Phenotypic Characterisation and Molecular Identification of Potentially Probiotic Lactobacillus sp. Isolated from Fermented Rice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Location
2.2. Materials
2.3. Preparation of Rice Samples
2.4. Phenotypic Characterisation
2.4.1. Morphological Characteristics of the Isolates
2.4.2. Physiological and Biochemical Characterisation
Culture Preparation
Growth at Different Temperatures
Growth at Different NaCl Concentrations
Gas Production with Glucose
Milk Coagulation Assay
2.5. Carbohydrate Fermentation Pattern of Isolates on the API 50 CHL System
2.5.1. Preparation of the Culture
2.5.2. Preparation of the Incubation Box
2.5.3. Preparation of the Strips
2.5.4. Inoculation of the Strips
2.5.5. Reading and Interpretation
2.6. Identification of Lactobacillus sp. by 16S rDNA Sequencing
2.6.1. Extraction of Genomic DNA
2.6.2. Polymerase Chain Reaction for the Amplification of the 16S rDNA Region
2.6.3. Separation of Amplified PCR Products
2.6.4. Sequencing and Phylogenetic Tree Development
3. Results
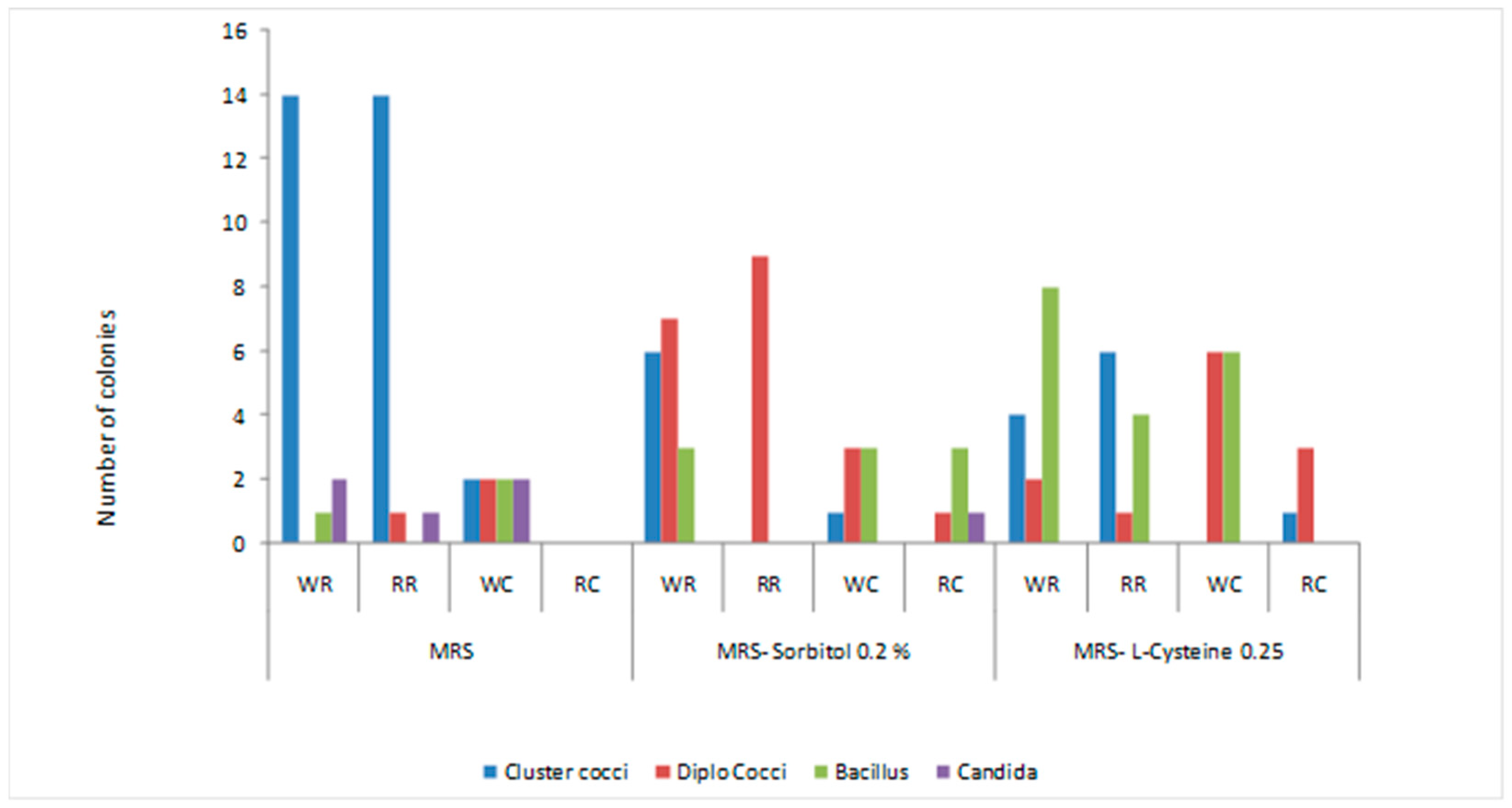
3.1. Morphological Characteristics of Isolates
3.2. Physiological and Biochemical Characterisation
3.2.1. Growth at Different Temperatures
3.2.2. Growth at Different NaCl Concentrations
3.2.3. Gas Production with Glucose
3.2.4. Milk Coagulation and Curd Formation
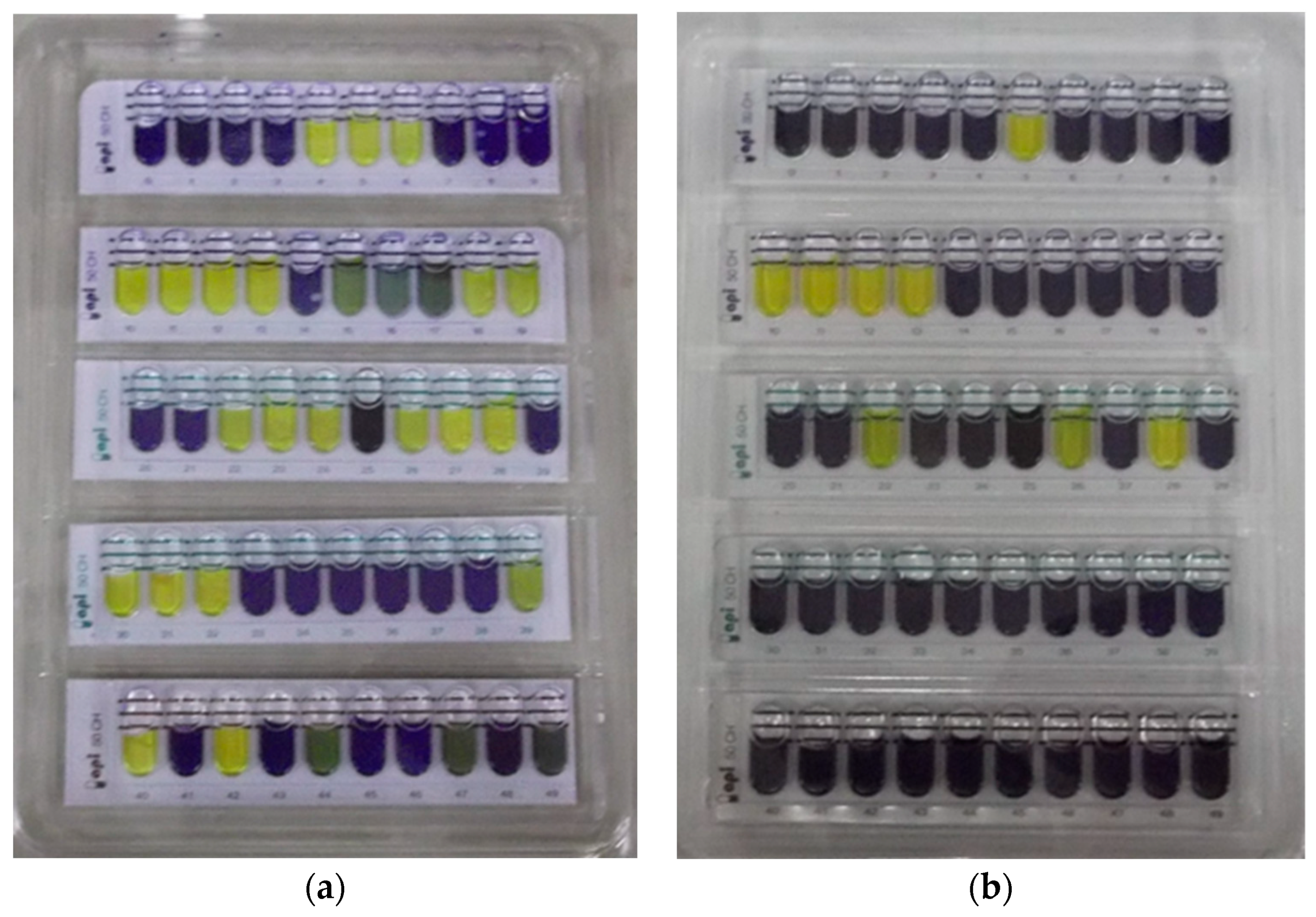
3.2.5. Carbohydrate Fermentation Pattern of Lactobacillus sp.
3.3. Molecular Identification and Genotypic Characteristics of Lactobacillus sp.
3.3.1. Genomic DNA Isolation
3.3.2. Amplification of 16S rDNA Region
3.3.3. Identification of LAB Based on Phylogenetic Analyses of 16S rDNA Sequences
3.3.4. Phytogenic Tree Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A


References
- Salovaara, H.; Simson, L. Fermented Cereal Based Functional Foods. In Hand Book of Food and Beverage Fermentation Technology; Hui, Y.H., Meunier-Goddik, L., Josephsen, J., Nip, W., Stanfield., P.S., Eds.; CRC Press: Boca Raton, FL, USA; Marcel Dekker: New York, NY, USA, 2004; p. 831. [Google Scholar]
- Blandinob, A.; Al-Aseeria, M.E.; Pandiellaa, S.S.; Canterob, D.; Webba, C. Review-Cereal-based fermented foods and beverages. Food Res. Intern. 2003, 36, 27–543. [Google Scholar] [CrossRef]
- Jagadeeswari, S.; Vidya, P.; MukeshKumar, D.J.; Balakumaran, M.D. Isolation and Characterization of Bacteriocin producing Lactobacillus sp. from Traditional Fermented Foods. Electron. J. Environ. Agric. Food 2010, 9, 575–581. [Google Scholar]
- Dahiya, D.; Nigam, P.S. Use of Characterized Microorganisms in Fermentation of Non-Dairy-Based Substrates to Produce Probiotic Food for Gut-Health and Nutrition. Fermentation 2023, 9, 1. [Google Scholar] [CrossRef]
- Heller, K.J. Probiotic bacteria in fermented foods: Product characteristics and starter organisms. Am. J. Clin. Nutr. 2001, 73, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Kauli, C.M.; Mathara, J.M.; Kutima, P.M. Probiotic potential of spontaneously fermented cereal-based foods—A review. Afr. J. Biotech. 2010, 9, 2490–2498. [Google Scholar]
- Kumar, P.; Begum, V.H.; Kumaravel, S. Mineral nutrients of ‘pazhaya sadham’: A traditional fermented food of Tamil Nadu, India. Intern. J. Nutr. Metab. 2012, 4, 151–152. [Google Scholar]
- Kowsalya, M.; Sudha, K.G.; Ali1, S.; Velmurugan, T.; Karunakaran, G.; Rajeshkumar, M.P. In-vitro assessment of probiotic properties of lactic acid bacteria isolated from naturally fermented rice gruel of south India. J. Microbiol. Biotech. Food Sci. 2022, 12, 4908. [Google Scholar] [CrossRef]
- Wikeramanayake, T.W. Legumes. In Food and Nutrition, 2nd ed.; Hector Kobbekaduwa Agrarian Training Institute: Colombo, Sri Lanka, 2002; ISBN 955-565-000-4168. [Google Scholar]
- Hatti-Kaul, R.; Chen, L.; Dishisha, T.; Enshasy, H.E. Lactic acid bacteria: From starter cultures to producers of chemicals. FEMS Microbiol. Lett. 2018, 365, fny213. [Google Scholar] [CrossRef]
- Axelsson, L. Lactic Acid Bacteria Classification and Physiology in Lactic Acid Bacteria Microbiological and Functional Aspects, 3rd ed.; Salminen, S., Wright, A.V., Ouwehand, A., Eds.; Marcel Dekker: New York, NY, USA, 2004; pp. 1–67. [Google Scholar] [CrossRef]
- Jeyagowri, N.; Parahitiyawa, N.B.; Jeyatilake, J.A.M.S.; Ranadheera, C.S.; Madhujith, M.M.T. Study on Isolation of potentially pro biotic Lactobacillus spp from fermented rice. Trop. Agric. Res. 2015, 26, 428–440. [Google Scholar] [CrossRef]
- Barrow, G.H.; Feltham, R.K.A. Cowan and Steel’s Manual for Identification of Medical Bacteria, 3rd ed.; Cambridge University Press: Cambridge, UK, 1993; p. 331. [Google Scholar]
- Hatice, Y. Isolation, Characterization and determination of probiotic properties of lactic acid bacteria from human milk. Master’s Thesis, Graduate School of Engineering and Sciences of Izmir Institute of Technology, Izmir, Turkey, 2007. Available online: https://www.academia.edu/3341345/ (accessed on 16 October 2011).
- Marrokil, A.; Zuniga, M.; Kihal, M.; Martinez, G. Characterization of Lactobacillus from Algeriangoats’s milk based on phenotypic, 16SrDNA Sequencing and their technological properties. Braz. J. Microbiol. 2011, 42, 58–171. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Sukara, E.; Salamah, A.; Dinoto, A.; Mangunwardoyo, W. Identification of lactic acid bacteria in sayur asin from Central Java (Indonesia) based on 16S rDNA sequence. Intern. Food Res. J. 2014, 21, 527–532. [Google Scholar]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef] [PubMed]
- Vaccalluzzo, A.; Pino, A.; Angelis, M.D.; Bautista-Gallego, J.; Romeo, F.V.; Foti, P.; Caggia, C.; Randazzo, C.L. Effects of Different Stress Parameters on Growth and on Oleuropein-Degrading Abilities of Lactobacillus plantarum Strains Selected as Tailored Starter Cultures for Naturally Table Olives. Microorganisms 2020, 8, 1607. [Google Scholar] [CrossRef] [PubMed]
- Kunji, E.R.S.; Mierau, I.A.; Poolman, B.; Konings, W.N. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek 1996, 70, 187–221. [Google Scholar] [CrossRef]
- Wang, H.; Yu, W.; Coolbear, T.; Sullivan, D.; Mckay, L.L. A deficiency in aspartate biosynthesis in Lactococcus lactis subsp. lactis C2 causes slow milk coagulation. Appl. Environ. Microbiol. 1998, 64, 1673. [Google Scholar] [CrossRef]
- Quere, F.; Deschamps, A.; Urdaci, M.C. DNA probe and PCR specific reaction for Lactobacillus plantarum. J. Appl. Microbiol. 1997, 82, 783–790. [Google Scholar] [CrossRef]
- Ahrne, S.; Molin, G.; Stahl, S. Plasmids in Lactobacillus strains isolated from meat and meat products. Syst. Appl. Microbiol. 1989, 11, 320–325. [Google Scholar] [CrossRef]
- Mirzaei, H.; Barzgari, A. Isolation and Molecular Study of Potentially Probiotic Lactobacilli in Traditional White Cheese of Tabriz in Iran. Ann. Biol. Res. 2012, 3, 2019–2022. [Google Scholar]
- Woo, P.C.; Fung, A.M.; Lau, S.K.; Yuen, K.Y. Identification by 16S rRNA Gene Sequencing of Lactobacillus salivarius Bacteremic Cholecystitis. J. Clin. Microbiol. 2002, 40, 265–267. [Google Scholar] [CrossRef]
- Tilahun, B.; Testaye, A.; Muleta, D.; Baihru, A.; Terefework, Z.; Wessel, G. Isolation and Molecular Identification of Lactic acid Bacteria using 16srRNA from fermented Teff (Eragrostis tef (Zucc.)) Dough. Intern. J. Food Sci. 2018, 8510620. [Google Scholar] [CrossRef]
- Holt, J.G.; Krieg, N.R.; Sneath, P.H.A.; Staley, J.T.; Williams, S.T. Bergey’s Manual of Determinative Bacteriology, 9th ed.; Williams and Wilkins: Baltimore, MD, USA, 1994; pp. 505–542. [Google Scholar]
- Hoque, M.Z.; Akter, F.; Hossain, K.M.; Rahman, M.S.M.; Billah, M.M.; Islam, K.M.D. Isolation, Identification and Analysis of Probiotic Properties of Lactobacillus Sp. From Selective Regional Yoghurts. World J. Dairy Food Sci. 2010, 5, 39–46. [Google Scholar]
- Mohammed, M.; Abd El-Aziz, H.; Omran, N.; Anwar, S.; Awad, S.; El-Soda, M. Rep-PCR characterization and biochemical selection of lactic acid bacteria isolated from the Delta area of Egypt. Intern. J. Food Microbiol. 2009, 128, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Khunajakr, N.; Wongwicharn, W.; Moonmangmee, D. Screening and Identification of Lactic acid bacteria producing antimicrobial compounds from Pig Gastrointestinal Tracts. KMITL Sci. Technol. J. 2008, 8, 10. [Google Scholar] [CrossRef]
- Hassan, Z.; Effat, B.; Magdoub, M.; Tawfik, N.; Sadek, Z.; Mabrouk, A. Molecular Identificati of lactic acis bacteria isolated from fermented dairy product. Intern. J. Biol. Pharm. Allied. Health Sci. 2016, 5, 3221–3230. [Google Scholar]
- Tajabadi, N.; Mardan, M.; Nazamid, S.; Shuhaimi, M.; Bahreini, R.; Manap, M.Y.A. Identification of Lactobacillus plantarum, Lactobacillus pentosus and Lactobacillus fermentum from honey stomach of honeybee. Braz. J. Microbiol. 2013, 44, 717–722. [Google Scholar] [CrossRef]
- Roos, S.; Engstrand, L.; Jonsson, H. Lactobacillus gastricus sp. nov., Lactobacillus antri sp. nov., Lactobacillus kalixensis sp. nov. and Lactobacillus ultunensis sp. nov., isolated from human stomach mucosa. Intern. J. Syst. Evol. Microbiol. 2005, 55, 77–82. [Google Scholar] [CrossRef]
- Kashiwagi, T.; Suzuki, T.; Kamakura, T. Lactobacillus nodensis sp. nov., isolated from rice bran. Intern. J. Syst. Evol. Microbiol. 2009, 59, 83–86. [Google Scholar] [CrossRef]
- Pedersen, C.; Roos, S. Lactobacillus saerimneri sp. nov isolated from pig faces. Intern. J. Syst. Evol. Microbiol. 2004, 54, 1365–1368. [Google Scholar] [CrossRef]
- Brolazo, E.M.; Leite, D.S.; Tiba, M.R.; Villarroel, M.; Marconi, C.; Simoes, J.A. Correlation between API 50CH and Multiple Polymerase Chain Reaction for the identification of Vaginal Lactobacillus in Isolates. Braz. J. Microbiol. 2011, 42, 225–232. [Google Scholar] [CrossRef]
- Satishkumar, R.; Ragu-Varman, R.; Kanmani, P.; Yuvaraj, N.; Paari, K.A.; Pattukumar, V.; Arul, V. Isolation, Characterization and Identification of a Potential Probiont from South Indian Fermented Foods (Kallappam, Koozh and MorKuzhambu) and Its Use as Biopreservative. Prob. Antimicrob. Proteins 2010, 2, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Srikanjana, K.; Siriporn, O. Antibacterial and Antioxidant actives of Acid and bile resistant strains of Lactobacillus fermentum isolated from Miang. Braz. J. Microbiol. 2009, 40, 757–766. [Google Scholar]
- Jeyagowri, N.; Parahitiyawa, N.B.; Jeyatilake, J.A.M.S.; Ranadheera, C.S.; Madhujith, W.M.T. Antimicrobial activity of potentially pro biotic Lactobacillus spp from fermented rice. In Proceedings of the Peradeniya University International Research Sessions (iPURSE 2014), Peradeniya, Sri Lanka, 4–5 July 2014; University of Peradeniya: Peradeniya, Sri Lanka, 2014; p. 182. [Google Scholar]
- Ahmadova, A.; Todorov, S.D.; Hadji-Sfaxi, I.; Choiset, Y.; Rabesona, H.; Messaoudi, S.; Kuliyev, A.; Franco, B.D.; Chobert, J.M.; Haertle, T. Antimicrobial and antifungal activities of Lactobacillus curvatus strain isolated from homemade Azerbaijani cheese. Anaerobe 2013, 20, 42–49. [Google Scholar] [CrossRef]
- Valerio, F.; Favilla, M.; De Bellis, P.; Sisto, A.; Valerio, F. Antifungal activity of strains of lactic acid bacteria isolated from a semolina ecosystem against Penicillium roqueforti, Aspergillus niger and Endomyces fibuliger contaminating bakery products. Syst. Appl. Microbiol. 2013, 32, 438–448. [Google Scholar] [CrossRef]
- Divisekera, D.M.W.D.; Gooneratne, J.; Jayawardana, D. Isolation and characterization and identification of Lactic acid bacteria and yeast from fermented organically grown rice and coconut milk. In Proceedings of the Peradeniya University International Research Sessions (iPURSE 2014), Peradeniya, Sri Lanka, 4–5 July 2014; University of Peradeniya: Peradeniya, Sri Lanka, 2014; p. 210. Available online: https://www.pdn.ac.lk/ipurse/2014/proceeding_book/FL/210.pdf (accessed on 22 May 2023).
- Sukumar, G.; Ghosh, A.R. Pediococcus spp.—A potential probiotic isolated from Khadi (an Indian fermented food) and identified by 16SrDNA sequence analysis. Afr. J. Food Sci. 2010, 4, 597–602. [Google Scholar]
- Jeon, H.; Kim, Y.-T.; Jang, W.Y.; Kim, J.-Y.; Heo, K.; Shim, J.-J.; Lee, J.-L.; Yang, D.-C.; Kang, S.C. Effects of Lactobacillus curvatus HY7602-Fermented Antlers in Dexamethasone-Induced Muscle Atrophy. Fermentation 2022, 8, 454. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, L.; Qiao, N.; Xiao, Y.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W.; Zhai, Q. Latilactobacillus curvatus: A Candidate Probiotic with Excellent Fermentation Properties and Health Benefits. Foods 2020, 9, 1366. [Google Scholar] [CrossRef]
- Bernard, D.; Jeyagowri, N.; Madhujith, T. Characterization of Lactic Acid Bacteria isolated from Idli Batter and their susceptibility to Antibiotics. Trop. Agric. Res. 2021, 32, 453–461. [Google Scholar] [CrossRef]
- Adikari, A.M.M.U.; Priyashantha, H.; Disanayaka, J.N.K.; Jayatileka, D.V.; Kodithuwakku, S.P.; Jayatilake, J.A.M.S.; Vidanarachchi, J.K. Isolation, identification and characterization of Lactobacillus speciesdiversity from Meekiri: Traditional fermented buffalo milk gels in Sri Lanka. Heliyon 2021, 7, e08136. [Google Scholar] [CrossRef]





| Isolates | Lactic Acid Bacteria | Morphological Description | Appearance under Light Microscope (×1000 Magnification) |
|---|---|---|---|
| Lb1 | Latilactobacillus curvatus | Medium size, rod (regular)-shaped bacteria, arranged as single/pair or as a group in a ‘V’ arrangement |  |
| Lb2 | Latilactobacillus curvatus | Medium size, rod (regular)-shaped bacteria, arranged as single/pair or as a group in a ‘V’ arrangement |  |
| Lb4 | Weissella confusa | Very long size, rod (regular)-shaped bacteria, arranged as single/pair or as a group in a ‘V’ arrangement |  |
| Lb8 | Latilactobacillus graminis | Small size, rod (regular)-shaped bacteria, arranged as single/pair or as a group in a ‘V’ arrangement |  |
| Lb10 | Latilactobacillus curvatus | Medium size, rod (regular)-shaped bacteria, arranged as single/pair or as a group in a ‘V’ arrangement |  |
| Lb11 | Latilactobacillus curvatus | Medium size, rod (regular)-shaped bacteria, arranged as single/pair or as a group in a ‘V’ arrangement |  |
| Lb17 | Limosilactobacillus fermentum | Long size, rod (regular)-shaped bacteria, arranged as single/pair or as a group in a ‘V’ arrangement |  |
| Lc1 | Pediococcus pentosaceus | Coccus-shaped bacteria, arranged as single, tetrad, or group/cluster |  |
| Physiological and Biochemical Characteristics | Lb-1 | Lb-2 | Lb-4 | Lb-8 | Lb-10 | Lb-11 | Lb-17 | Positive | Negative |
|---|---|---|---|---|---|---|---|---|---|
| Gram test | + | + | + | + | + | + | + | NA | NA |
| Catalase test | − | − | − | − | − | − | − | NA | NA |
| Motility | − | − | − | − | − | − | − | NA | NA |
| Spore formation | − | − | − | − | − | − | − | NA | NA |
| Gas from glucose | − | − | − | + | − | − | − | ND | − |
| Growth at 10 °C | ++ | ++ | ++ | + | ++ | ++ | ++ | NA | − |
| Growth at 37 °C | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | − |
| Growth at 40 °C | ++ | ++ | ++ | ++ | ++ | ++ | ++ | NA | − |
| Growth at 45 °C | + | + | + | + | + | + | + | NA | − |
| Growth at 55 °C | − | − | − | − | − | − | − | NA | − |
| 0% Nacl | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ | |
| 2% NaCl | ++ | ++ | ++ | ++ | ++ | ++ | ++ | NA | − |
| 4% NaCl | ++ | ++ | ++ | ++ | ++ | ++ | ++ | NA | − |
| 6.5% NaCl | ++ | ++ | ++ | ++ | ++ | ++ | ++ | NA | − |
| 10% NaCl | − | − | − | − | − | − | − | NA | − |
| Milk coagulation | + | + | + | + | + | + | + | + | − |
| Isolate Code | Species of Lactobacillus | * Identity (%) |
|---|---|---|
| Lb-1 | Lactobacillus curvatus ssp.curvatus | 98.9 |
| Lb-2 | Lactobacillus curvatus ssp.curvatus | 99.3 |
| Lb-4 | Lactobacillus helveticus | 86.3 |
| Lb-8 | Lactobacillus delbrueckii ssp. delbrueckii | 95.5 |
| Lb-10 | Lactobacillus pentosus | 63.3 |
| Lb-11 | Lactobacillus curvatus ssp.curvatus | 99.3 |
| Lb-17 | Lactobacillus plantrum | 91.3 |
| Lc-1 | Pediococcus pentosaceus | 99.9 |
| Sequence ID | Microorganism | Accession Number | Sequence Length |
|---|---|---|---|
| Lb1 | Latilactobacillus curvatus GRLb1 | OQ733261 | 1475 bp |
| Lb2 | Latilactobacillus curvatus GRLb2 | OQ733262 | 1465 bp |
| Lb4 | Weissella confusa strain GRLb4 | OQ733263 | 1486 bp |
| Lb8 | Latilactobacillus graminis GRLb8 | OQ861079 | 1097 bp |
| Lb10 | Latilactobacillus curvatus GRLb10 | OQ733264 | 1469 bp |
| Lb11 | Latilactobacillus curvatus GRLb11 | OQ733265 | 1456 bp |
| Lb17 | Limosilactobacillus fermentum GRLb17 | OQ861078 | 819 bp |
| Lc1 | Pediococcus pentosaceus GRLc1 | OQ733260 | 1480 bp |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeyagowri, N.; Ranadheera, C.S.; Manap, M.Y.; Gamage, A.; Merah, O.; Madhujith, T. Phenotypic Characterisation and Molecular Identification of Potentially Probiotic Lactobacillus sp. Isolated from Fermented Rice. Fermentation 2023, 9, 807. https://doi.org/10.3390/fermentation9090807
Jeyagowri N, Ranadheera CS, Manap MY, Gamage A, Merah O, Madhujith T. Phenotypic Characterisation and Molecular Identification of Potentially Probiotic Lactobacillus sp. Isolated from Fermented Rice. Fermentation. 2023; 9(9):807. https://doi.org/10.3390/fermentation9090807
Chicago/Turabian StyleJeyagowri, Nimalan, Chaminda Senaka Ranadheera, Mohd Yazid Manap, Ashoka Gamage, Othmane Merah, and Terrence Madhujith. 2023. "Phenotypic Characterisation and Molecular Identification of Potentially Probiotic Lactobacillus sp. Isolated from Fermented Rice" Fermentation 9, no. 9: 807. https://doi.org/10.3390/fermentation9090807
APA StyleJeyagowri, N., Ranadheera, C. S., Manap, M. Y., Gamage, A., Merah, O., & Madhujith, T. (2023). Phenotypic Characterisation and Molecular Identification of Potentially Probiotic Lactobacillus sp. Isolated from Fermented Rice. Fermentation, 9(9), 807. https://doi.org/10.3390/fermentation9090807










