Distribution of Plasmopara viticola Causing Downy Mildew in Russian Far East Grapevines
Abstract
1. Introduction
2. Materials and Methods
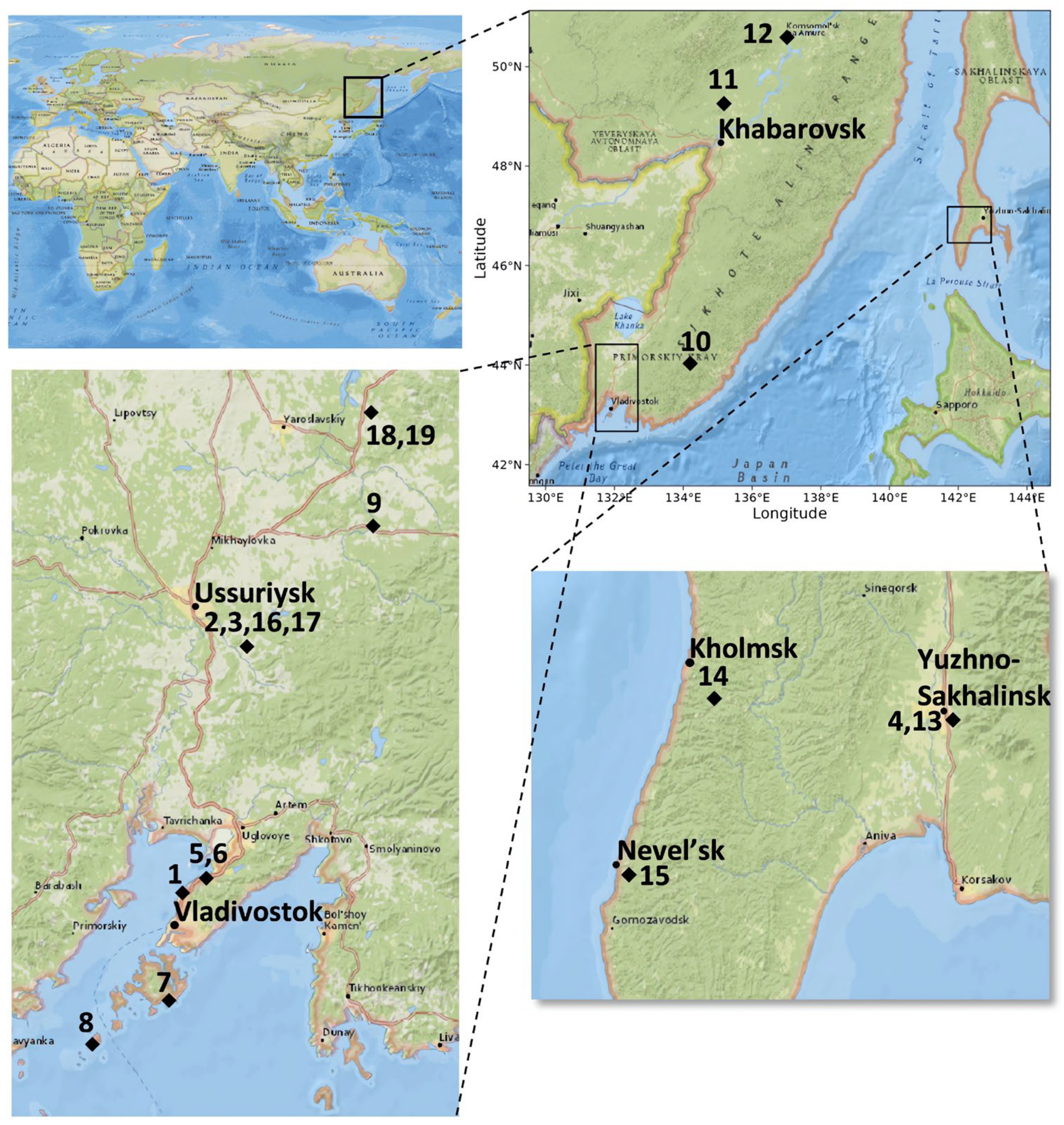
2.1. Plant Material and Surface Sterilization of Samples
2.2. DNA Isolation, Library Preparation, and Illumina MiSeq Sequencing
2.3. Data Processing
3. Results
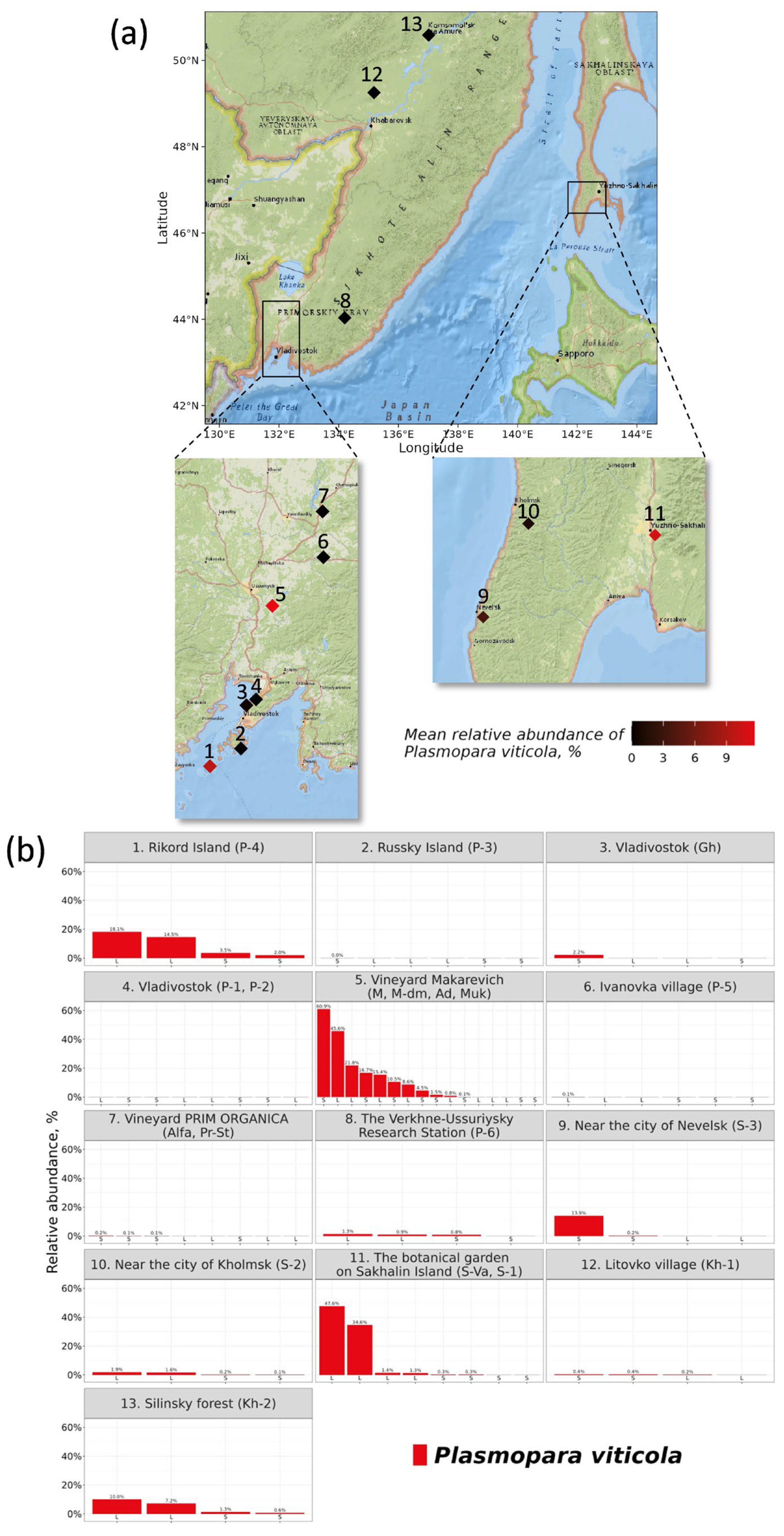
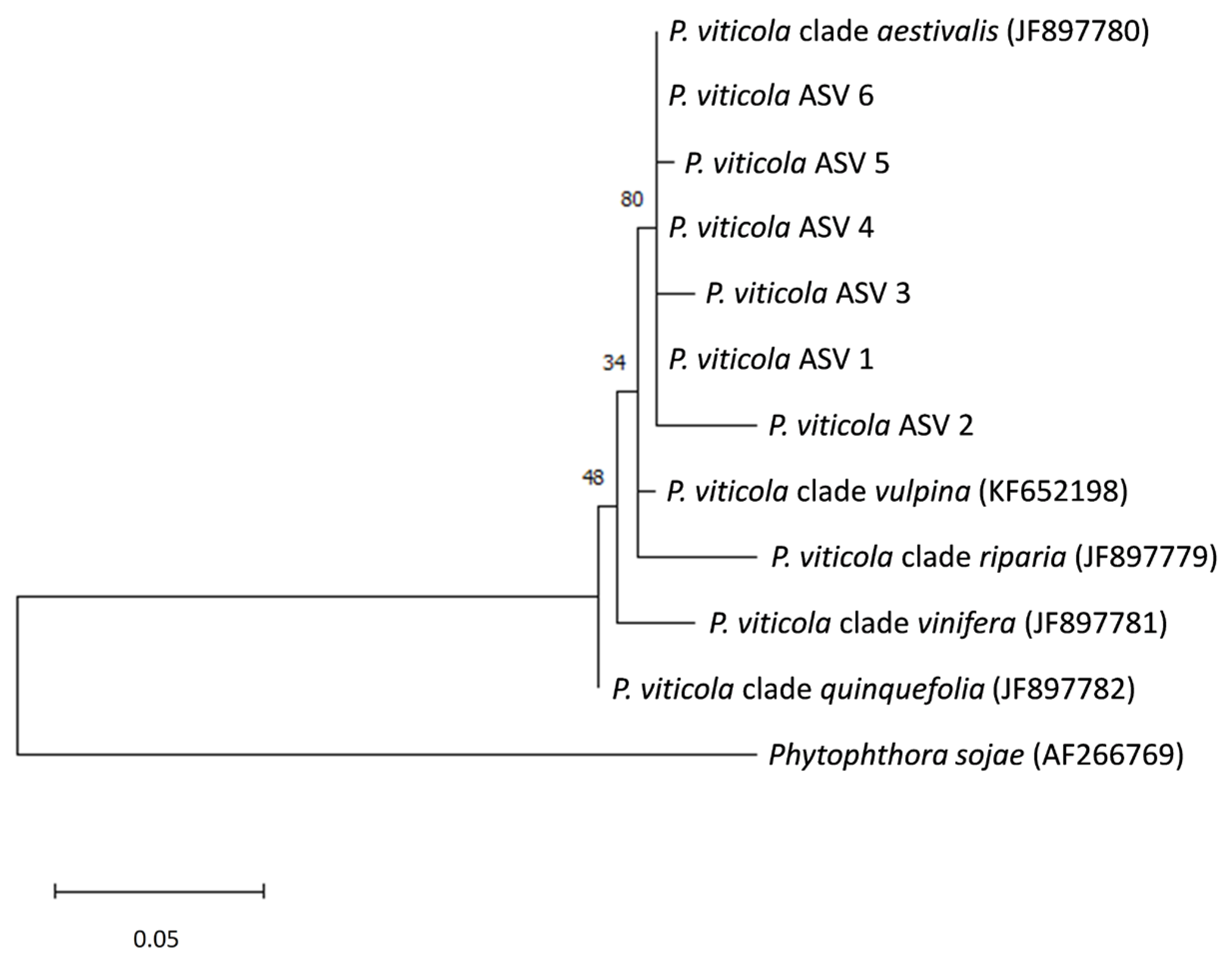
3.1. Distribution of the Plasmopara Viticola ITS1 Sequences in Grape Samples
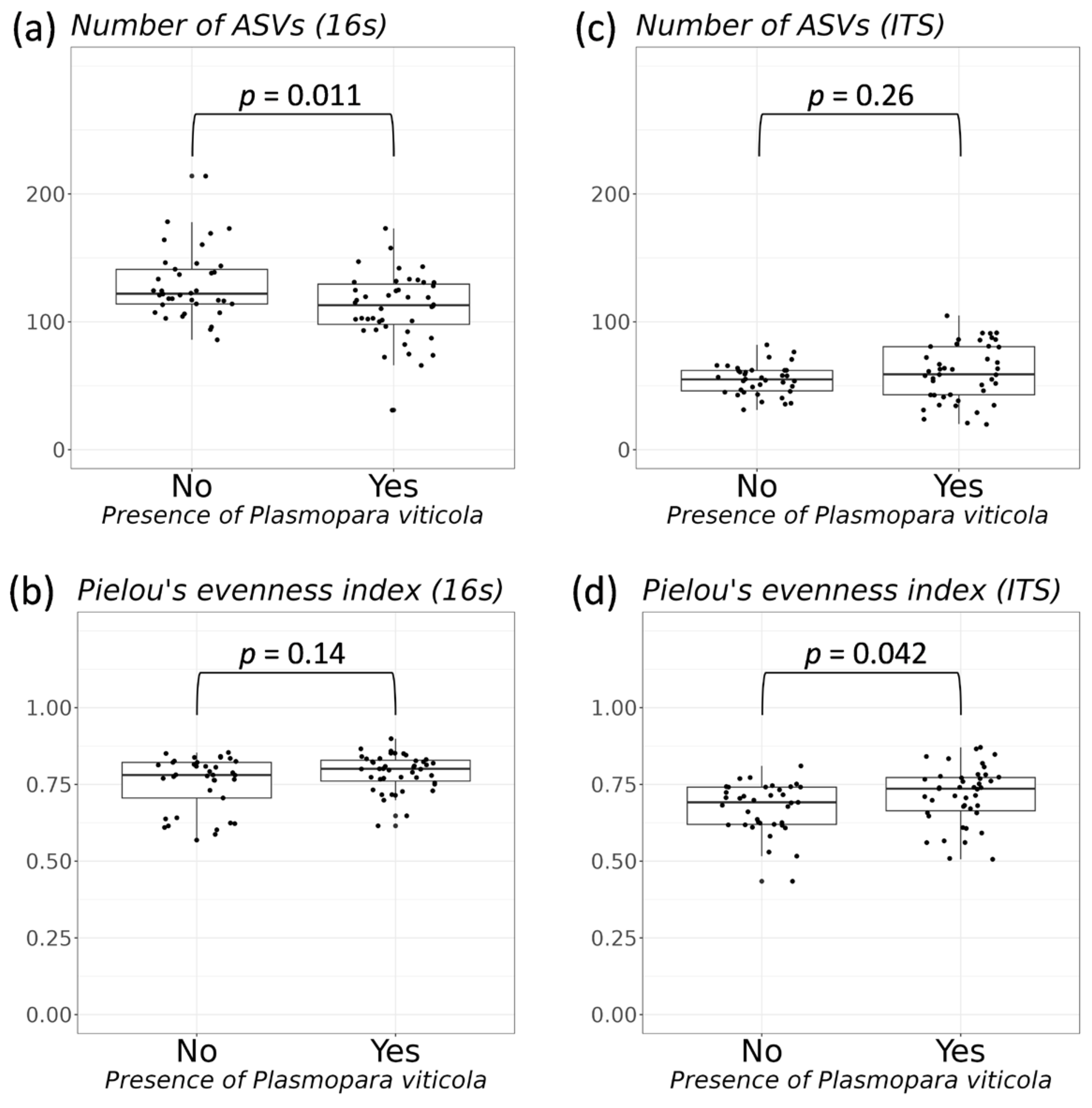
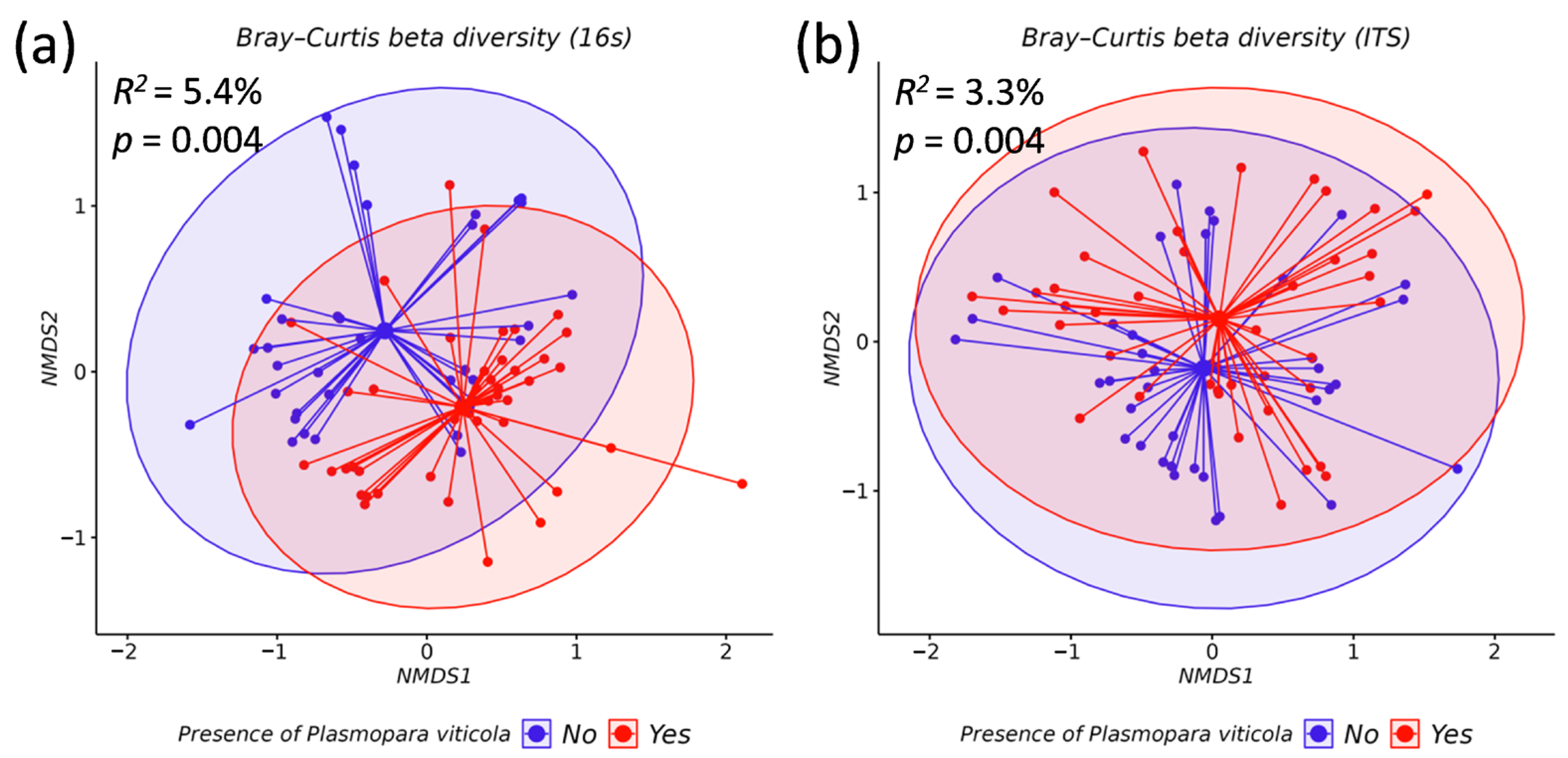
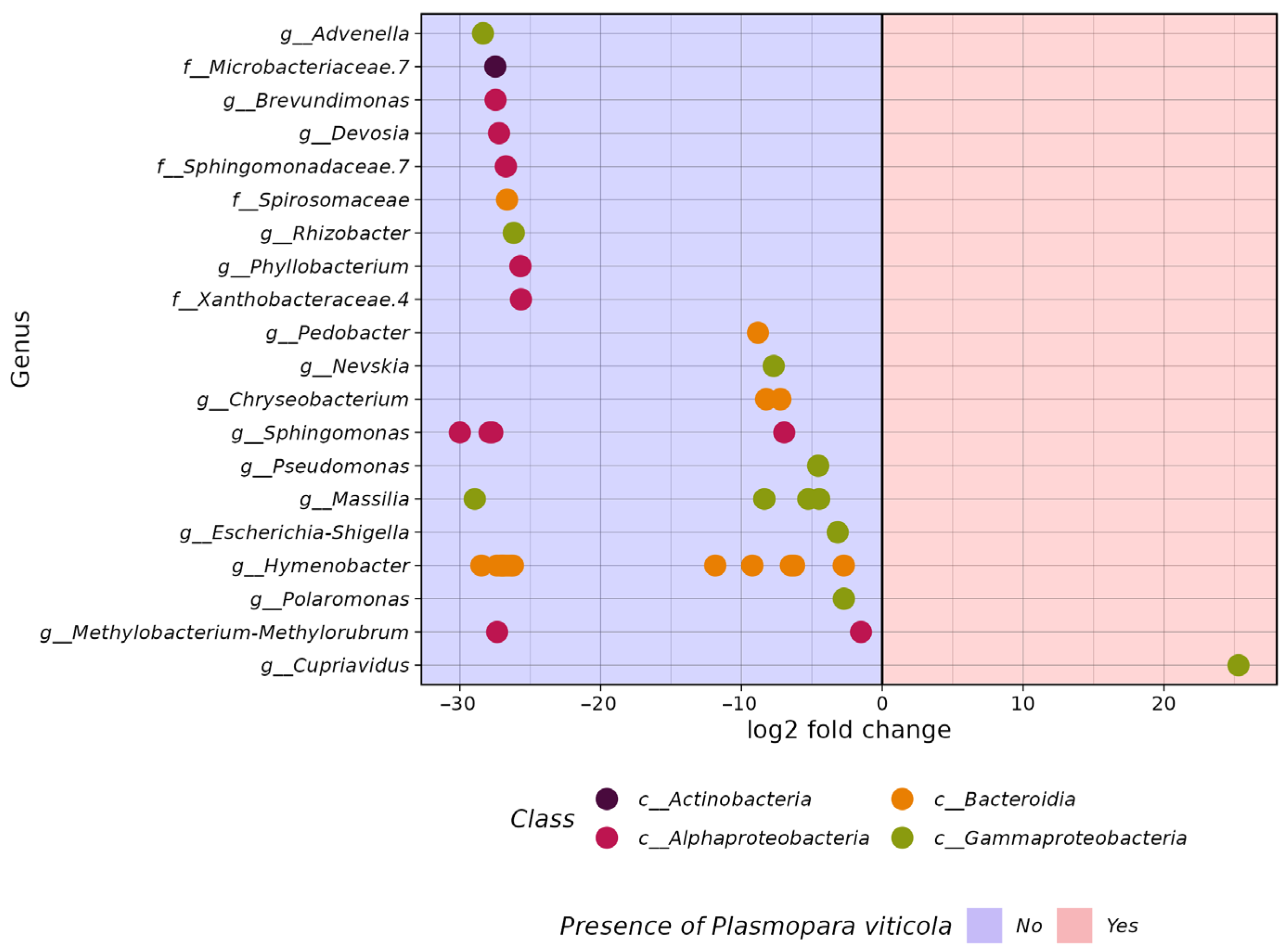
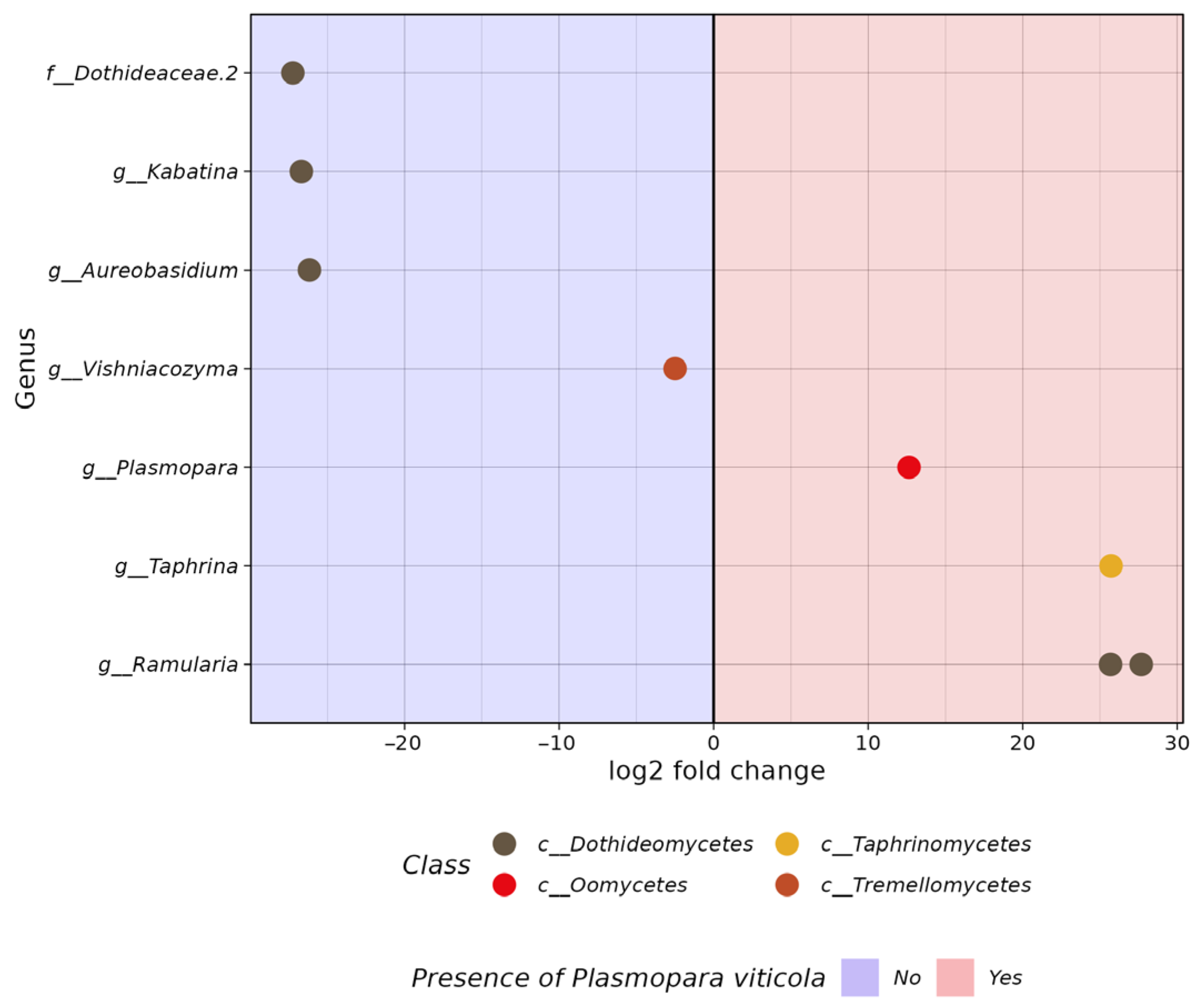
3.2. Comparative Analysis of the Biodiversity of Grape Endophytes in Grape Samples Affected by P. viticola
3.3. The In Silico Analysis of Potential Microorganisms Plasmopara viticola Antagonists
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lafon, R.; Clerjeau, M. Downy Mildew. In Compendium of Grape Diseases; APS Press: St. Paul, MN, USA, 1988; pp. 11–13. [Google Scholar]
- Koledenkova, K.; Esmaeel, Q.; Jacquard, C.; Nowak, J.; Clément, C.; Ait Barka, E. Plasmopara viticola the Causal Agent of Downy Mildew of Grapevine: From Its Taxonomy to Disease Management. Front. Microbiol. 2022, 13, 889472. [Google Scholar] [CrossRef]
- Massi, F.; Torriani, S.F.F.; Borghi, L.; Toffolatti, S.L. Fungicide Resistance Evolution and Detection in Plant Pathogens: Plasmopara viticola as a Case Study. Microorganisms 2021, 9, 119. [Google Scholar] [CrossRef]
- Feechan, A.; Anderson, C.; Torregrosa, L.; Jermakow, A.; Mestre, P.; Wiedemann-Merdinoglu, S.; Merdinoglu, D.; Walker, A.R.; Cadle-Davidson, L.; Reisch, B.; et al. Genetic Dissection of a TIR-NB-LRR Locus from the Wild North American Grapevine Species Muscadinia rotundifolia Identifies Paralogous Genes Conferring Resistance to Major Fungal and Oomycete Pathogens in Cultivated Grapevine. Plant J. 2013, 76, 661–674. [Google Scholar] [CrossRef]
- Yin, L.; An, Y.; Qu, J.; Li, X.; Zhang, Y.; Dry, I.; Wu, H.; Lu, J. Genome Sequence of Plasmopara viticola and Insight into the Pathogenic Mechanism. Sci. Rep. 2017, 7, 46553. [Google Scholar] [CrossRef]
- Delbac, L.; Delière, L.; Schneider, C.; Delmotte, F. Downy Mildew Is Able to Carry out Its Sexual Cycle on Resistant Grape Varieties: Sourced from the Research Article “Evidence for Sexual Reproduction and Fertile Oospore Production by Plasmopara viticola on the Leaves of Partially Resistant Grapevine Varieties” (ACTA Horticulturae, 1248. ISHS, 2019). Original Language of the Article: French. IVES Tech. Rev. Vine Wine 2020. [Google Scholar] [CrossRef]
- Olmo, H.P. Vinifera rotundifolia Hybrids as Wine Grapes. Am. J. Enol. Vitic. 1971, 22, 87–91. [Google Scholar] [CrossRef]
- Staudt, G.; Kassemeyer, H.H. Evaluation of Downy Mildew Resistance in Various Accessions of Wild Vitis Species. Vitis 1995, 34, 225–228. [Google Scholar]
- Díez-Navajas, A.M.; Wiedemann-Merdinoglu, S.; Greif, C.; Merdinoglu, D. Nonhost Versus Host Resistance to the Grapevine Downy Mildew, Plasmopara viticola, Studied at the Tissue Level. Phytopathology 2008, 98, 776–780. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, Y.; Zhang, H.; Huang, H.; Folta, K.M.; Lu, J. Whole Genome Wide Expression Profiles of Vitis amurensis Grape Responding to Downy Mildew by Using Solexa Sequencing Technology. BMC Plant Biol. 2010, 10, 234. [Google Scholar] [CrossRef]
- Liu, L.; Li, H. Review: Research Progress in Amur Grape, Vitis amurensis Rupr. Can. J. Plant Sci. 2013, 93, 565–575. [Google Scholar] [CrossRef]
- Ma, Y.Y.; Zhang, Y.L.; Shao, H.; Lu, J. Differential Physio-Biochemical Responses to Cold Stress of Cold-Tolerant and Non-Tolerant Grapes (Vitis L.) from China. J. Agron. Crop Sci. 2010, 196, 212–219. [Google Scholar] [CrossRef]
- Chen, Q.; Diao, L.; Song, H.; Zhu, X. Vitis amurensis Rupr: A Review of Chemistry and Pharmacology. Phytomedicine 2018, 49, 111–122. [Google Scholar] [CrossRef]
- Blasi, P.; Blanc, S.; Wiedemann-Merdinoglu, S.; Prado, E.; Rühl, E.H.; Mestre, P.; Merdinoglu, D. Construction of a Reference Linkage Map of Vitis amurensis and Genetic Mapping of Rpv8, a Locus Conferring Resistance to Grapevine Downy Mildew. Theor. Appl. Genet. 2011, 123, 43–53. [Google Scholar] [CrossRef]
- Schwander, F.; Eibach, R.; Fechter, I.; Hausmann, L.; Zyprian, E.; Töpfer, R. Rpv10: A New Locus from the Asian Vitis Gene Pool for Pyramiding Downy Mildew Resistance Loci in Grapevine. Theor. Appl. Genet. 2012, 124, 163–176. [Google Scholar] [CrossRef]
- Venuti, S.; Copetti, D.; Foria, S.; Falginella, L.; Hoffmann, S.; Bellin, D.; Cindrić, P.; Kozma, P.; Scalabrin, S.; Morgante, M.; et al. Historical Introgression of the Downy Mildew Resistance Gene Rpv12 from the Asian Species Vitis amurensis into Grapevine Varieties. PLoS ONE 2013, 8, e61228. [Google Scholar] [CrossRef]
- Wan, Y.; Schwaninger, H.; He, P.; Wang, Y. Comparison of Resistance to Powdery Mildew and Downy Mildew in Chinese Wild Grapes. Vitis 2007, 46, 132–136. [Google Scholar]
- Takayama, F.; Nakamoto, K.; Kawasaki, H.; Mankura, M.; Egashira, T.; Ueki, K.; Hasegawa, A.; Okada, S.; Mori, A. Beneficial Effects of Vitis coignetiae Pulliat Leaves on Nonalcoholic Steatohepatitis in a Rat Model. Acta Medica Okayama 2009, 63, 105–111. [Google Scholar]
- Kim, M.J.; Paramanantham, A.; Lee, W.S.; Yun, J.W.; Chang, S.H.; Kim, D.C.; Park, H.S.; Choi, Y.H.; Kim, G.S.; Ryu, C.H.; et al. Anthocyanins Derived from Vitis coignetiae Pulliat Contributes Anti-Cancer Effects by Suppressing NF-κB Pathways in Hep3B Human Hepatocellular Carcinoma Cells and in vivo. Molecules 2020, 25, 5445. [Google Scholar] [CrossRef]
- Kim, B.R.; Kim, I.H.; Lee, J.S.; Choi, Y.J. First Report of Downy Mildew Caused by Plasmopara viticola on Vitis coignetiae in Korea. Plant Dis. 2019, 103, 1793. [Google Scholar] [CrossRef]
- Hamaoka, K.; Aoki, Y.; Suzuki, S. Isolation and Characterization of Endophyte Bacillus velezensis KOF112 from Grapevine Shoot Xylem as Biological Control Agent for Fungal Diseases. Plants 2021, 10, 1815. [Google Scholar] [CrossRef]
- Lo Piccolo, S.; Alfonzo, A.; Giambra, S.; Conigliaro, G.; Lopez-Llorca, L.V.; Burruano, S. Identification of Acremonium Isolates from Grapevines and Evaluation of Their Antagonism towards Plasmopara viticola. Ann. Microbiol. 2015, 65, 2393–2403. [Google Scholar] [CrossRef]
- Bruisson, S.; Zufferey, M.; L’Haridon, F.; Trutmann, E.; Anand, A.; Dutartre, A.; De Vrieze, M.; Weisskopf, L. Endophytes and Epiphytes from the Grapevine Leaf Microbiome as Potential Biocontrol Agents Against Phytopathogens. Front. Microbiol. 2019, 10, 491406. [Google Scholar] [CrossRef] [PubMed]
- Musetti, R.; Polizzotto, R.; Vecchione, A.; Borselli, S.; Zulini, L.; D’Ambrosio, M.; di Toppi, L.S.; Pertot, I. Antifungal Activity of Diketopiperazines Extracted from Alternaria alternata against Plasmopara viticola: An Ultrastructural Study. Micron 2007, 38, 643–650. [Google Scholar] [CrossRef]
- National Geographic Basemap. Available online: https://www.esri.com/news/arcuser/0312/national-geographic-basemap.html (accessed on 21 March 2024).
- Aleynova, O.A.; Nityagovsky, N.N.; Dubrovina, A.S.; Kiselev, K.V. The Biodiversity of Grapevine Bacterial Endophytes of Vitis amurensis Rupr. Plants 2022, 11, 1128. [Google Scholar] [CrossRef] [PubMed]
- Aleynova, O.A.; Nityagovsky, N.N.; Suprun, A.R.; Ananev, A.A.; Dubrovina, A.S.; Kiselev, K.V. The Diversity of Fungal Endophytes from Wild Grape Vitis amurensis Rupr. Plants 2022, 11, 2897. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, K.V.; Nityagovsky, N.N.; Aleynova, O.A. A Method of DNA Extraction from Plants for Metagenomic Analysis Based on the Example of Grape Vitis amurensis Rupr. Appl. Biochem. Microbiol. 2023, 59, 361–367. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME Release for Eukaryotes; Version 29.11.2022; UNITE Community: London, UK, 2022. [Google Scholar] [CrossRef]
- Bisanz, J. qiime2R: Importing QIIME2 Artifacts and Associated Data into R Sessions. 2018. Available online: https://github.com/jbisanz/qiime2R (accessed on 21 March 2024).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 21 March 2024).
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hernangómez, D. Using the Tidyverse with Terra Objects: The Tidyterra Package. J. Open Source Softw. 2023, 8, 5751. [Google Scholar] [CrossRef]
- Pebesma, E. Simple Features for R: Standardized Support for Spatial Vector Data. R J. 2018, 10, 439–446. [Google Scholar] [CrossRef]
- Giraud, T. Maptiles: Download and Display Map Tiles. 2023. Available online: https://github.com/riatelab/maptiles (accessed on 21 March 2024).
- Hugh-Jones, D. Ggmagnify: Create a Magnified Inset of Part of a Ggplot Object. 2023. Available online: https://github.com/hughjonesd/ggmagnify (accessed on 21 March 2024).
- Rouxel, M.; Mestre, P.; Comont, G.; Lehman, B.L.; Schilder, A.; Delmotte, F. Phylogenetic and Experimental Evidence for Host-Specialized Cryptic Species in a Biotrophic Oomycete. New Phytol. 2013, 197, 251–263. [Google Scholar] [CrossRef]
- Rouxel, M.; Mestre, P.; Baudoin, A.; Carisse, O.; Delière, L.; Ellis, M.A.; Gadoury, D.; Lu, J.; Nita, M.; Richard-Cervera, S.; et al. Geographic Distribution of Cryptic Species of Plasmopara viticola Causing Downy Mildew on Wild and Cultivated Grape in Eastern North America. Phytopathology 2014, 104, 692–701. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000; ISBN 978-0-19-513585-5. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Toffolatti, S.L.; Serrati, L.; Sierotzki, H.; Gisi, U.; Vercesi, A. Assessment of QoI Resistance in Plasmopara viticola Oospores. Pest Manag. Sci. 2007, 63, 194–201. [Google Scholar] [CrossRef]
- Huang, X.; Wang, X.; Kong, F.; van der Lee, T.; Wang, Z.; Zhang, H. Detection and Characterization of Carboxylic Acid Amide-Resistant Plasmopara viticola in China Using a TaqMan-MGB Real-Time PCR. Plant Dis. 2020, 104, 2338–2345. [Google Scholar] [CrossRef]
- Yang, L.; Chu, B.; Deng, J.; Yuan, K.; Sun, Q.; Jiang, C.; Ma, Z. Use of a Real-Time PCR Method to Quantify the Primary Infection of Plasmopara viticola in Commercial Vineyards. Phytopathol. Res. 2023, 5, 19. [Google Scholar] [CrossRef]
- Mouafo-Tchinda, R.A.; Beaulieu, C.; Fall, M.L.; Carisse, O. Effect of Temperature on Aggressiveness of Plasmopara viticola f. sp. aestivalis and P. viticola f. sp. riparia from Eastern Canada. Can. J. Plant Pathol. 2021, 43, 73–87. [Google Scholar] [CrossRef]
- Pezet, R.; Pont, V.; Cuenat, P. Method to Determine Resveratrol and Pterostilbene in Grape Berries and Wines Using High-Performance Liquid Chromatography and Highly Sensitive Fluorimetric Detection. J. Chromatogr. A 1994, 663, 191–197. [Google Scholar] [CrossRef]
- Langcake, P. Disease Resistance of Vitis spp. and the Production of the Stress Metabolites Resveratrol, ε-Viniferin, α-Viniferin and Pterostilbene. Physiol. Plant Pathol. 1981, 18, 213–226. [Google Scholar] [CrossRef]
- Dercks, W.; Creasy, L.L. The Significance of Stilbene Phytoalexins in the Plasmopara viticola-Grapevine Interaction. Physiol. Mol. Plant Pathol. 1989, 34, 189–202. [Google Scholar] [CrossRef]
- Pezet, R.; Perret, C.; Jean-Denis, J.B.; Tabacchi, R.; Gindro, K.; Viret, O. δ-Viniferin, a Resveratrol Dehydrodimer: One of the Major Stilbenes Synthesized by Stressed Grapevine Leaves. J. Agric. Food Chem. 2003, 51, 5488–5492. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, K.V.; Aleynova, O.A.; Grigorchuk, V.P.; Dubrovina, A.S. Stilbene Accumulation and Expression of Stilbene Biosynthesis Pathway Genes in Wild Grapevine Vitis amurensis Rupr. Planta 2017, 245, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.-F.; Ma, L.; Wang, L.-N.; Li, S.-H.; Wang, L.-J. Differential Response of the Biosynthesis of Resveratrols and Flavonoids to UV-C Irradiation in Grape Leaves. N. Z. J. Crop Hortic. Sci. 2015, 43, 163–172. [Google Scholar] [CrossRef]
- Possamai, T.; Wiedemann-Merdinoglu, S. Phenotyping for QTL Identification: A Case Study of Resistance to Plasmopara viticola and Erysiphe necator in Grapevine. Front. Plant Sci. 2022, 13, 930954. [Google Scholar] [CrossRef]
- Fontaine, M.C.; Labbé, F.; Dussert, Y.; Delière, L.; Richart-Cervera, S.; Giraud, T.; Delmotte, F. Europe as a Bridgehead in the Worldwide Invasion History of Grapevine Downy Mildew, Plasmopara viticola. Curr. Biol. 2021, 31, 2155–2166.e4. [Google Scholar] [CrossRef]
- Chiapello, M.; Rodríguez-Romero, J.; Nerva, L.; Forgia, M.; Chitarra, W.; Ayllón, M.A.; Turina, M. Putative New Plant Viruses Associated with Plasmopara viticola-Infected Grapevine Samples. Ann. Appl. Biol. 2020, 176, 180–191. [Google Scholar] [CrossRef]
- Estrada-de los Santos, P.; Vacaseydel-Aceves, N.B.; Martínez-Aguilar, L.; Cruz-Hernández, M.A.; Mendoza-Herrera, A.; Caballero-Mellado, J. Cupriavidus and Burkholderia Species Associated with Agricultural Plants That Grow in Alkaline Soils. J. Microbiol. 2011, 49, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Videira, S.I.R.; Groenewald, J.Z.; Braun, U.; Shin, H.D.; Crous, P.W. All That Glitters Is Not Ramularia. Stud. Mycol. 2016, 83, 49–163. [Google Scholar] [CrossRef] [PubMed]
- McGrann, G.; Stavrinides, A.; Russell, J.; Corbitt, M.; Booth, A.; Chartrain, L.; Thomas, W.; Brown, J. A Trade off between mlo Resistance to Powdery Mildew and Increased Susceptibility of Barley to a Newly Important Disease, Ramularia Leaf Spot. J. Exp. Bot. 2014, 65, 1025–1037. [Google Scholar] [CrossRef] [PubMed]
- Tsai, I.J.; Tanaka, E.; Masuya, H.; Tanaka, R.; Hirooka, Y.; Endoh, R.; Sahashi, N.; Kikuchi, T. Comparative Genomics of Taphrina Fungi Causing Varying Degrees of Tumorous Deformity in Plants. Genome Biol. Evol. 2014, 6, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Maeng, S.; Kim, M.K.; Subramani, G. Hymenobacter jejuensis sp. nov., a UV Radiation-Tolerant Bacterium Isolated from Jeju Island. Antonie Van Leeuwenhoek 2020, 113, 553–561. [Google Scholar] [CrossRef]
- Gadoury, D.M.; Sapkota, S.; Cadle-Davidson, L.; Underhill, A.; McCann, T.; Gold, K.M.; Gambhir, N.; Combs, D.B. Effects of Nighttime Applications of Germicidal Ultraviolet Light Upon Powdery Mildew (Erysiphe necator), Downy Mildew (Plasmopara viticola), and Sour Rot of Grapevine. Plant Dis. 2023, 107, 1452–1462. [Google Scholar] [CrossRef]
- Wassermann, B.; Korsten, L.; Berg, G. Plant Health and Sound Vibration: Analyzing Implications of the Microbiome in Grape Wine Leaves. Pathogens 2021, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Lee, D.; Lee, S.; Kong, H.J.; Park, J.; Seo, Y.-S. Comparative Genomic Analysis of Chryseobacterium Species: Deep Insights into Plant-Growth-Promoting and Halotolerant Capacities. Microb. Genom. 2023, 9, 001108. [Google Scholar] [CrossRef]
- Peláez, F.; Cabello, A.; Platas, G.; Díez, M.T.; del Val, A.G.; Basilio, A.; Martán, I.; Vicente, F.; Bills, G.F.; Giacobbe, R.A.; et al. The Discovery of Enfumafungin, a Novel Antifungal Compound Produced by an Endophytic Hormonema Species Biological Activity and Taxonomy of the Producing Organisms. Syst. Appl. Microbiol. 2000, 23, 333–343. [Google Scholar] [CrossRef]
- Di Francesco, A.; Zajc, J.; Gunde-Cimerman, N.; Aprea, E.; Gasperi, F.; Placì, N.; Caruso, F.; Baraldi, E. Bioactivity of Volatile Organic Compounds by Aureobasidium Species against Gray Mold of Tomato and Table Grape. World J. Microbiol. Biotechnol. 2020, 36, 171. [Google Scholar] [CrossRef] [PubMed]
- Yalage Don, S.M.; Schmidtke, L.M.; Gambetta, J.M.; Steel, C.C. Aureobasidium pullulans Volatilome Identified by a Novel, Quantitative Approach Employing SPME-GC-MS, Suppressed Botrytis cinerea and Alternaria alternata In Vitro. Sci. Rep. 2020, 10, 4498. [Google Scholar] [CrossRef] [PubMed]
- Lutz, M.C.; Lopes, C.A.; Rodriguez, M.E.; Sosa, M.C.; Sangorrín, M.P. Efficacy and Putative Mode of Action of Native and Commercial Antagonistic Yeasts against Postharvest Pathogens of Pear. Int. J. Food Microbiol. 2013, 164, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, T.; Xu, X.; Shi, X.; Wang, B. Succession of Fungal Communities at Different Developmental Stages of Cabernet Sauvignon Grapes from an Organic Vineyard in Xinjiang. Front. Microbiol. 2021, 12, 718261. [Google Scholar] [CrossRef]
| Name of ASV | Occurrence in the Plants | Mean Relative Abundance, % | Number of ASV-Affected Samples | Total P. viticola-Affected Samples |
|---|---|---|---|---|
| P. viticola ASV 1 | P-4, P-5, P-6, Kh-1, Kh-2, M, M-dm, Ad, Muk, Pr-St, Alfa, S-Va, S-1, S-2, S-3 | 8.84 | 37 | 43 |
| P. viticola ASV 2 | P-3, P-4, P-6, M-dm, Ad, S-1, S-2 | 0.76 | 9 | |
| P. viticola ASV 3 | S-1, S-3, P-4, Kh-2 | 0.50 | 7 | |
| P. viticola ASV 4 | Gh, Kh-1, Muk, Ad, S-1 | 2.46 | 5 | |
| P. viticola ASV 5 | M-dm, S-1 | 1.75 | 2 | |
| P. viticola ASV 6 | S-1 | 0.28 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nityagovsky, N.N.; Ananev, A.A.; Suprun, A.R.; Ogneva, Z.V.; Dneprovskaya, A.A.; Tyunin, A.P.; Dubrovina, A.S.; Kiselev, K.V.; Sanina, N.M.; Aleynova, O.A. Distribution of Plasmopara viticola Causing Downy Mildew in Russian Far East Grapevines. Horticulturae 2024, 10, 326. https://doi.org/10.3390/horticulturae10040326
Nityagovsky NN, Ananev AA, Suprun AR, Ogneva ZV, Dneprovskaya AA, Tyunin AP, Dubrovina AS, Kiselev KV, Sanina NM, Aleynova OA. Distribution of Plasmopara viticola Causing Downy Mildew in Russian Far East Grapevines. Horticulturae. 2024; 10(4):326. https://doi.org/10.3390/horticulturae10040326
Chicago/Turabian StyleNityagovsky, Nikolay N., Alexey A. Ananev, Andrey R. Suprun, Zlata V. Ogneva, Alina A. Dneprovskaya, Alexey P. Tyunin, Alexandra S. Dubrovina, Konstantin V. Kiselev, Nina M. Sanina, and Olga A. Aleynova. 2024. "Distribution of Plasmopara viticola Causing Downy Mildew in Russian Far East Grapevines" Horticulturae 10, no. 4: 326. https://doi.org/10.3390/horticulturae10040326
APA StyleNityagovsky, N. N., Ananev, A. A., Suprun, A. R., Ogneva, Z. V., Dneprovskaya, A. A., Tyunin, A. P., Dubrovina, A. S., Kiselev, K. V., Sanina, N. M., & Aleynova, O. A. (2024). Distribution of Plasmopara viticola Causing Downy Mildew in Russian Far East Grapevines. Horticulturae, 10(4), 326. https://doi.org/10.3390/horticulturae10040326










