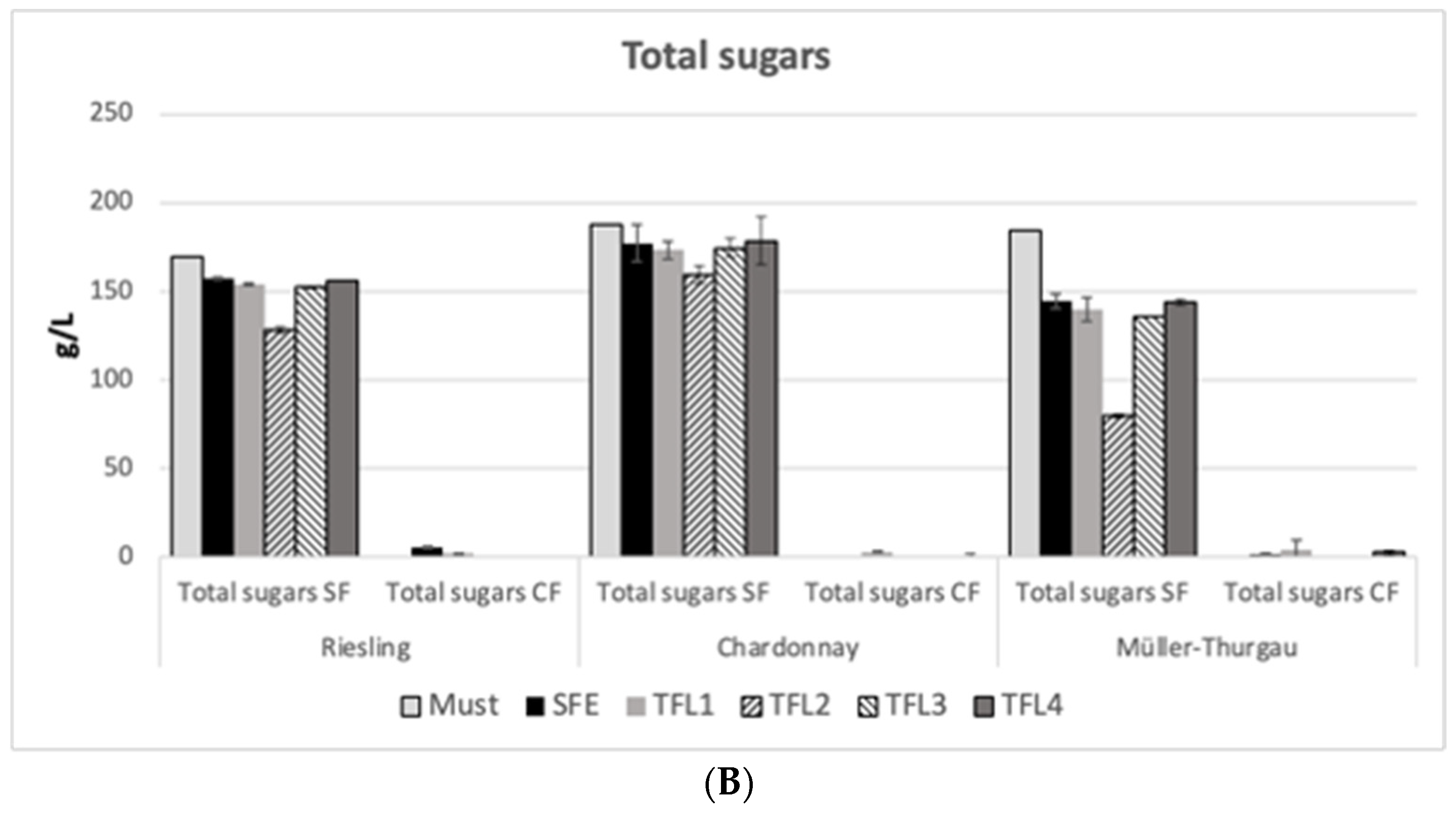
3.5. HPLC Results of Ethanol and Sugars
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pretorius, I.S.; Curtin, C.; Chambers, P. The Winemaker’s Bug. Bioengineered 2012, 3, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Maicas, S. The Role of Yeasts in Fermentation Processes. Microorganisms 2020, 8, 1142. [Google Scholar] [CrossRef] [PubMed]
- Lambrechts, M.G.; Pretorius, I.S. Yeast and Its Importance to Wine Aroma—A Review. S. Afr. J. Enol. Vitic. 2019, 21, 97–129. [Google Scholar] [CrossRef]
- Goldner, M.C.; Zamora, M.C.; Di Leo Lira, P.; Gianninoto, H.; Bandoni, A.L. Effect of ethanol level in the perception of aroma attributes and the detection of volatile compounds in red wine. J. Sens. Stud. 2009, 24, 243–257. [Google Scholar] [CrossRef]
- Pinto, C.; Pinho, D.; Cardoso, R.; Custódio, V.; Fernandes, J.O.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.M.P. Wine Fermentation Microbiome: A Landscape from Different Portuguese Wine Appellations. Front. Microbiol. 2015, 6, 905. [Google Scholar] [CrossRef] [PubMed]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not Your Ordinary Yeast: Non-Saccharomyces Yeasts in Wine Production Uncovered. FEMS Yeast Res. 2013, 14, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Hansen, E.; Nissen, P.; Sommer, P.; Nielsen, J.; Arneborg, N. The Effect of Oxygen on the Survival of Non-Saccharomyces Yeasts during Mixed Culture Fermentations of Grape Juice with Saccharomyces cerevisiae. J. Appl. Microbiol. 2001, 91, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Fleet, G.H. Yeast Interactions and Wine Flavour. Int. J. Food Microbiol. 2003, 86, 11–22. [Google Scholar] [CrossRef]
- Hazelwood, L.A.; Daran, J.-M.; Van Maris, A.J.A.; Pronk, J.T.; Dickinson, J.R. The Ehrlich Pathway for Fusel Alcohol Production: A Century of Research on Saccharomyces cerevisiae Metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef]
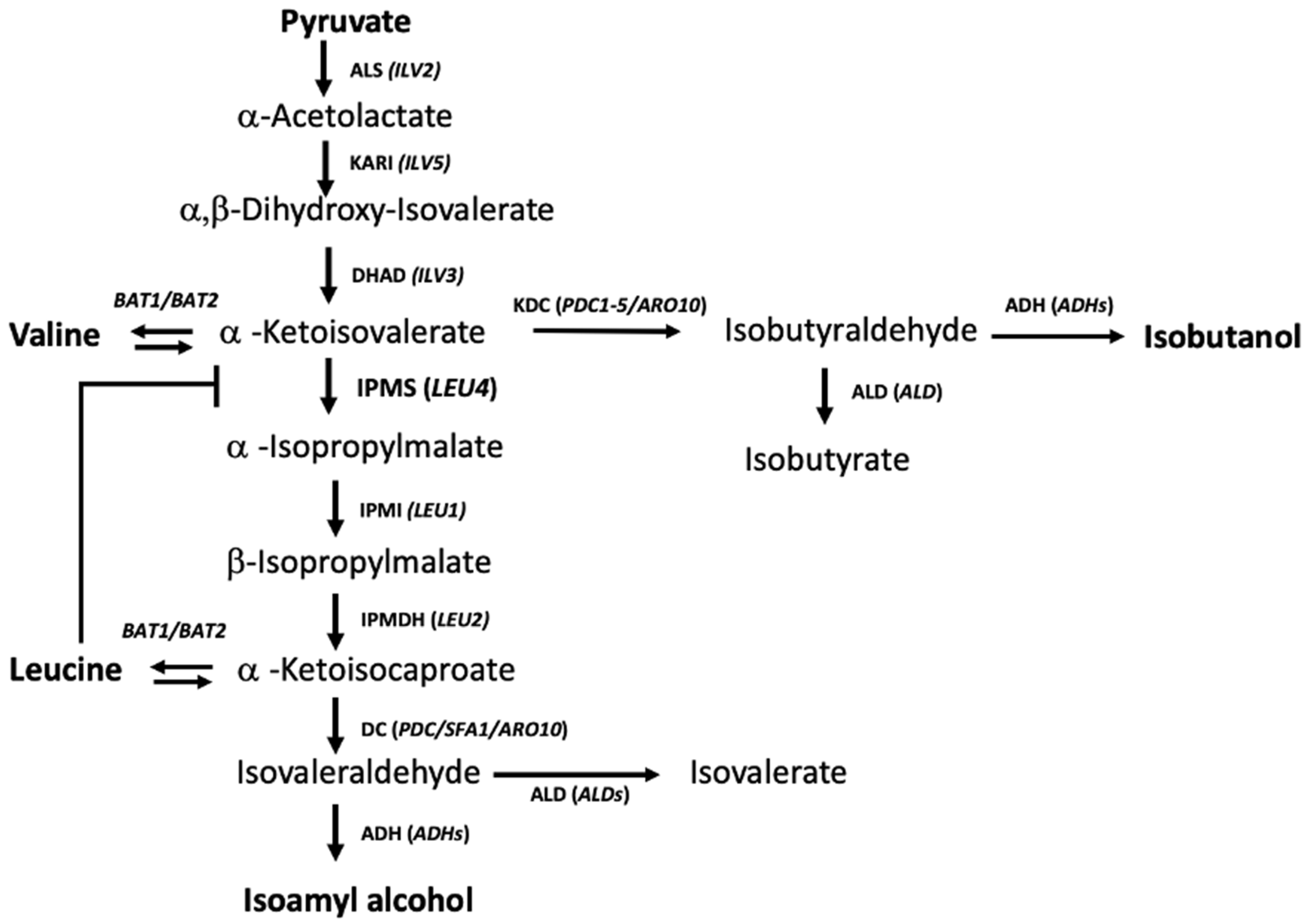
- Kohlhaw, G.B. Leucine Biosynthesis in Fungi: Entering Metabolism through the Back Door. Microbiol. Mol. Biol. Rev. 2003, 67, 1–15. [Google Scholar] [CrossRef]
- Takagi, H.; Yamamoto, K.; Matsuo, Y.; Furuie, M.; Kasayuki, Y.; Ohtani, R.; Shiotani, M.; Hasegawa, T.; Ohnishi, T.; Ohashi, M.; et al. Influence of Mutation in the Regulatory Domain of α-Isopropylmalate Synthase from Saccharomyces cerevisiae on Its Activity and Feedback Inhibition. Biosci. Biotechnol. Biochem. 2022, 86, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Hirata, D.; Aoki, S.; Watanabe, K.; Tsukioka, M.; Suzuki, T. Stable Overproduction of Isoamyl Alcohol by Saccharomyces cerevisiae with Chromosome-Integrated Multicopy LEU4 Genes. Biosci. Biotechnol. Biochem. 1992, 56, 1682–1683. [Google Scholar] [CrossRef]
- Baichwal, V.; Cunningham, T.J.; Gatzek, P.R.; Kohlhaw, G.B. Leucine Biosynthesis in Yeast. Curr. Genet. 1983, 7, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Cavalieri, D.J.; Casalone, E.; Bendoni, B.; Fia, G.; Polsinelli, M.; Barberio, C. Trifluoroleucine Resistance and Regulation of α-Isopropyl Malate Synthase in Saccharomyces cerevisiae. Mol. Genet. Genom. 1999, 261, 152–160. [Google Scholar] [CrossRef]
- Oba, T.; Yamamoto, Y.; Nomiyama, S.; Suenaga, H.; Muta, S.; Tashiro, K.; Kuhara, S. Properties of a Trifluoroleucine-Resistant Mutant Of Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2006, 70, 1776–1779. [Google Scholar] [CrossRef] [PubMed]
- Canonico, L.; Galli, E.; Ciani, E.; Comitini, F.; Ciani, M. Exploitation of Three Non-Conventional Yeast Species in the Brewing Process. Microorganisms 2019, 7, 11. [Google Scholar] [CrossRef]
- Van Wyk, N.; Grossmann, M.; Wendland, J.; Von Wallbrunn, C.; Pretorius, I.S. The Whiff of Wine Yeast Innovation: Strategies for Enhancing Aroma Production by Yeast during Wine Fermentation. J. Agric. Food Chem. 2019, 67, 13496–13505. [Google Scholar] [CrossRef]
- Badura, J.; Kiene, F.; Brezina, S.; Fritsch, S.; Semmler, H.; Rauhut, D.; Pretorius, I.S.; Von Wallbrunn, C.; van Wyk, N. Aroma Profiles of Vitis Vinifera L. Cv. Gewürztraminer Must Fermented with Co-Cultures of Saccharomyces cerevisiae and Seven Hanseniaspora Spp. Fermentation 2023, 9, 109. [Google Scholar] [CrossRef]
- Lachance, M.-A.; Pupovac-Velikonja, A.; Natarajan, S.; Schlag-Edler, B. Nutrition and Phylogeny of Predacious Yeasts. Can. J. Microbiol. 2000, 46, 495–505. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Smith, M.T. Saccharomycopsis Schiönning (1903). In The Yeasts; Elsevier: Amsterdam, The Netherlands, 2011; pp. 751–763. [Google Scholar]
- Hesselbart, A.; Junker, K.; Wendland, J. Draft Genome Sequence of Saccharomycopsis fermentans CBS 7830, a Predacious Yeast Belonging to the Saccharomycetales. Genome Announc. 2018, 6, e01445-17. [Google Scholar] [CrossRef]
- Kayacan, Y.; Griffiths, A.; Wendland, J. A Script for Initiating Molecular Biology Studies with Non-Conventional Yeasts Based on Saccharomycopsis schoenii. Microbiol. Res. 2019, 229, 126342. [Google Scholar] [CrossRef] [PubMed]
- Dutra-Silva, L.; Pereira, G.E.; Batista, L.R.; Matteoli, F.P. Fungal Diversity and Occurrence of Mycotoxin Producing Fungi in Tropical Vineyards. World J. Microbiol. Biotechnol. 2021, 37, 112. [Google Scholar] [CrossRef] [PubMed]
- Park, E.Y.; Kim, M.-D. Draft Genome Sequence Data of the Starch-Utilizing Yeast Saccharomycopsis fibuligera MBY1320 Isolated from Nuruk. Data Brief 2021, 35, 106888. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.-B.; Zhang, K.; Kang, Z.; Yang, J. Saccharomycopsis fibuligera in Liquor Production: A Review. Eur. Food Res. Technol. 2021, 247, 1569–1577. [Google Scholar] [CrossRef]
- Rensburg, P.; Stidwell, T.; Lambrechts, M.; Cordero-Otero, R.; Pretorius, I. Development and assessment of a recombinant Saccharomyces cerevisiae wine yeast producing two aroma-enhancing β-glucosidase encoded by the Saccharomycopsis fibuligera BGL1 and BGL2 genes. Ann. Microbiol. 2005, 55, 33–42. [Google Scholar]
- Methner, Y.; Magalhães, F.; Raihofer, L.; Zarnkow, M.; Jacob, F.; Hutzler, M. Beer Fermentation Performance and Sugar Uptake of Saccharomycopsis fibuligera–A Novel Option for Low-Alcohol Beer. Front. Microbiol. 2022, 13, 1011155. [Google Scholar] [CrossRef]
- Methner, Y.; Hutzler, M.; Matoulková, D.; Jacob, F.; Michel, M. Screening for the Brewing Ability of Different Non-Saccharomyces Yeasts. Fermentation 2019, 5, 101. [Google Scholar] [CrossRef]
- Methner, Y.; Hutzler, M.; Zarnkow, M.; Prowald, A.; Endres, F.; Jacob, F. Investigation of non-Saccharomyces yeast strains for their suitability for the production of non-alcoholic beers with novel flavor profiles. J. Am. Soc. Brew. Chem. 2022, 80, 341–355. [Google Scholar] [CrossRef]
- Takagi, H.; Hashida, K.; Watanabe, D.; Nasuno, R.; Ohashi, M.; Iha, T.; Nezuo, M.; Tsukahara, M. Isolation and Characterization of Awamori Yeast Mutants with L-Leucine Accumulation That Overproduce Isoamyl Alcohol. J. Biosci. Bioeng. 2015, 119, 140–147. [Google Scholar] [CrossRef]
- Schneider, A.; Gerbi, V.; Redoglia, M. A Rapid HPLC Method for Separation and Determination of Major Organic Acids in Grape Musts and Wines. Am. J. Enol. Vitic. 1987, 38, 151–155. [Google Scholar] [CrossRef]
- Scansani, S.; van Wyk, N.; Bou Nader, K.; Beisert, B.; Brezina, S.; Fritsch, S.; Semmler, H.; Pasch, L.; Pretorius, I.S.; von Wallbrunn, C.; et al. The film-forming Pichia spp. in a winemaker’s toolbox: A simple isolation procedure and their performance in a mixed-culture fermentation of Vitis vinifera L. cv. Gewürztraminer must. Int. J. Food Microbiol. 2022, 365, 109549. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, A.; Garzelli, F.; Schuessler, C.; Fritsch, S.; Loisel, C.; Pons, A.; Patz, C.-D.; Rauhut, D.; Jung, R. Wine Storage at Cellar vs. Room Conditions: Changes in the Aroma Composition of Riesling Wine. Molecules 2021, 26, 6256. [Google Scholar] [CrossRef]
- Krause, B.; Löhnertz, O. Saccharopin und Pipecolinsäure—Diagnostische Biomarker in der klassischen Aminosäureanalytik. MTA Dialog 2017, 18, 316–321. [Google Scholar] [CrossRef]
- Oba, T.; Nomiyama, S.; Hirakawa, H.; Tashiro, K.; Kuhara, S. Asp578 in LEU4p Is One of the Key Residues for Leucine Feedback Inhibition Release in Sake Yeast. Biosci. Biotechnol. Biochem. 2005, 69, 1270–1273. [Google Scholar] [CrossRef]
- Abe, T.; Toyokawa, Y.; Sugimoto, Y.; Azuma, H.; Tsukahara, K.; Nasuno, R.; Watanabe, D.; Tsukahara, M.; Takagi, H. Characterization of a New Saccharomyces cerevisiae Isolated From Hibiscus Flower and Its Mutant With L-Leucine Accumulation for Awamori Brewing. Front. Genet. 2019, 10, 490. [Google Scholar] [CrossRef] [PubMed]
- Link, A.J.; Mock, M.L.; Tirrell, D.A. Non-Canonical Amino Acids in Protein Engineering. Curr. Opin. Biotechnol. 2003, 14, 603–609. [Google Scholar] [CrossRef]
- Shen, L.; Nishimura, Y.; Matsuda, F.; Ishii, J.; Kondo, A. Overexpressing Enzymes of the Ehrlich Pathway and Deleting Genes of the Competing Pathway in Saccharomyces cerevisiae for Increasing 2-Phenylethanol Production from Glucose. J. Biosci. Bioeng. 2016, 122, 34–39. [Google Scholar] [CrossRef]
- Atsumi, S.; Hanai, T.; Liao, J.K. Non-Fermentative Pathways for Synthesis of Branched-Chain Higher Alcohols as Biofuels. Nature 2008, 451, 86–89. [Google Scholar] [CrossRef]
- Vuralhan, Z.; Luttik, M.a.H.; Tai, S.L.; Boer, V.M.; De Morais, M.A.; Schipper, D.; Almering, M.J.H.; Kötter, P.; Dickinson, J.R.; Daran, J.-M.; et al. Physiological Characterization of the ARO10-Dependent, Broad-Substrate-Specificity 2-Oxo Acid Decarboxylase Activity of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 3276–3284. [Google Scholar] [CrossRef]
- Yuan, J.; Mishra, P.; Ching, C.B. Engineering the Leucine Biosynthetic Pathway for Isoamyl Alcohol Overproduction in Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2017, 44, 107–117. [Google Scholar] [CrossRef]
- Herschorn, S.; Kim, S.-K.; Lee, Y.-G.; Park, K.W.; Seo, J.-H. Elimination of Biosynthetic Pathways for L-Valine and l-Isoleucine in Mitochondria Enhances Isobutanol Production in Engineered Saccharomyces cerevisiae. Bioresour. Technol. 2018, 268, 271–277. [Google Scholar] [CrossRef]
- Wess, J.; Brinek, M.; Boles, E. Improving Isobutanol Production with the Yeast Saccharomyces cerevisiae by Successively Blocking Competing Metabolic Pathways as Well as Ethanol and Glycerol Formation. Biotechnol. Biofuels 2019, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Saliba, A.; Ovington, L.; Moran, C.C. Consumer Demand for Low-Alcohol Wine in an Australian Sample. Int. J. Wine Res. 2013, 1, 1–8. [Google Scholar] [CrossRef]



| Code Name | Strain Name | Description | Nucleotide Exchange | Amino Acid Residue in S. cerevisiae |
|---|---|---|---|---|
| EC1118 | Saccharomyces cerevisiae | Wild-type wine strain | — | — |
| SFE | Saccharomycopsis fermentans | Wild type | — | — |
| G058, TFL1 | Saccharomycopsis fermentans | Thr514Lys | ACA–AAA | Val522 |
| G059, TFL2 | Saccharomycopsis fermentans | His534Pro and Ala545Thr | GCT–ACT, CAC–CCC | His541, Ala551 |
| G236, TFL3 | Saccharomycopsis fermentans | Ser541Tyr | TCC–TAC | Ser547 |
| G060, TFL4 | Saccharomycopsis fermentans | Ser511Tyr | TCT–TAT | Ser519 |
| Valine | Leucine | Isoleucine | Phenylalanine | Alanine | Methionine | Glycine | Glutamine | Threonine | Tyrosine | Tryptophan | Serine | Total | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC1118 | 0.2 | 0.4 | 0.3 | 0.3 | 6.2 | 0.2 | 0.7 | 0.3 | 1.1 | 0.2 | 0.1 | 0.7 | 21.9 |
| SFE | 1.6 | 0.4 | 0.5 | 2.1 | 2.6 | 0.1 | 1.2 | 0.1 | 1.2 | 0.1 | <0.1 | 0.3 | 126.1 |
| TFL1 | <0.1 | 3.2 | 0.6 | 1.3 | 5.4 | 0.4 | 1.9 | 0.7 | 1.7 | 0.1 | <0.1 | 0.3 | 171.2 |
| TFL2 | 0.2 | 3.2 | 2.2 | 7.7 | 2.6 | 0.3 | 5.8 | 1.1 | 10.0 | 3.2 | 1.7 | 0.6 | 196.9 |
| TFL3 | 0.1 | 2.8 | 0.4 | 0.4 | 5.1 | 0.2 | 1.9 | 0.7 | 1.3 | 0.1 | <0.1 | 0.7 | 172.3 |
| TFL4 | 1.5 | 2.9 | 0.7 | 2.3 | 4.1 | 0.8 | 2.2 | 1.2 | 3.5 | 0.6 | <0.1 | 0.8 | 201.2 |
| Riesling Must | 9.2 | 7.2 | 4.9 | 6.6 | 77.3 | 0.9 | 2.4 | <0.1 | 30.3 | 3.9 | <0.1 | 27.5 | 589.7 |
| Must Type | Strain | Tartaric Acid (g/L) | Malic Acid (g/L) | Shikimic Acid (mg/L) | Lactic Acid (g/L) | Acetic Acid (g/L) | Citric Acid (g/L) | Isovaleric Acid ** (mg/L) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SF | CF | SF | CF | SF | CF | SF | CF | SF | CF | SF | CF | SF | CF | ||
| Riesling | EC1118 | 4.5 | 2.7 | 23.9 | <0.1 | 5.4 | <0.1 | 3 | |||||||
| SFE | 6.8 | 6.1 | 1.9 | <0.1 | 28.6 | 23.3 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | 0.4 | 20 | 116 | |
| TFL1 | 6.7 | 6.2 | 1.6 | <0.1 | 29.9 | 23.3 | <0.1 | <0.1 | <0.1 | <0.1 | 0.1 | 0.3 | 76 | 302 | |
| TFL2 | 6.8 | 6.2 | <0.1 | <0.1 | 29.4 | 21.9 | <0.1 | <0.1 | <0.1 | 0.1 | 0.2 | 0.3 | 118 | 486 | |
| TFL3 | 6.8 | 6.0 | 1.4 | <0.1 | 29.0 | 22.5 | <0.1 | <0.1 | <0.1 | <0.1 | 0.1 | 0.3 | 80 | 419 | |
| TFL4 | 5.4 | 5.6 | 1.9 | <0.1 | 28.3 | 23.5 | <0.1 | <0.1 | <0.1 | <0.1 | 0.1 | 0.3 | 73 | 329 | |
| Must | 6.2 | 6.2 | 3.9 | 3.9 | 29 | 29.3 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | nd | nd | |
| Chardonnay | EC1118 | 3.6 | 3.5 | 46 | <0.1 | 6.9 | <0.1 | 4 | |||||||
| SFE | 3.8 | 3.5 | 3.7 | <0.1 | 55 | 50.8 | 0.17 | <0.1 | <0.1 | 0.3 | <0.1 | 0.2 | 9 | 80 | |
| TFL1 | 3.5 | 3.6 | 3.5 | 0.8 | 54 | 40.4 | 0.14 | <0.1 | <0.1 | 0.1 | <0.1 | 0.2 | 34 | 71 | |
| TFL2 | 3.5 | 3.3 | 3.1 | <0.1 | 48 | 51.0 | 0.18 | <0.1 | <0.1 | <0.1 | <0.1 | 0.2 | 33 | 289 | |
| TFL3 | 3.4 | 3.5 | 1.6 | <0.1 | 55 | 49.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | 0.2 | 29 | 253 | |
| TFL4 | 3.4 | 3.5 | 2.4 | 0.6 | 54 | 51.2 | <0.1 | <0.1 | <0.1 | 0.2 | <0.1 | 0.2 | 50 | 206 | |
| Must | 3.3 | 3.3 | 4.6 | 3.8 | 54 | 52.9 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | 0.2 | nd | nd | |
| Müller-Thurgau | EC1118 | 4.4 | 1.8 | 25 | 0.16 | 0.36 | <0.1 | 5 | |||||||
| SFE | 5.4 | 5.1 | 1.6 | 1.5 | 16 | 13.4 | 0.48 | 0.3 | <0.1 | 0.1 | <0.1 | 0.1 | 3 | 9 | |
| TFL1 | 5.5 | 5.2 | 1.3 | 1.2 | 26 | 15.1 | 0.44 | 0.3 | <0.1 | 0.4 | <0.1 | 0.1 | 11 | 19 | |
| TFL2 | 5.4 | 5.1 | 1.1 | 1.2 | 25 | 6.7 | <0.1 | 0.2 | 0.36 | 0.1 | <0.1 | <0.1 | 20 | 21 | |
| TFL3 | 5.4 | 5.1 | 1.3 | 1.6 | 25 | 14.2 | 0.69 | 0.4 | <0.1 | 0.2 | <0.1 | 0.1 | 8 | 14 | |
| TFL4 | 5.3 | 5.1 | 1.3 | 1.5 | 25 | 14.8 | 0.40 | 0.4 | <0.1 | 0.1 | <0.1 | 0.1 | 10 | 19 | |
| Must | 5.6 | 5.0 | 1.6 | 1.6 | 28 | 26.0 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | <0.1 | nd | nd | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akan, M.; Gudiksen, A.; Baran, Y.; Semmler, H.; Brezina, S.; Fritsch, S.; Rauhut, D.; Wendland, J. Exploring the Potential of Non-Conventional Yeasts in Wine Fermentation with a Focus on Saccharomycopsis fermentans. Fermentation 2023, 9, 786. https://doi.org/10.3390/fermentation9090786
Akan M, Gudiksen A, Baran Y, Semmler H, Brezina S, Fritsch S, Rauhut D, Wendland J. Exploring the Potential of Non-Conventional Yeasts in Wine Fermentation with a Focus on Saccharomycopsis fermentans. Fermentation. 2023; 9(9):786. https://doi.org/10.3390/fermentation9090786
Chicago/Turabian StyleAkan, Madina, Andreas Gudiksen, Yasemin Baran, Heike Semmler, Silvia Brezina, Stefanie Fritsch, Doris Rauhut, and Jürgen Wendland. 2023. "Exploring the Potential of Non-Conventional Yeasts in Wine Fermentation with a Focus on Saccharomycopsis fermentans" Fermentation 9, no. 9: 786. https://doi.org/10.3390/fermentation9090786
APA StyleAkan, M., Gudiksen, A., Baran, Y., Semmler, H., Brezina, S., Fritsch, S., Rauhut, D., & Wendland, J. (2023). Exploring the Potential of Non-Conventional Yeasts in Wine Fermentation with a Focus on Saccharomycopsis fermentans. Fermentation, 9(9), 786. https://doi.org/10.3390/fermentation9090786







