Abstract
Fusarium basal rot (FBR) places a significant limitation on Allium production worldwide. The damage caused by the disease can be observed throughout the entire crop cycle. This research aimed to further our understanding of the impact of FBR on the cultivation of onions (Allium cepa) in northeast Israel. It focused on studying the composition and incidence of Fusarium species involved in disease outbursts in two representative fields, one in Galilee (Hula Valley) and the second in the Golan Heights, where the disease incidences reached 8%. Using colony morphology, microscopic taxonomic keys, and molecular methods, a new, unreported Neocosmospora (previously Fusarium solani) species complex (SC, mostly N. falciformis) was discovered as a wildly spread member of the Fusarium pathobiome community. This species complex appeared more generalist in its nature since it was found in all three onion cultivars’ samples. It was also less virulent in seed germination (42–52% higher sprout biomass, p < 0.05) and bulb pathogenicity tests (41–45% less necrotic) than Fusarium acutatum. Whereas the Galilee yellow Orlando (Riverside) onion cultivar bulbs sampled were colonized by Neocosmospora SC (70%) and two other, less abundant species, F. oxysporum f. sp. cepae and F. acutatum (15% each), the Golan Heights field’s Fusarium community showed host specificity. In the Golan Heights field, F. oxysporum f. sp. cepae inhabited the red Ha2 onion cultivar bulbs, whereas F. acutatum colonized the yellow Ha1 cultivar (40% and 50% prevalence along with Neocosmospora SC). A better understanding of the complexity of this disease caused by different Fusarium species and with a divergence in host susceptibility and virulence is critical for developing disease management strategies. Since each Fusarium species reacts differently to pest control treatments, changes in the species composition may require specifically adapted management solutions.
1. Introduction
Onion (Allium cepa L.) is an important agricultural crop globally. In 2022, the global planted area of onions and shallots was estimated to be 5,967,491 ha, representing a production of ca. 110,616,270 tons of dry cultivars (FAO 2022, available at: https://www.fao.org/faostat/en/#data/QCL, accessed 1 April 2024). Fusarium basal rot (FBR, also known as Fusarium rot, Fusarium wilt, or basal plate rot of Allium spp.) is a serious fungal disease affecting onion crops in many parts of the world [1]. Notwithstanding the ubiquitous incidence of FBR across various Allium species, knowledge pertaining to this disease remains fragmented and incomplete [2].
The FBR disease is caused by various species of the genus Fusarium, with Fusarium oxysporum f. sp. cepae (formae speciales in Allium cepa) being the most commonly reported species [2]. The pathogen infects the onion plant through its roots and causes rotting of the basal plate (the roots and stem connection portion). Infected plants may show delayed growth, yellowing, and wilting. Young seedlings and dormant mature onion bulbs are the two most valuable plant phenological stages of FBR. Yet, the disease can occur throughout the entire crop cycle [3,4,5,6]. It leads to seedlings’ pre- and post-emergence mortality (damping off) and can cause significant crop losses. Yield losses in the field can reach 39% (Turkey [7]) and 50% (Nigeria [8]), depending on the inoculum pressure and the cultivar’s susceptibility. FBR pre- and post-harvest results in significant crop reduction due to reduced bulb size (growth suppression), bulb rot, and decreased onion shelf-life [9].
Interspecies variation in virulence is a common feature in Fusarium populations, and its impact on the developmental stage susceptibility to FBR is well-documented [10]. In most cases, onion susceptibility to FBR decreases with seedling age but increases at bulb development until post-harvest [5]. Nowadays, studies are attempting to uncover the mechanisms behind those differential responses and the role of mycotoxins in determining disease outcomes. These are particularly important knowledge gains since the Fusarium species produce various mycotoxins that may harm human health and animals [11]. Defense-related genes in seedlings and bulbs are expressed depending on the Fusarium isolates’ aggressiveness as part of the plant defense [1]. Yet, in susceptible plants, they may not be effective in ensuring a resistance response against FBR. Meanwhile, secondary fungal metabolites play a differential role during colonization at the respective stages. For example, the toxin fumonisin B1 appears to be a virulence factor specific to the seedling phase [1].
Notwithstanding the extensive implementation of various control measures, the disease continues to pose a significant challenge for Allium producers on a global scale [2]. Early detection and rapid disease management action are essential to minimize the impact on onion crops. FBR control toolkits should utilize a combination of cultural, chemical, and biological control measures. These could include planting disease-free onion sets, using crop rotation to reduce the buildup of the fungus in the soil, applying fungicides, and using biocontrol agents such as Trichoderma and Bacillus [2]. In addition, it is crucial to apply good sanitation practices to prevent the spread of fungus. Such hygiene practices could include removing infected plants and crop residuals from the field and disinfecting tools and equipment between uses.
Recent research has also explored other control strategies, such as breeding for resistance and RNA interference (RNAi) to silence specific fungal genes [12,13]. For example, it was demonstrated that spray application of a long dsRNA (791 nt CYP3-dsRNA), that targets F. graminearum cytochrome P450, lanosterol, and C-14α-demethylase genes (required for fungal ergosterol biosynthesis) significantly inhibited fungal growth [14]. Furthermore, synthetic siRNAs proved to down-regulate key fungal genes involved in Fusarium toxin production [15,16]. In banana (Musa sp.), significant resistance (70–85% reduction in disease symptom) to F. oxysporum was observed at eight months post-inoculation in the RNAi plants that could silence the Fusarium velvet protein complex (which regulates fungal development and secondary metabolism) and transcription factor 1 gene [17].
In 2022, onion and shallot bulb (dry, excluded dehydrated) production in Israel covered an area of 3865 ha throughout the country, and the commercial production reached 82,503 tons (FAO 2022, available at: https://www.fao.org/faostat/en/#data/QCL, accessed 1 April 2024). Fusarium basal rot has been reported as a significant disease of onion crops in Israel, with outbreaks occurring in various onion-growing regions in the country [18,19]. Despite the latest scientific efforts [9,20,21,22], information regarding Israel’s FBR prevalence is scarce. In a recent study, four distinct Fusarium species were successfully isolated from onion bulbs sampled from infected fields in northeastern Israel’s Golan Heights region [21]. The isolated species, F. proliferatum, F. oxysporum f. sp. cepae, F. acutatum, and F. anthophilium, were identified and characterized. The latter two species are lesser-known species implicated as FBR causal agents. Despite these findings, other pathogenic Fusarium species may also contribute to FBR. Significant knowledge gaps remain pertaining to the nature and distribution of the disease in Israel and the control measures employed to combat it. Specifically, no structured data have been reported concerning the disease’s historical prevalence or its rate of spread over time, and there is no current map of its distribution. Furthermore, no onion cultivars demonstrating resistance to FBR have been identified. Only recently have fungicides that can effectively target the disease’s causal pathogens been established [9], and their application on a commercial field scale has not yet been accomplished.
The objectives of the present study were to improve our understanding of the Fusarium species involved in FBR epidemics in two representative commercial fields in northeastern Israel, one in Galilee (Hula Valley) and the other in the Golan Heights. Based on the accumulating scientific data [9,20,21,22], our research hypothesis was that the variety of Fusarium species involved in FBR would be found to be much greater than that so far discovered and that they would be found to thrive in complex compositions depending on the host plant and the environment. We also hypothesized that the impact of the disease in some commercial fields would be found to be more significant than previously thought. The disease damage was evaluated in a field survey at the flowering stage by determining the dry inflorescence percentage in a sample of 2000 plants. Fungal species were isolated from bulb samples and identified using colony morphology, microscopic evaluation, and molecular targeting of the Fusarium translation elongation factor-1 alpha gene (TEF1), the RNA polymerase largest or second-largest subunit (RPB1 or RPB2), and the F. oxysporum f.sp. cepae species-specific putative effector secreted in xylem genes 3 (SIX3). Using a phylogenetic tree and the inter simple sequence repeat (ISSR)-polymerase chain reaction (PCR) molecular method, the Fusarium microflora of each onion cultivar was uncovered, leading to a curious new insight. The newly identified Neocosmospora (previously F. solani) species complex (SC) was tested for its pathogenicity towards onion seedlings and bulbs, and Koch’s postulates were accomplished. The results of this study provide deep insights into the pathosystem associated with Fusarium basal rot and its significant implications for disease management decision-making.
2. Materials and Methods
2.1. Evaluation of Disease Severity
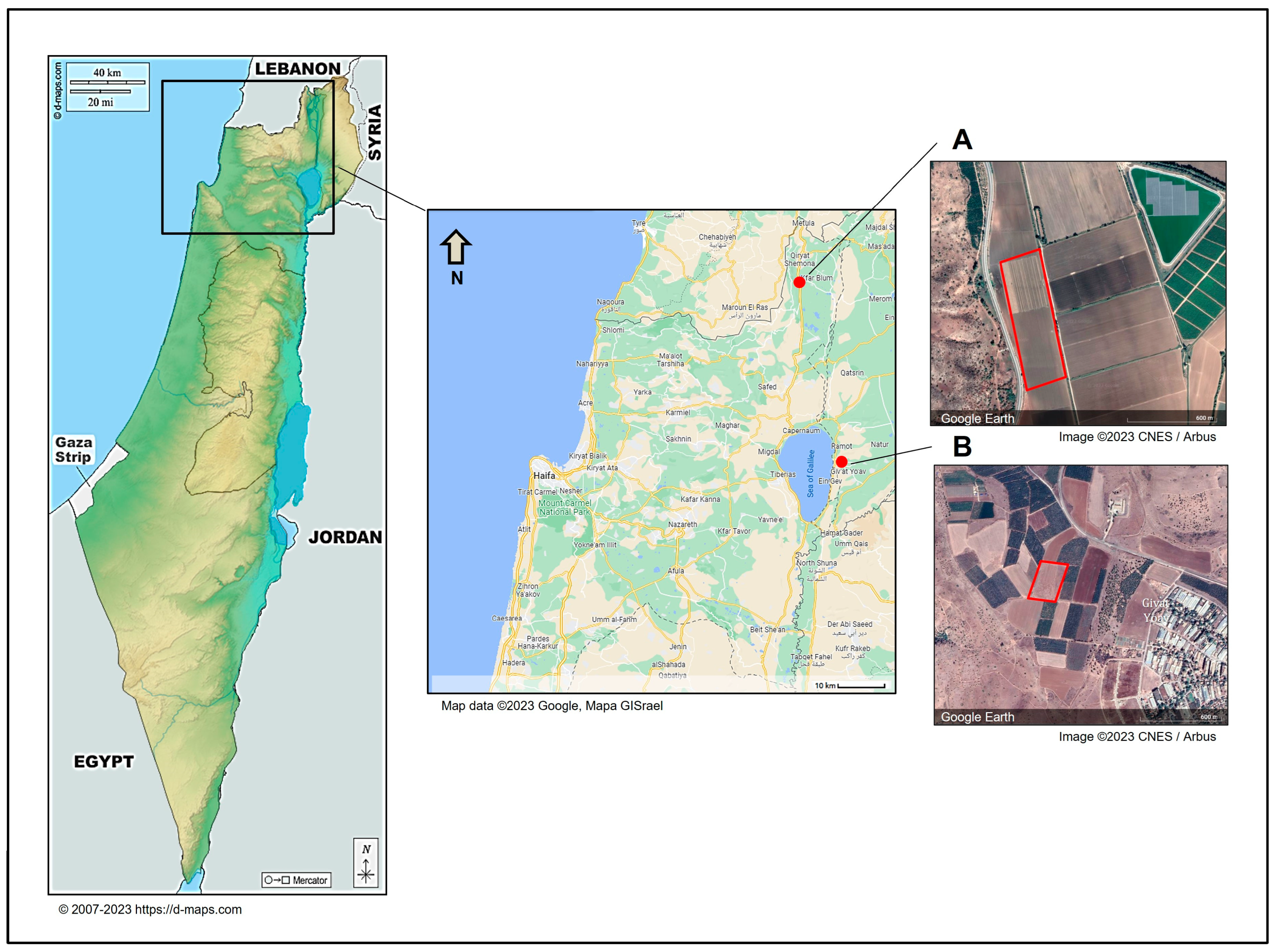
This study aimed to enhance our understanding of FBR in onion cultivars infected with toxigenic Fusarium species in northeastern Israel. The FBR disease incidence was evaluated in a commercial field in northeast Israel’s Golan Heights, part of the Givat Yoav farm, termed plot 8 (32°48′16.7″ N 35°40′16.2″ E, Figure 1). The area was sown on 7 October 2021, and sampling was carried out on day 237 from sowing (1 June 2022). We utilized our fieldwork to sample onion bulbs for the Fusarium species identification study and conducted a disease incidence survey, although this was not the primary focus of the current work. The Givat Yoav field, which was included in the survey, consisted of inbred plants that had undergone vernalization and were in the flowering stage. This enabled us to easily identify diseased dry inflorescences and conduct the survey without reducing crop yield. In contrast, the second field, Yiron’s field, was seeded with a hybrid yellow Orlando (Riverside) cultivar and was cultivated for bulb production, thus denying the flowering stage. Consequently, identifying diseased bulbs near the end of the season (before harvest) in 2000 plants could have reduced the yield, and we decided to avoid this. Moreover, the different cultivation purposes (seeds or bulbs) rendered these two fields noncomparable.
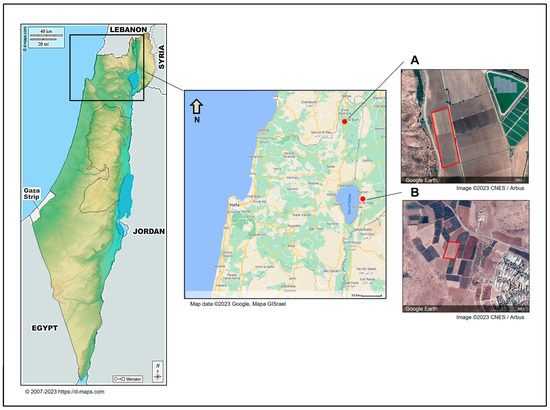
Figure 1.
The locations of sampling sites in northeastern Israel. Onion samples were collected from two representative commercial fields in northeast Israel. The sampled fields were Kibbutz Yiron (Galilee, Hula Valley, (A)) and Givat Yoav (Golan Heights, (B)), highlighted by red boxes. The Givat Yoav field was also used for a survey evaluating disease severity. According to d-maps.com (accessed on 1 April 2024) and the Google Maps/Google Earth (https://www.google.com/intl/en_GB/help/terms_maps/, accessed on 1 April 2024) Terms of Service, the maps and photos can be used and adapted. The map of Israel is from https://d-maps.com/pays.php?num_pay=88&lang=en (accessed on 1 April 2024).
The FBR assessment survey was conducted in the eastern part of the plot. Along the 300 m row, ten inflorescences were counted at every two meters. Each blossom was gently drawn. The diseased plants were easily uprooted. In the red onion varieties (Ha2—a male breed cultivar, and Ha3—a female cultivar), 500 inflorescences were sampled per cultivar. In the yellow variety (Ha1—a female cultivar), 1000 inflorescences were tested. All three cultivars were from Hazera Seeds Ltd. (Berurim, Israel). The disease incidence (number of diseased plants/total plants sampled) was calculated according to [21].
2.2. Isolation of Pathogens from Diseased Onion Plants
Near the time of onion collection (the season’s end), onion samples were collected from two representative commercial fields of onion production in northeast Israel (Galilee and Golan Heights). The two fields were typical of Israel’s northeast agriculture region. The tested fields were Givat Yoav and Yiron (Figure 1). Yiron’s field (33°09′19.4″ N 35°34′23.1″ E) was sown with the yellow Orlando cv. on 9 January 2022 and was sampled on day 228 of sowing (25 August 2022). The Givat Yoav field (described in Section 2.1) included three onion cultivars: red female (Ha3 cv.), red male (Ha2 cv.), and yellow female (Ha1 cv.) A total of 67 onion bulbs were sampled on day 237 from sowing (1 June 2022). About 20 samples were taken arbitrarily from each onion variety grown in the Sde Yoav and Yiron fields to isolate possible FBR causal agents. These samples included healthy-looking onions (about one-third of the bulbs) and infected onions. Infected onion plants had typical FBR-characteristic symptoms of discoloration of the roots to brown, rotting in the basal plate (change in color to brown and the appearance of moist or dry necrotic areas), and separation of the scales (fleshy leaves). Some of the samples were infected by secondary parasites, such as maggots of the onion fly (Delia antiqua).
For the pathogens’ isolation, the onion’s basal plate was cut from each bulb, about 7 mm from the lower tip (all roots, if present, were removed). Each basal plate was divided into 3–4 pieces of tissue. All the pieces were disinfected with 3.5% commercial bleach (NaOCl) for about 30 s and then thoroughly washed for 1 min with tap water. Each tissue was placed in the center of a 90 mm Petri dish that contained potato dextrose agar (PDA). The PDA medium was made by dissolving 39 g of PDA powder in 1 L of double-distilled water (DDW).
The isolates (228 plates in total) were incubated in the dark at 28 ± 1 °C. After two days, the developed isolates were transferred to new PDA media. Transfer to new plates was performed for isolates with colony characteristics and spores corresponding to the Fusarium genus, and the process continued until pure colonies were obtained. The series of transformations for each colony to a new plate was performed each time by taking a colony agar disk from the young margins of the old colony and carefully tracking the colony morphology. Additionally, molecular verification using the ISSR and sequencing was performed to ensure that a single species was obtained.
The isolates were classified according to their colony characteristics. Of these, 31 were selected according to their morphological and spore characteristics resembling the Fusarium genus and subjected to molecular identification (PCR, gel electrophoresis, sequence determination, and homology search against the GenBank and the Fusarioid-ID databases).
2.3. DNA Extraction and Molecular Identification of Fusarium spp.
Molecular identification was conducted for each of the Fusarium species involved, according to the methods described previously [21]. DNA extraction was performed using the Master Pure™ Yeast DNA Purification Kit (Epicentre, Madison, WI, USA). The DNA concentration and purity test after extraction was carried out using a NanoDrop™ One spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) via the microvolume spectrophotometric UV–vis method. The test ensures a high DNA quality (median value of 1.8 or above of absorbance ratio at wavelength 260/280 nm) and at least 30 nanograms/µL DNA. The samples’ average DNA concentration was 83 nanograms/µL. Molecular identification (PCR followed by sequencing) was performed using the Fa/R8 or 7cF/11aR primers targeting RPB1 or RPB2—the RNA polymerase largest or second-largest subunit (Table 1). Additionally, we used the primers E1/E2 (specific to the genus Fusarium, targeting the Fusarium translation elongation factor-1 alpha gene, TEF1) (Table 1). DNA fragments were amplified by PCR and detected by gel electrophoresis.
The PCR was conducted using the T-100 Thermal Cycler (Bio-Rad, Hercules, CA, USA) with a total volume of 25 μL per reaction. Each of the reaction mixtures tested here included 1 μL of a primer mix (each primer in the solution was at a concentration of 10 μM), 12.5 μL of the commercial reaction mixture (PCRBIO Hot Start VeriFi™ or PCRBIO Taq Mix Red, PCR Biosystems, London, UK), 1 μL of template DNA, and 10.5 µL of UltraPure water (Bio-Lab, Jerusalem, Israel). For the TEF1 gene amplification, the PCR conditions consisted of an initial denaturation at 95 °C for 1 min, followed by 35 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 15 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min. For the RPB1 or RPB2 gene amplification, the PCR protocol was initial denaturation at 95 °C for 2 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 15 s, extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min.
Microsatellite-sequence-based primers were used for the ISSR-PCR molecular technique (Table 1). This approach generates multilocus markers for DNA fingerprinting [23]. PCR reactions for ISSR amplification were conducted with the following parameters: an initial denaturation step at 95 °C for 2 min followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 43 or 50 °C for 15 s, and extension at 72 °C for 30 s. The samples were then cooled to 12 °C for sample retrieval.

Table 1.
Primers used for the Fusarium spp. detection.
Table 1.
Primers used for the Fusarium spp. detection.
| Primer | Gene | Sequence a | Fragment Length | Reference |
|---|---|---|---|---|
| E1/E2 | TEF1—Fusarium-specific | F-ATGGGTAAGGAGGACAAG | 680 bp | [24] |
| R-GGAAGTACCAGTGATCAT | ||||
| 7cF/11aR | RPB2—RNA polymerase second-largest subunit | F-ATGGGYAARCAAGCYATGGG | ~970 bp | [25] |
| R-GCRTGGATCTTRTCRTCSACC | ||||
| Fa/R8 | RPB1—RNA polymerase largest subunit | F-CAYAARGARTCYATGATGGGWC | 1607 bp | [26] |
| R-CAATGAGACCTTCTCGACCAGC | ||||
| SIX3 F/R | Fusarium oxysporum f. sp. cepae secreted in xylem genes 3 | F-ATGCGTTTCCTTCTGCTTATC | 306 bp | [21] |
| R-AGGTGCGACATCAATGACAG | ||||
| ISSR1 | Inter simple sequence repeat | F + R-AGAGAGAGAGAGAGA | Multiple lengths | [23] |
a Y = C or T, R = A or G.
2.4. Identification of the Fusarium Species and Phylogenetic Relationships
A total of 31 isolates amplified with TEF1 or the RPB1/RPB2 genes were sent for sequencing (Macrogen Europe, Amsterdam, The Netherlands). Similarity percentages between sequences compared to other already recognized species were determined online using the identification database Fusarioid-ID (accessible at www.fusarium.org, accessed on 25 March 2024) and an NCBI GenBank BLASTN search (National Center for Biotechnology Information, Bethesda, MD, USA, at: http://www.ncbi.nlm.nih.gov, accessed on 25 March 2024). Sequence comparison of the TEF1 gene-conserved regions was conducted using the Clone Manager 11.0 program (Sci Ed Software, Durham, NC, USA). Sequences were aligned, and phylogenetic tree construction was performed using the SeaView version 5.0 software (http://doua.prabi.fr/software/seaview, accessed on 25 March 2024) [27]. The trees were constructed using a distance-based method with the default parameters (BioNJ (neighbor-joining algorithm), distance (maximum likelihood), Jukes–Cantor (J-C), ignore positions with gaps, and bootstrap based on 1000 tree replications). The phylogenetic trees that presented similarity percentages between sequences, were generated with the TEF1 Fusarium-specific sequences. The analysis also used the TEF1 gene from reference strains to assist with taxonomic assignment. These included previously identified Fusarium species in Israel’s Golan Heights (isolates B1, B5, B7, B8, B14, and B16 [21]) and four reference Neocosmospora (F. solani) SC species that were taken from the GenBank: isolate cc41W (HQ731052.1), strain hc0001 (KP143718.1), isolate DB-C2 (KY486693.1), and strain gss53 (MH341207.1). Also, an outgroup was set using the onion pathogen Rhizopus arrhizus (E12 strain, MK174988.1), isolated and identified as part of the current research.
2.5. Colony Morphology and Identification of the Fusarium Species
The Fusarium species involved in FBR were isolated from onion bulbs and identified using the microscopic characteristics of the spores and colony morphology, according to [21]. All isolates were grown on PDA plates for four days to allow hyphae and spore formation. Mycelial mats or conidia were carefully scraped off the plates, and a small amount was suspended in 10 µL of potato dextrose broth (PDB) or DDW. The suspensions were then placed on sterile glass slides for microscopic observations using a light microscope at a magnification of 250× without staining.
2.6. Germination Pathogenicity Assay
The pathogenicity test was conducted according to [21] with modifications and aimed at assessing the virulence level of the Neocosmospora SC isolates on onion seedlings. The experiment was performed with four replicates using Petri dishes, each containing ten onion seeds. The two onion cultivars selected for this seedling test (and the bulb assay described below) were the yellow onion cultivar Orlando and the red onion cultivar Noam (supplied by Hazera Seeds Ltd., Berurim, Israel). These varieties were selected because they are widely grown in Israel and are common in the markets. The yellow Orlando cv. is the same as the one sampled from the Kibbutz Yiron (Galilee, Hula Valley) field, and the red Noam cv. is very similar in its characteristics to the red cultivars tested in the Givat Yoav (Golan Heights) field. Seeds were washed to remove their commercial coating (Thiram, Captan, Carboxin, Metalaxyl-M, manufactured by Rogers/Syngenta Seeds, Boise, ID, USA, supplied by CTS, Tel Aviv, Israel). This procedure was carried out by dipping them in tap water for 15 min and then replacing the water ten times. Seeds were disinfected with 70% ethanol for 1 min, rinsed three times with DDW, soaked for 3 min in disinfection solution (2% NaOCl, 12 µL dish soap, 150 mL tap water), and washed vigorously six times with DDW.
For this seedling assay and the following bulb test, six of the Neocosmospora SC isolates were selected (Nos. E3, E7, E8, E9, E14, and E21). These isolates were tested against a non-infected control group and a positive control group, which was inoculated with F. acutatum (isolate B5 [21]). After drying the seeds on a sterile paper towel, each group of onion seeds was transferred onto a Petri dish with sterile Whatman paper soaked in sterile DDW. A 6 mm diameter disc was cut from a selected 5-day-old Fusarium sp. colony (grown previously on PDA in the dark at 28 ± 1 °C) and placed onto each onion seed group plate (in the middle). The control group was grown without inoculation. After nine days of incubation under gentle rotation in a rotary shaker (to assist hyphae and spores’ dispersion) in the dark at 28 ± 1 °C, the seeds were photographed and washed, and their germination percentages, biomasses, and the epicotyl emergence numbers were measured and compared to the mock uninfected control group. A germinating seed was defined as one in which the radicle had broken the seed coat.
2.7. Onion Bulb Pathogenicity Assay
An onion bulb pathogenicity assay was conducted on two cultivars, Orlando cv. and Noam cv. (yellow and red onions), as previously described [21] but with modifications. The experiment included five repetitions per isolate (40 bulbs per cultivar). The same six Neocosmospora SC isolates (Nos. E3, E7, E8, E9, E14, and E21) tested in the seed assay were evaluated here. As in the seedling assay, these isolates were tested against a non-infected control and a positive control group inoculated with F. acutatum (isolate B5) [21]. The experiment was performed in a sterile environment inside a biological hood. A stock of ca. 2 × 106 spores/mL (in sterile water) was prepared from five-day-old colonies, previously grown on PDA at 28 ± 1 °C in the dark.
The bulbs were sterilized in 70% ethanol for 1 min, and the outer scales were removed. A sterile pipette tip (10 mm in diameter) was used to stab the basal plate once, and 50 µL spores were pipetted into each puncture. For control bulbs, a similar volume of sterile water was injected. Every bulb was stored separately within a sealed sterilized plastic bag to maintain a moist environment and prevent contamination. The bulbs were incubated in a temperature-controlled incubator in the dark at 22 ± 1 °C for two weeks. The appearance of early decay symptoms two weeks post-infection on the bulbs’ basal plate exterior and interior tissue and mycelial growth emergence on the bulb surface were assessed. The necrotic lesion dimension was measured as the length from an onion’s lower (root) tip to the scales. The necrotic lesion severity was evaluated using categories where 5 indicates severe rotting and 1 is healthy tissue. Finally, the fungus from selected infected onions was re-isolated on PDA and identified to fulfill Koch’s postulates.
2.8. Statistical Analysis
The seedlings’ and bulbs’ pathogenicity assay data were analyzed using Microsoft Excel (Microsoft 365 MSO, version 2401 Build 16.0.17231.20290) and GraphPad Prism software, version 9.5.1.733 (GraphPad Software Inc., San Diego, CA, USA). The data were analyzed using a one-way analysis of variance (ANOVA), the Brown–Forsythe test, and a posterior Dunnett’s test (which is restricted to comparing the experimental groups against a single control group) at a significance level of p < 0.05.
3. Results
3.1. Evaluation of Disease Incidence
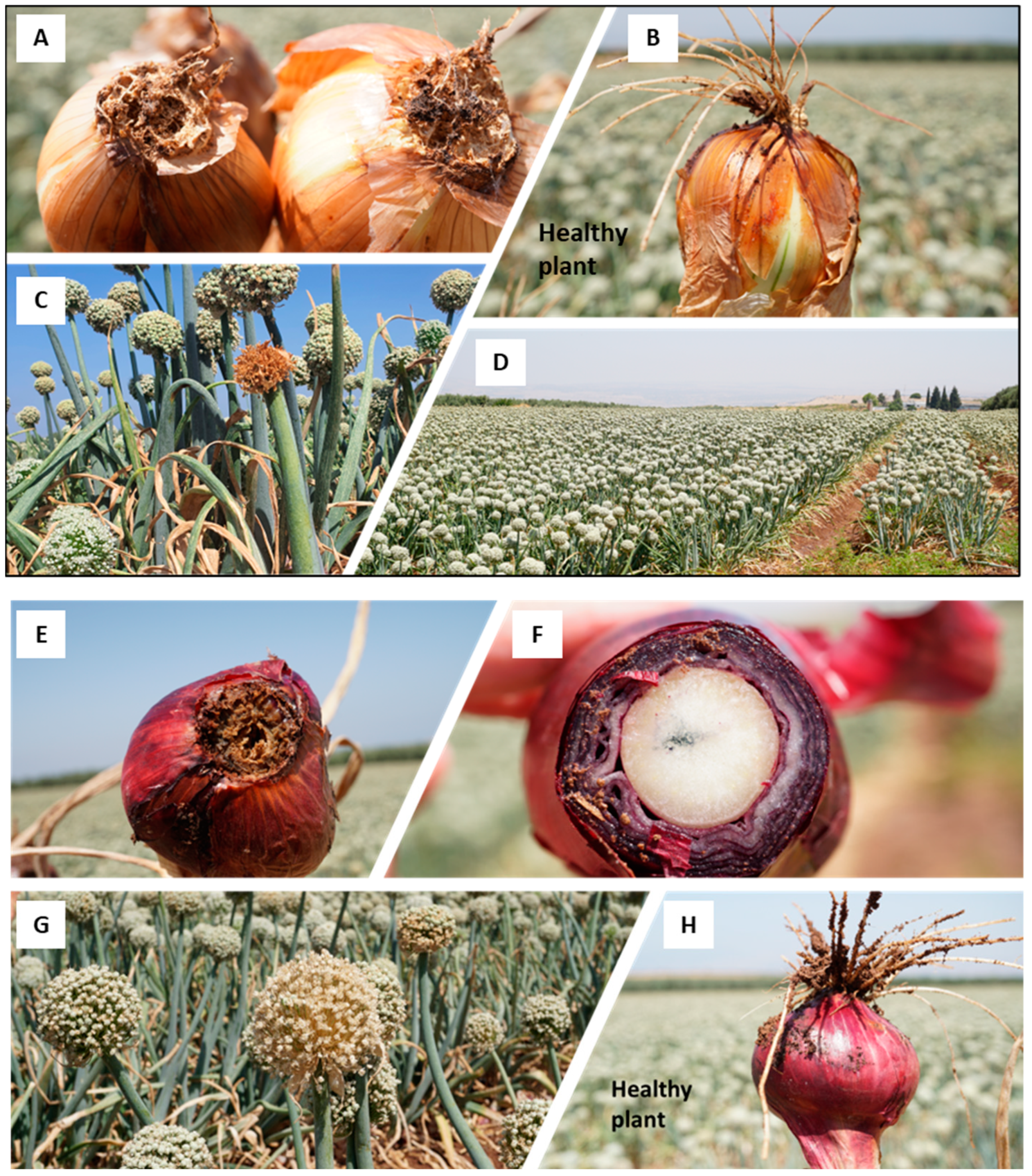
The Givat Yoav field (Golan Heights) was chosen for the FBR field’s crop loss estimation. Typical symptoms in red (variety Ha3) and yellow (variety Ha1) female plants were documented (Figure 2). The estimated incidence in the field is shown in Table 2. The disease prevalence in the red onion variety ranged from 2.4% in the male (Ha2 cv.) plants to 8% in the female (Ha3 cv.) plants. For comparison, in female plants of the yellow onion (variety Ha1), an infection level of 2.4% was determined. The red female (Ha3 cv.) plants had statistically significantly higher FBR incidence than the other two varieties (p < 0.05).

Figure 2.
The Allium cepa Fusarium basal rot (FBR) disease outcome in the yellow onion (Ha1 cv., (A–D)) and the red onion (Ha3 cv., (E–H)) in the Givat Yoav commercial field in northeastern Israel’s Golan Heights (location in Figure 1). (A,E,F)—bulb samples with typical FBR disease symptoms. (B,H)—a healthy-looking bulb sample. (C,G)—dehydrated yellowish-to-brown color inflorescences scattered in the field. (D)—an overview of the field.

Table 2.
Disease incidence in the Givat Yoav field, Golan Heights a.
3.2. Isolation and Identification of the Fusarium Species from the Collected Onions
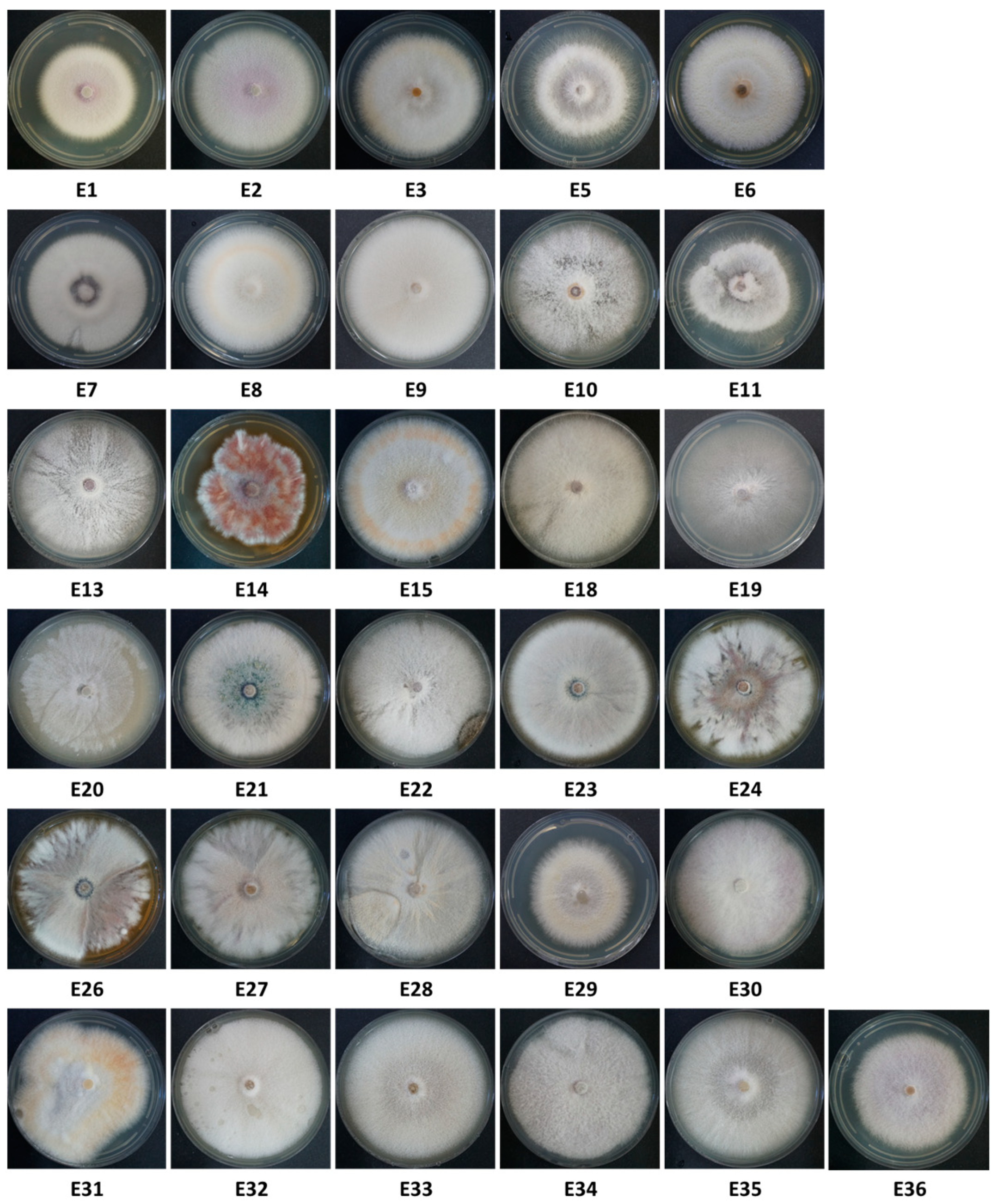
About 20 samples (onion bulbs) were taken from each of the onion varieties grown in the Givat Yoav and Kibbutz Yiron fields (Figure 1) to isolate possible disease agents. The onions were in different degrees of decay, and secondary parasites were seen in some of them, such as maggots of the onion fly. In addition, ten onions that looked healthy were sampled from the plants. A total of 67 onions were sampled. From the collected onions, 228 fungal isolates were isolated. Of these, 31 were selected according to their microscopic (spores characteristics) and colony morphological traits resembling the Fusarium genus (Figure 3).
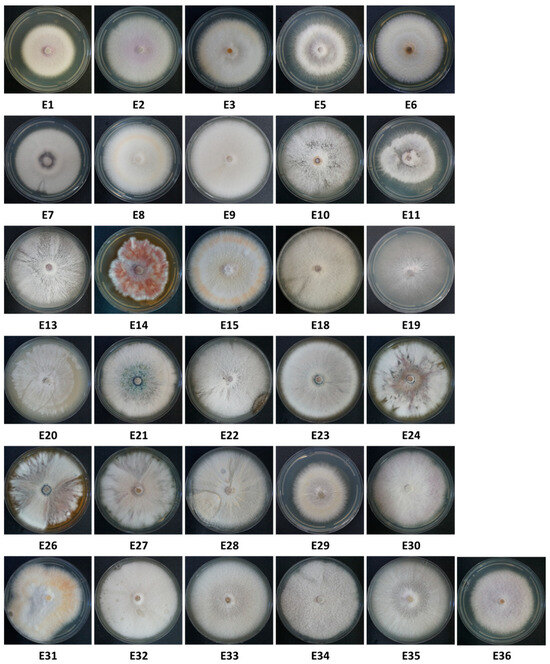
Figure 3.
Thirty-one Fusarium isolate colonies from onions collected from the Givat Yoav (Golan Heights, E1–E11) and Kibbutz Yiron (Galilee, Hula Valley, E13–E36) fields (see Figure 1). Isolates were selected from 228 according to colony and spore characteristics. Some isolate numbers are missing because they were identified as different species than Fusarium. That is why 31 isolates are presented, while some have a higher number (E32–E36). Colonies were grown in the dark for six days on a rich solid medium (PDA) at 28 ± 1 °C.
The sequenced isolates were compared to the Fusarioid-ID (Table 3) and GenBank (NCBI, nucleotide blast BLASTN, Supplementary file—Table S1) databases. The Fusarium Pairwise ID alignment search resulted in a high similarity (>99% in most cases) of the new sequences to Fusarium species in this GenBank. Likewise, BLASTN identification results in all analyzed species showed a significant similarity (ranging from 99.10% to 99.98%, except for E3, which reached 91.02%) to the previously reported Fusarium spp. sequences in the GenBank database. Interestingly, the new analysis using the Fusarioid-ID database and the phylogenetic analysis described below confirmed that isolate B16, previously thought to be F. anthophilium [21], is actually F. acutatum. The identity of all other isolates of that previous work was confirmed. The B1 isolate (F. proliferatum) is named by the synonym F. annulatum.

Table 3.
FUSARIOID-ID database identification of the Fusarium isolates from this study a.
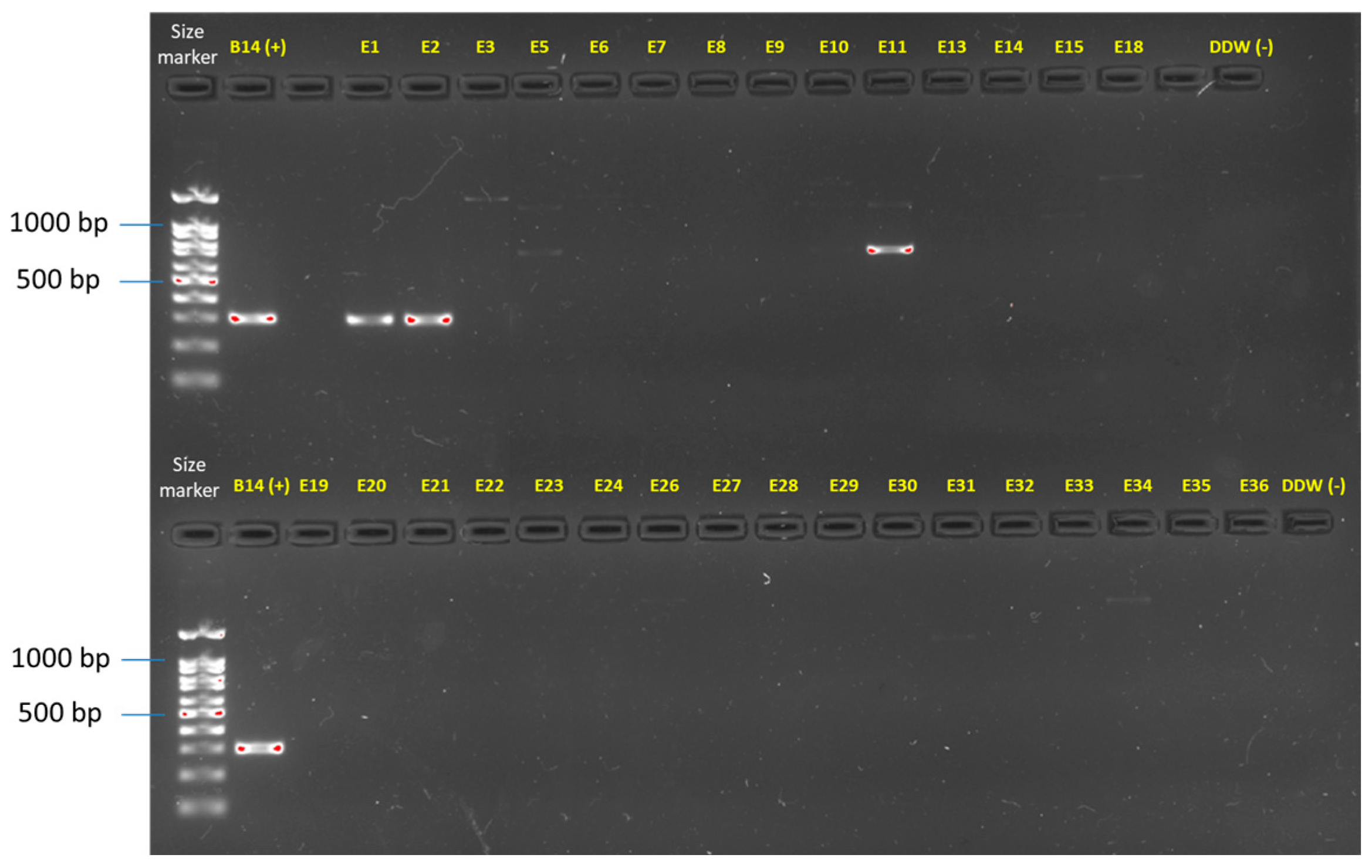
Final verification of the Fusarium species identity was performed by amplifying the F. oxysporum f. sp. cepae secreted in xylem genes 3 (Table 1, Figure 4). Identification of the Fusarium isolates was further reinforced by assessing the similarities between the isolates using the inter simple sequence repeat (ISSR)-PCR molecular technique. The DNA fingerprinting analysis revealed that isolates of the same species exhibited highly similar DNA profiles (Figure 5). Also, some species were subdivided into subspecies families according to shared homology. The species found included F. oxysporum f. sp. cepae and F. acutatum, which were already found in a previous study [21], but also a new species complex that has to date not been identified—Neocosmospora (previously the F. solani). The last is dominated mainly by N. falciformis. Intriguingly, as detailed below, this species complex was much more prevalent in the isolates’ samples than the other two species.
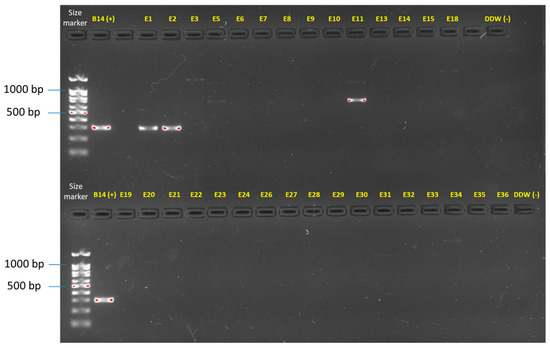
Figure 4.
Molecular identification of the F. oxysporum f. sp. cepae secreted in xylem genes 3 (SIX3, primer sets and references in Table 1). The gel presented is uncropped. This analysis was performed on all the Fusarium isolates (detailed in Table 3). Only two isolates (E1 and E2) were identified using this specific approach as F. oxysporum f. sp. cepae. Isolate E11 and some other isolates present an unspecific band product. The F. oxysporum f. sp. cepae (isolate B14, [21]) was used as a positive control. Sterile double-distilled water (DDW) was used as a template in the PCR reaction to provide a negative control.
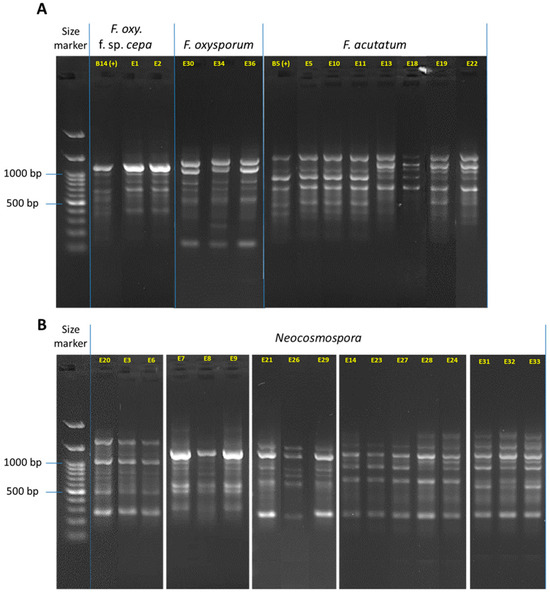
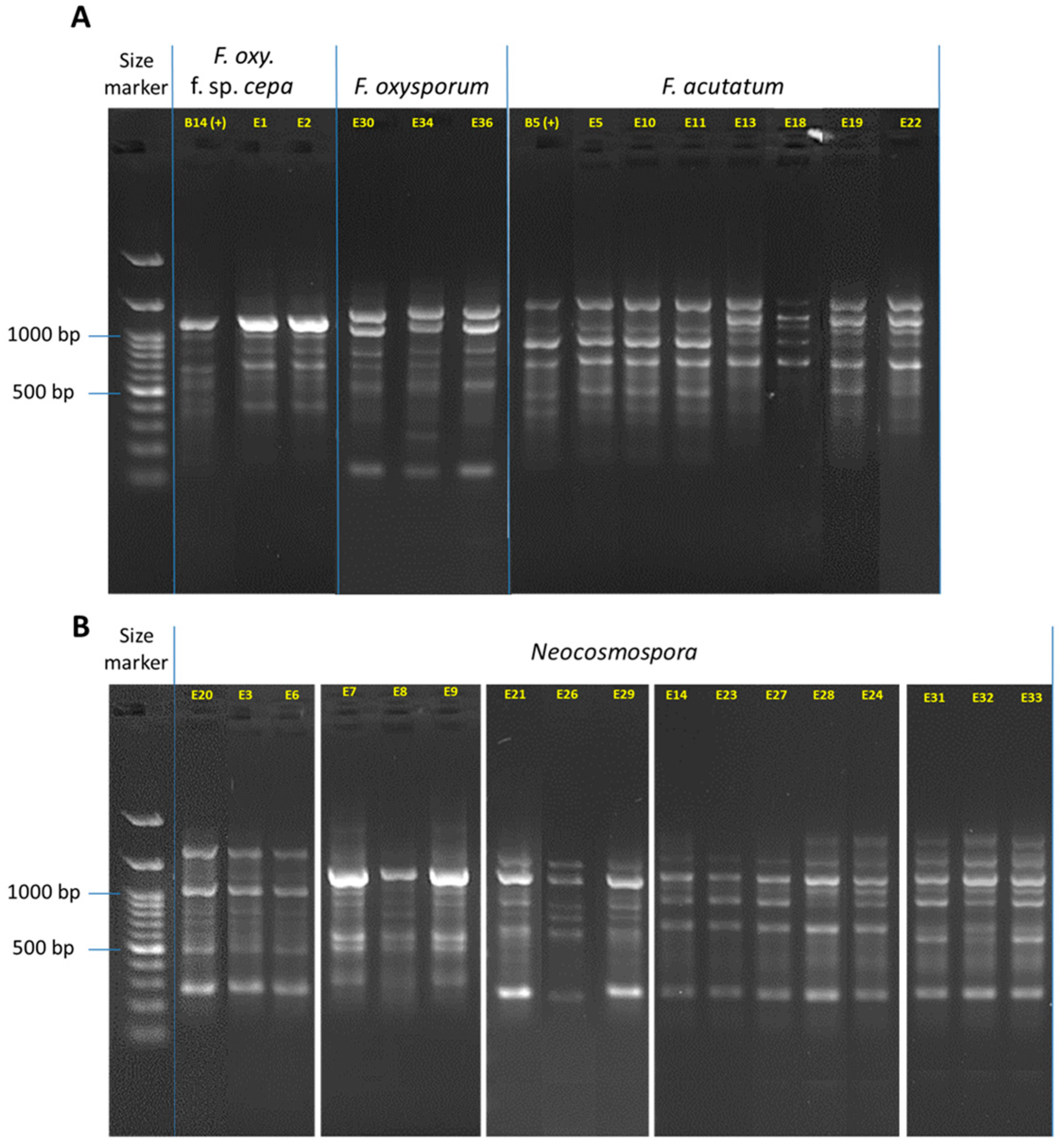
Figure 5.
DNA fingerprinting for the Fusarium isolates (detailed in Table 3). The complex band profile was generated using the inter simple sequence repeat (ISSR)-PCR molecular method. The ISSR primers are listed in Table 1. The gels were cropped and rearranged to improve the presentation’s clarity and conciseness. Full-length gels are presented in Supplementary Figure S2. (A) F. oxysporum f. sp. cepae, F. oxysporum species complex (SC, F. inflexum), and F. acutatum (F. fujikuroi SC). (B) Neocosmospora (F. solani) SC. The F. oxysporum f. sp. cepae (isolate B14, [21]) was used as a positive control.
3.3. Phylogenetic Relationships between the Fusarium Species
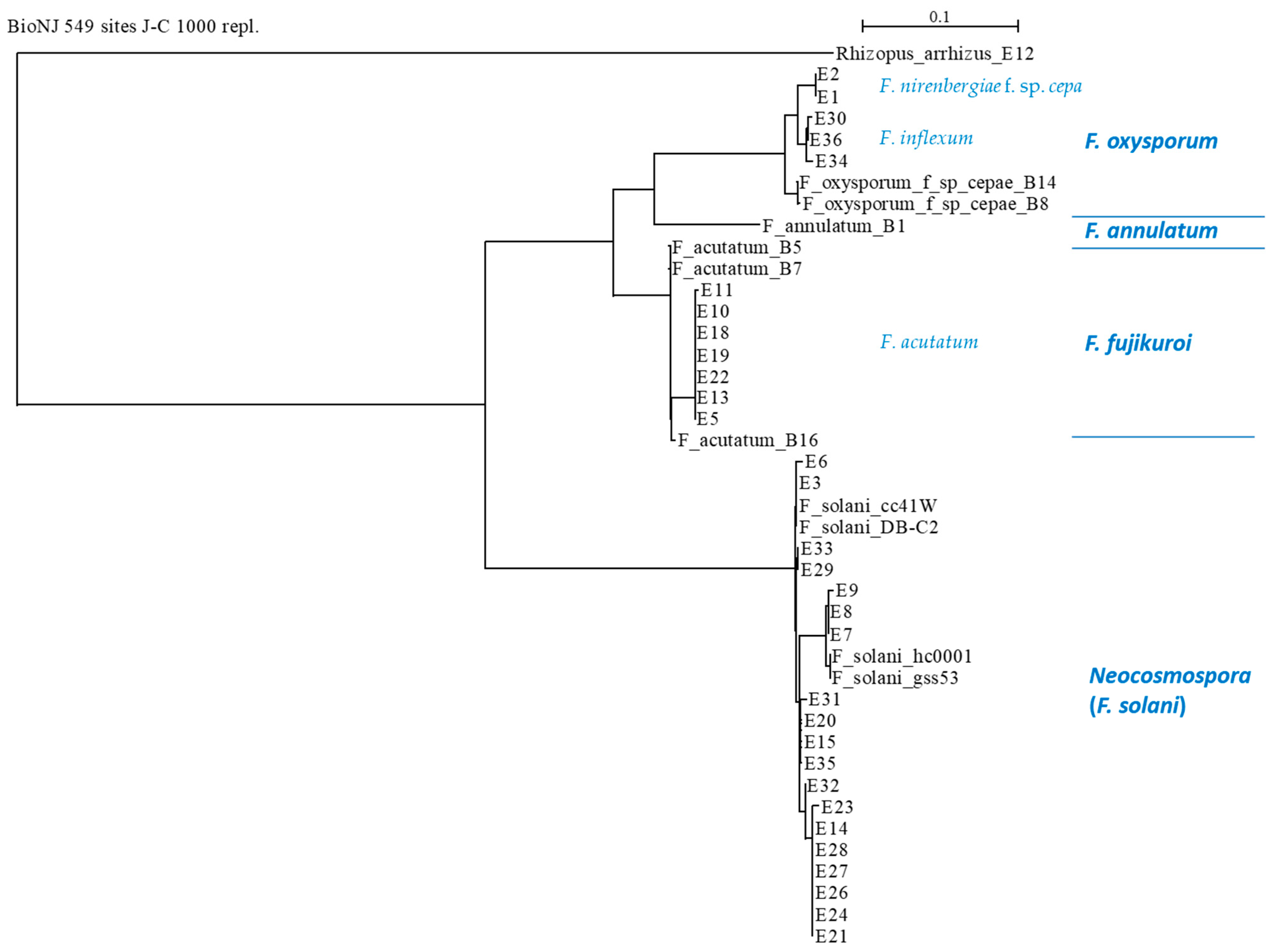
The similarity between the sequences (Supplementary file—Figure S1) showed the conserved and distinct DNA regions for all species. Phylogenetic analysis for the identified species (Figure 6 and Figure 7) demonstrated the relatively close relationships between F. oxysporum f. sp. cepae, F. acutatum species, and the separate branch of Neocosmospora isolates. Interestingly, F. oxysporum f. sp. cepae was found only in the Givat Yoav (Golan Heights) field. At the same time, F. inflexum (F. oxysporum SC) was isolated only from the Kibbutz Yiron (Galilee, Hula Valley) area. In contrast, F. acutatum species were dispersed in both fields. Furthermore, as in the ISSR DNA fingerprinting, it could be seen that all species included sub-branches (clades), which indicates that they undergo further division into subspecies.
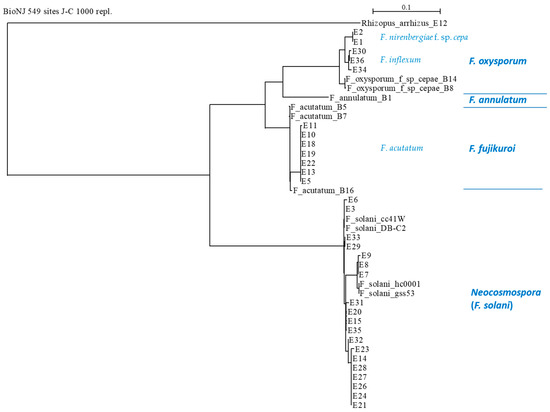
Figure 6.
Phylogenetic analysis of the TEF1 gene of the Fusarium isolates (presented in Table 3). The upper part’s scale describes the genetic resemblance of the isolates. The SeaView version 5.0 program (http://doua.prabi.fr/software/seaview, accessed on 1 April 2024) generated the phylogenetic tree. The alignment was performed using the distance-based method with the default parameters (BioNJ (neighbor-joining algorithm), distance (maximum likelihood), Jukes–Cantor (J-C), bootstrap with 1000 replicates, and excluded positions with gaps). The phylogenetic tree was charted with the forward TEF1-Fusarium-specific (E1/E2) primers. The analysis contained the TEF1 gene from the reference strains to assist with the taxonomic assignment. These included previously identified Fusarium species in the Golan Heights, Israel (isolates B1, B5, B7, B8, B14, and B16 [21]) and four reference Neocosmospora (F. solani) species that were taken from the GenBank: isolate cc41W (HQ731052.1), strain hc0001 (KP143718.1), isolate DB-C2 (KY486693.1), and strain gss53 (MH341207.1). Also, an outgroup was set using the onion pathogen Rhizopus arrhizus (E12 strain, MK174988.1), isolated and identified as part of the current research.
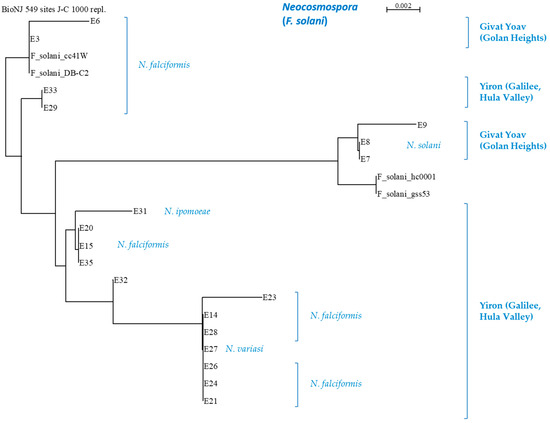
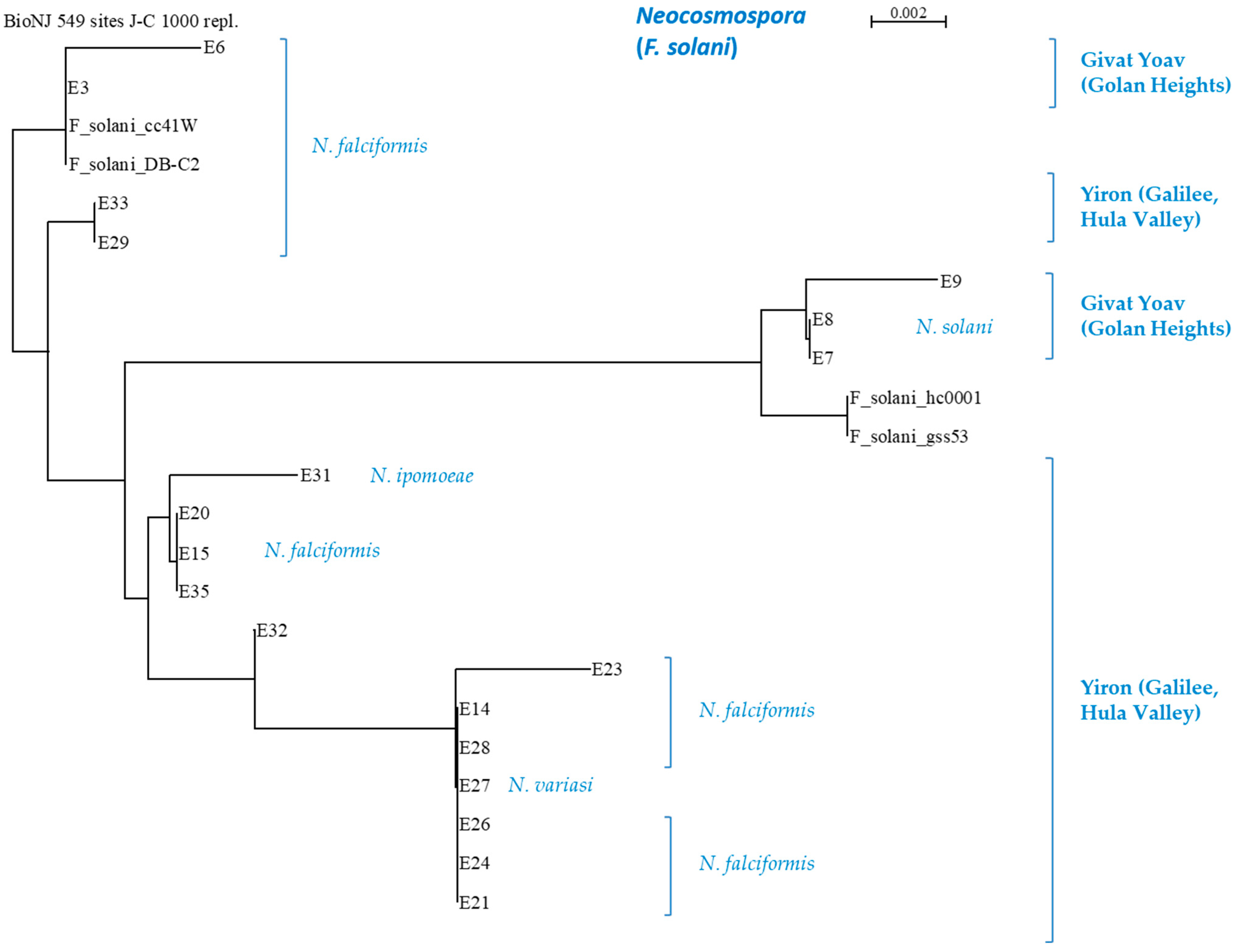
Such subspecies families can be easily noted in the Neocosmospora branch (Figure 7), where the subdivision is related to the collection site and/or the onion cultivar source. An agreement exists between the ISSR and the phylogenetic analysis regarding some of the sub-branches. For example, the E7, E8, and E9 isolates (N. falciformis) are differentiated as separate branches of Neocosmospora SC in the phylogenetic tree and share a unique DNA profiling in the ISSR results. This compatibility between the two methods also exists with respect to the E14, E21, E23, E24, E26, E27, and E28 isolate groups. Yet, some isolates are associated differently (i.e., belong to different subspecies families) by the two methods (for example, E21, E24, and E26).
3.4. Geographic Distribution, Composition, and Incidence of the Fusarium Species Involved in Onion Basal Rot Disease in Northeastern Israel
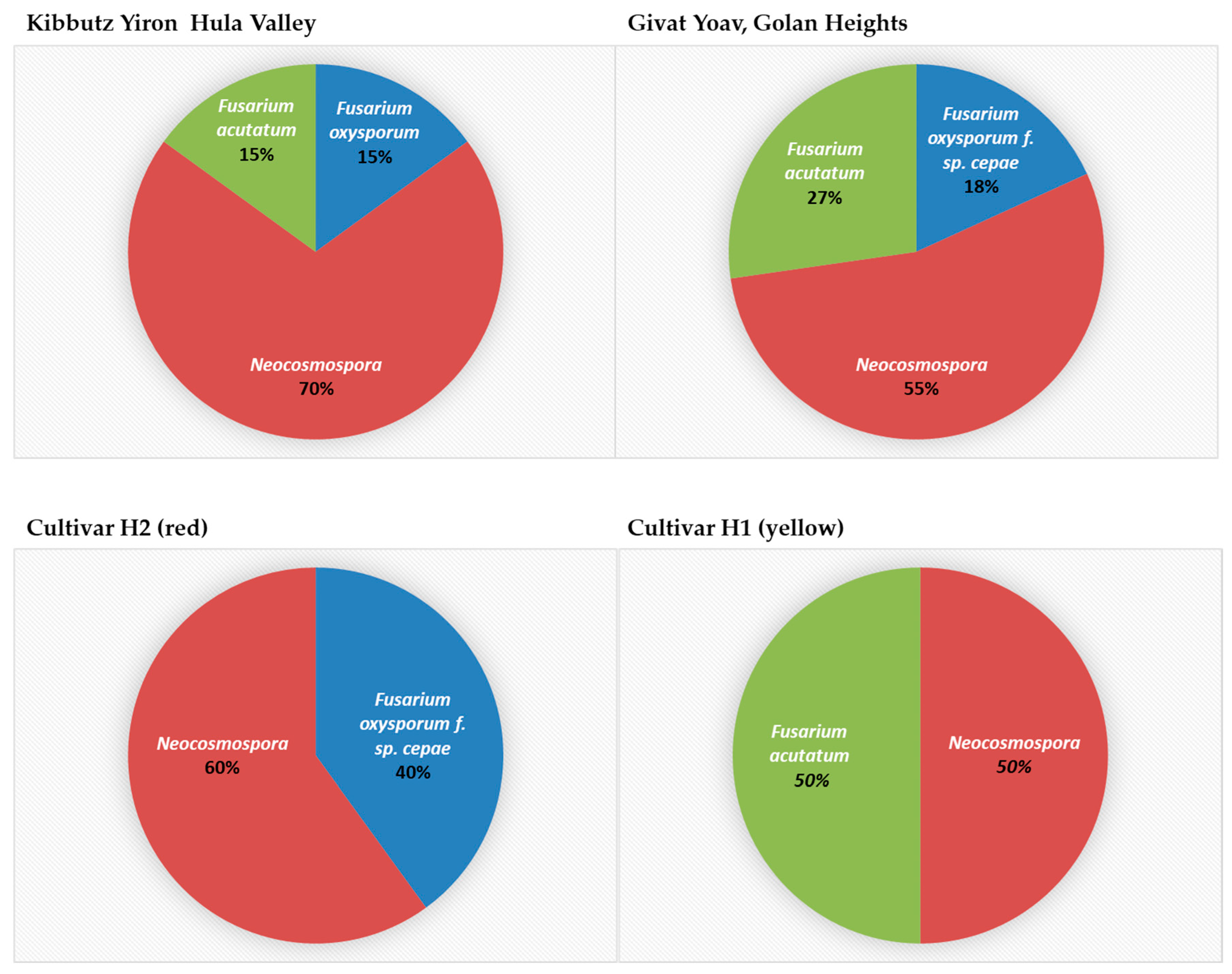
The segmentation of isolates according to their place of origin and the host species revealed an interesting pattern (Figure 8). In yellow onions of the Orlando cv. grown in Galilee (Hula Valley), Neocosmospora SC was found with the other two species, F. inflexum (F. oxysporum SC) and F. acutatum. In contrast, the Golan Heights bulb sample Fusarium spp.’s colonization was divided between species populating red or yellow onion cultivars. The Golan Heights results supported the findings of the work by Kalman et al. [21]. According to the former and current reports, F. oxysporum f. sp. cepae is abundant in red onions (Ha4/Ha2 cv.), while F. acutatum dominates yellow onions (Ha1 cv.). It was curious to discover that Neocosmospora SC inhabited both onion cultivars in the Golan Heights.
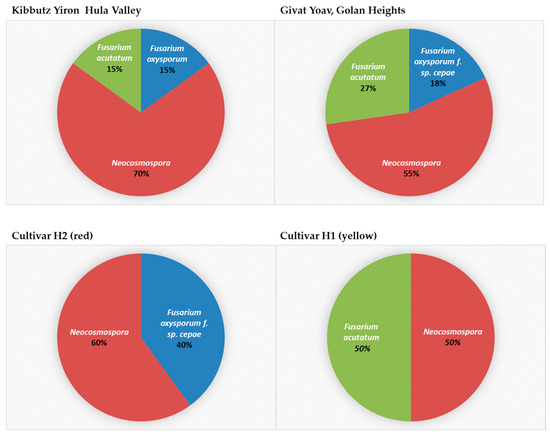
Figure 8.
The composition and incidence of Fusarium species involved in onion basal rot disease in northeastern Israel. Thirty-one Fusarium isolate colonies were collected from onions originating from the Givat Yoav (Golan Heights) and Kibbutz Yiron (Galilee, Hula Valley) fields (see Figure 1). These isolates were grouped according to their collection area (upper panel) or onion cultivar source (lower panel). Their prevalence (in percentages) is presented.
3.5. Pathogenicity Tests
Fusarium spp. can infect onion plants in various ways, leading to observable symptoms in small seedlings and different plant organs of mature plants. These include the roots, basal stem plate, leaves, and bulb scales [28,29]. Onion seedling and bulb inoculation assays were used to determine the virulence of selected Neocosmospora (F. solani) isolates. These pathogenicity trials were also aimed at completing Koch’s postulates. The onion seedling assay was performed as previously described [21] (Figure 9, Figure 10 and Figure 11, Supplementary file—Table S2). After nine days of incubation, white mycelia grew on or near the onion seeds as a food source while reducing their developmental rate (Figure 9, see, for example, isolates E3 and E21). The germination percentages were similar (p = 0.17–0.56) in all treatments, ranging between 90 and 100% (Supplementary file—Table S2).
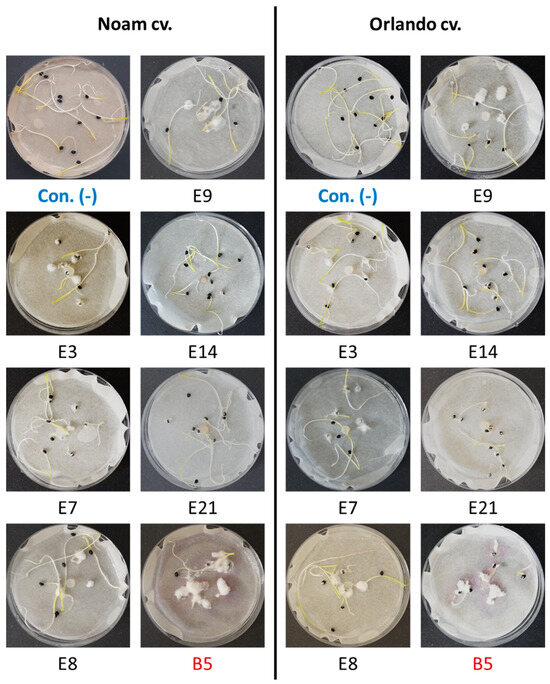
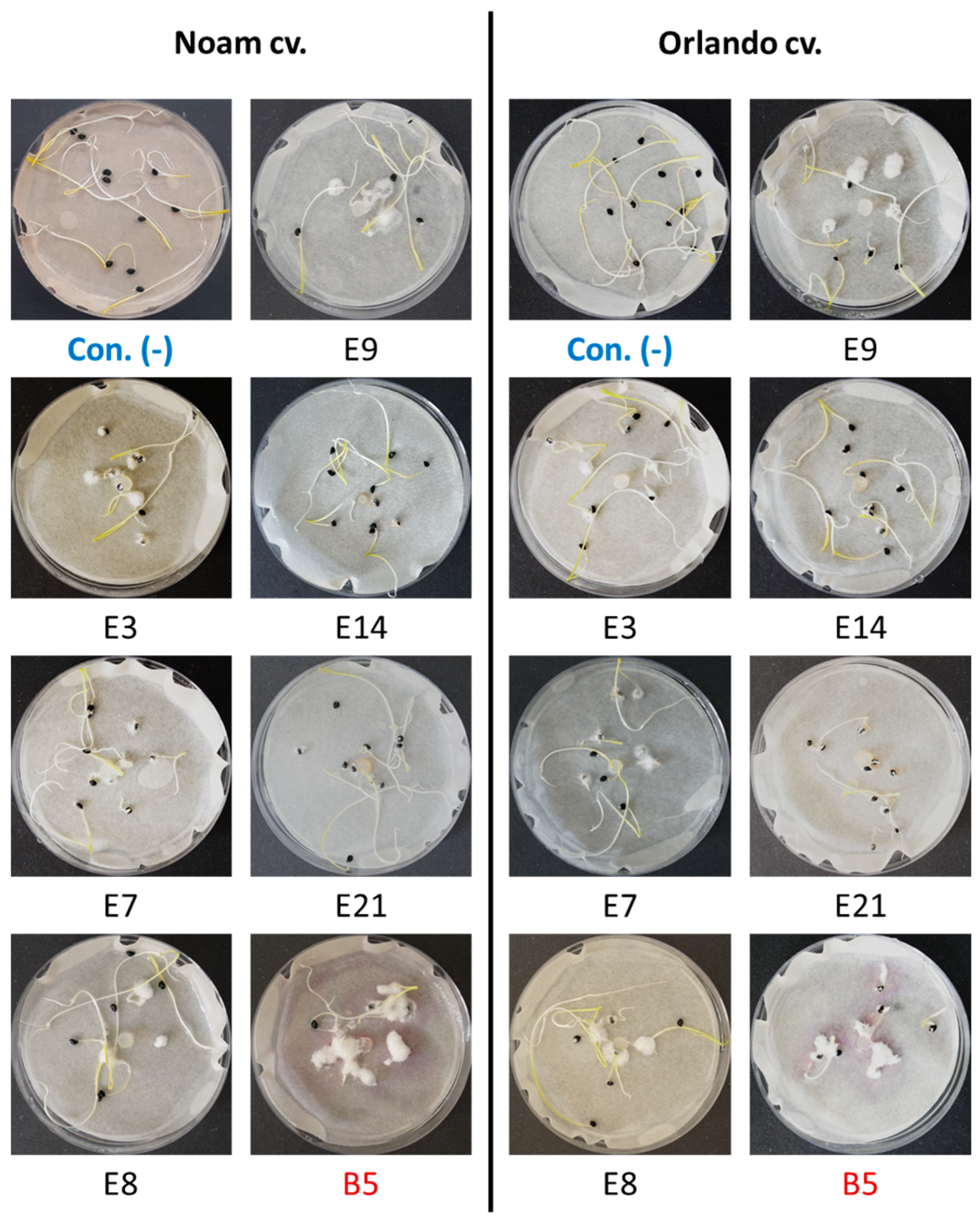
Figure 9.
The onion germination pathogenicity assay. Onion seeds from the Orlando (Riverside) and Noam cultivars were used for the pathogenicity test of selected Neocosmospora (F. solani) isolates (see Table 3). Each group of 10 germinating sprouts was inoculated with a 6 mm diameter disc from a 5-day Fusarium sp. colony in the center of a Petri dish. The F. acutatum (isolate B5 [21], highlighted in red) was used as a positive control. The control group (Con. [-], highlighted in blue) was grown without inoculation. Photos of representative seeds’ plates, taken nine days post incubation at 28 ± 1 °C in the dark, show minor or massive Fusarium white mycelial growth on or near the onion seeds, attributed to their growth suppression.
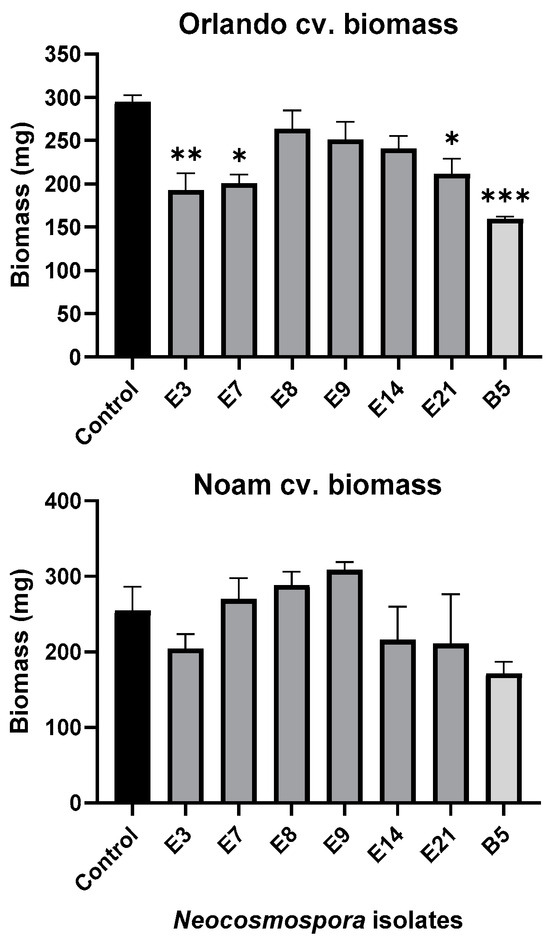
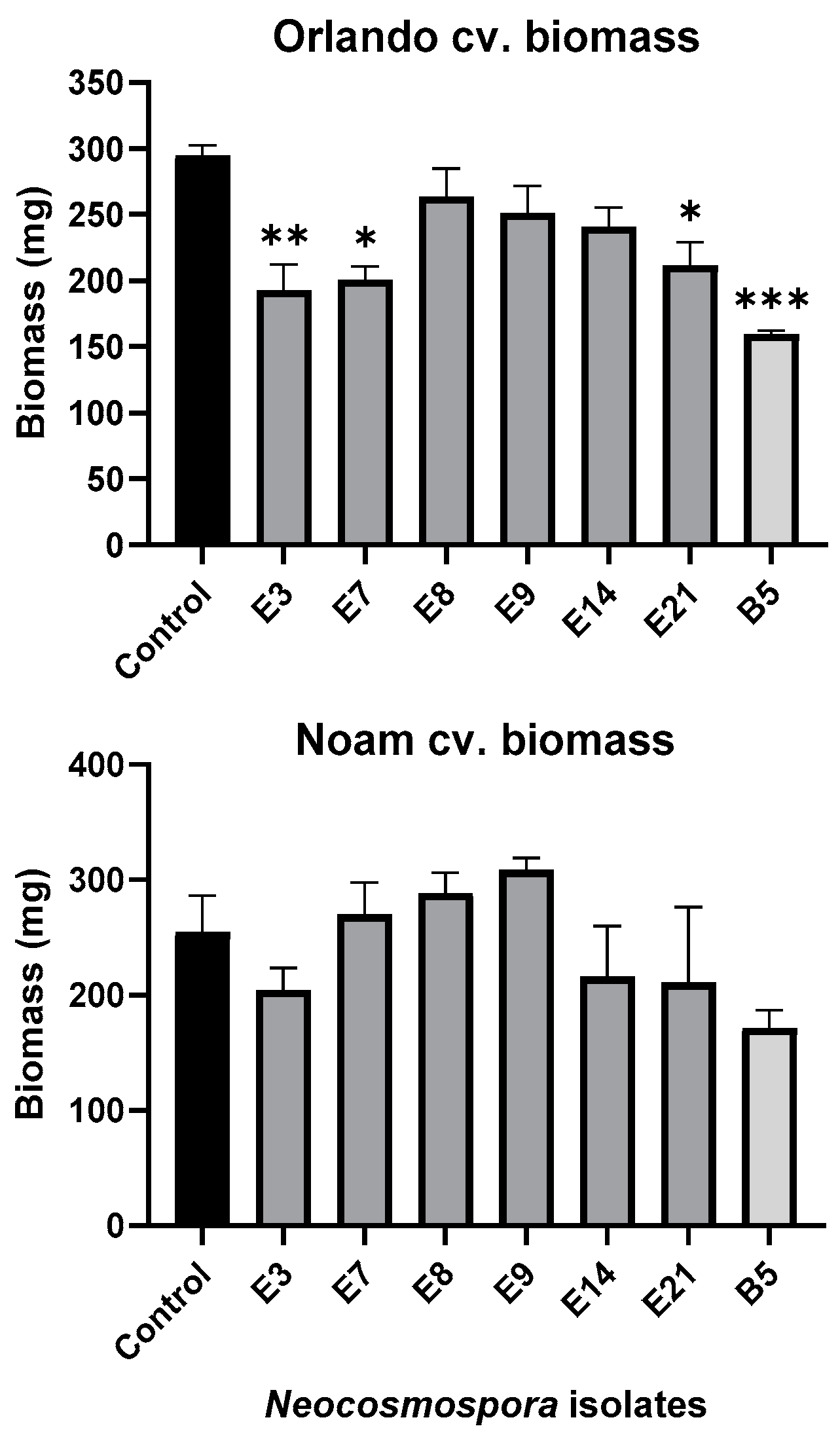
Figure 10.
Seedlings’ fresh biomass in the pathogenicity experiment (presented in Figure 9). The wet biomass of the resulting seedlings was measured after a nine-day incubation period. The standard error of the mean of four replicates is shown by the vertical upper bars, with asterisks above the error bars indicating a significant difference (* p < 0.05, ** p < 0.005, *** p < 0.0005) between each group and the control in the one-way analysis of variance (ANOVA).
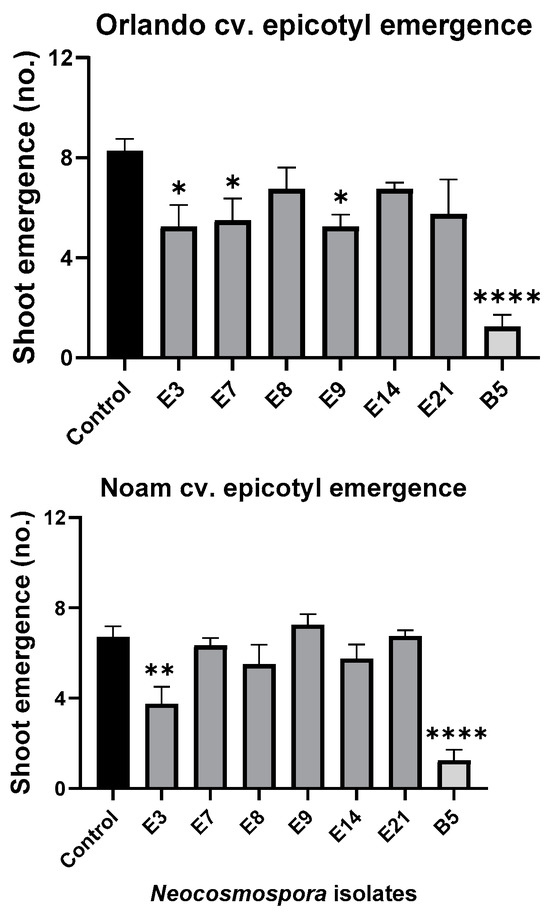
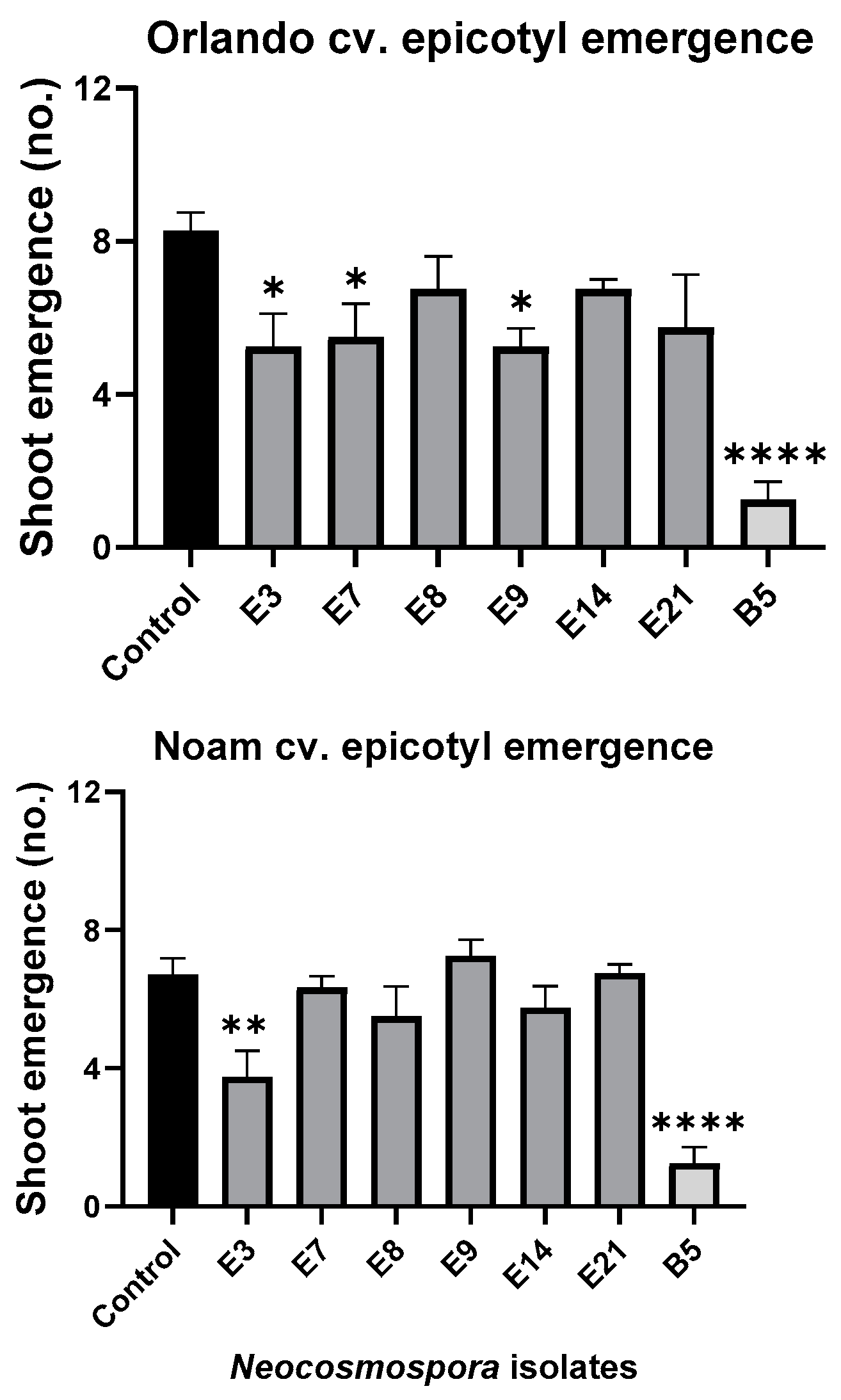
Figure 11.
Seedlings’ epicotyl emergence number (out of ten per plate) in the pathogenicity experiment (presented in Figure 9). The standard error of the mean of four replicates is shown by the vertical upper bars, with asterisks above the error bars indicating a significant difference (* p < 0.05, ** p < 0.005, **** p < 0.00005) between each group and the control in ANOVA test.
Assessing the sprouts’ fresh biomass (Figure 10) and epicotyl emergence number (Figure 11) in each assay plate allows for a more accurate evaluation of the isolates virulence. The Neocosmospora isolates E3, E7, and E21 (and, to a lesser extent, E14) caused significant (28–35% reduced biomass, p < 0.05) sprouting development repression in the yellow Orlando cv. This growth suppression is expressed in significantly lower shoot emergence percentages in isolates E3, E7, and E9. Isolates E3, E14, and E21 were also the most aggressive toward the red Noam cv. seedlings, though with statistical significance (p < 0.05), reached only in the E3 epicotyl emergence evaluation (biomass reduction was 17–21%, p = 0.62–0.90). Compared to the positive control species F. acutatum (B5 isolate), the Neocosmospora SC isolates were (in most cases) less aggressive in these tests (11–54% higher sprout biomass and improved epicotyl emergence, p < 0.05).
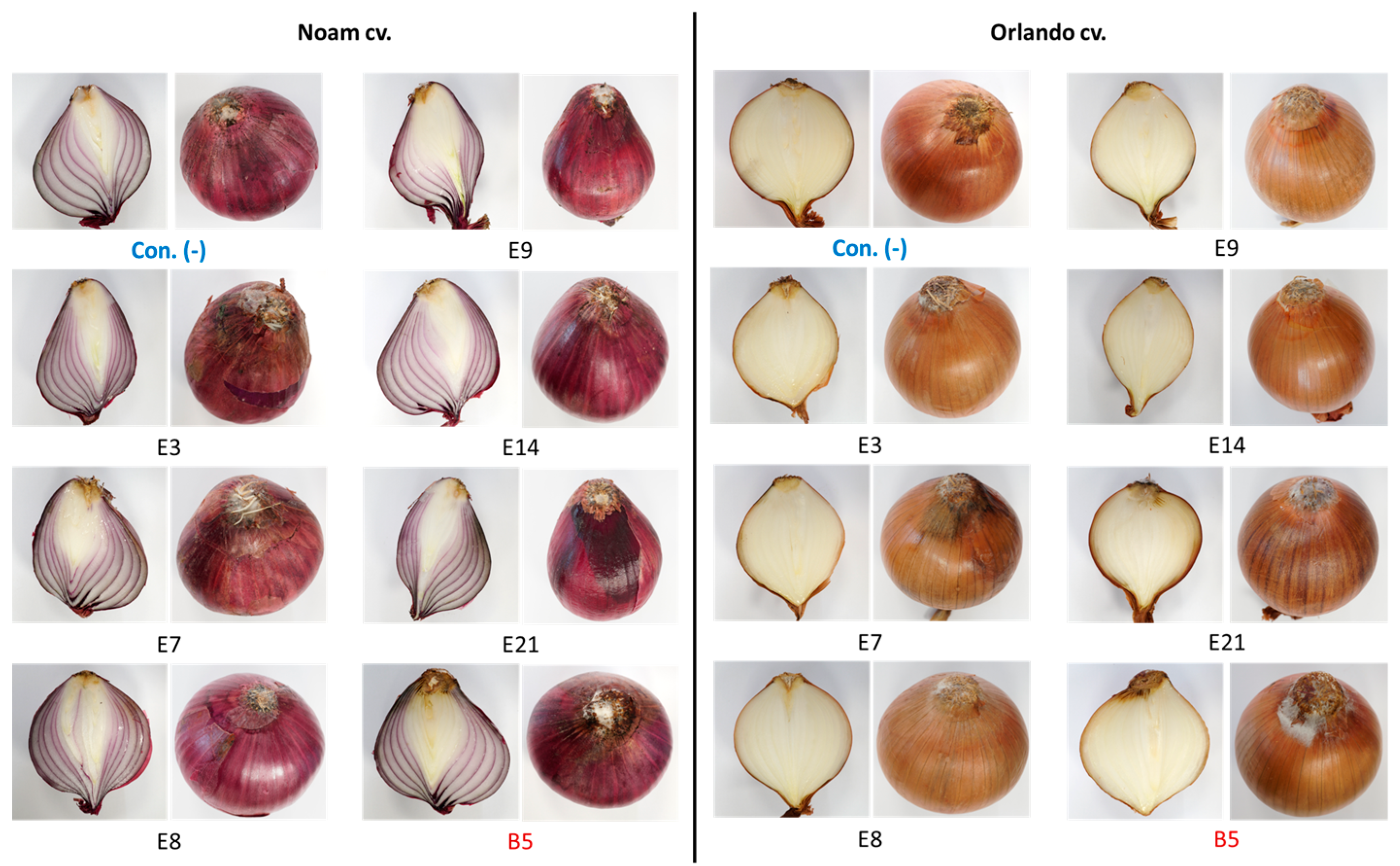
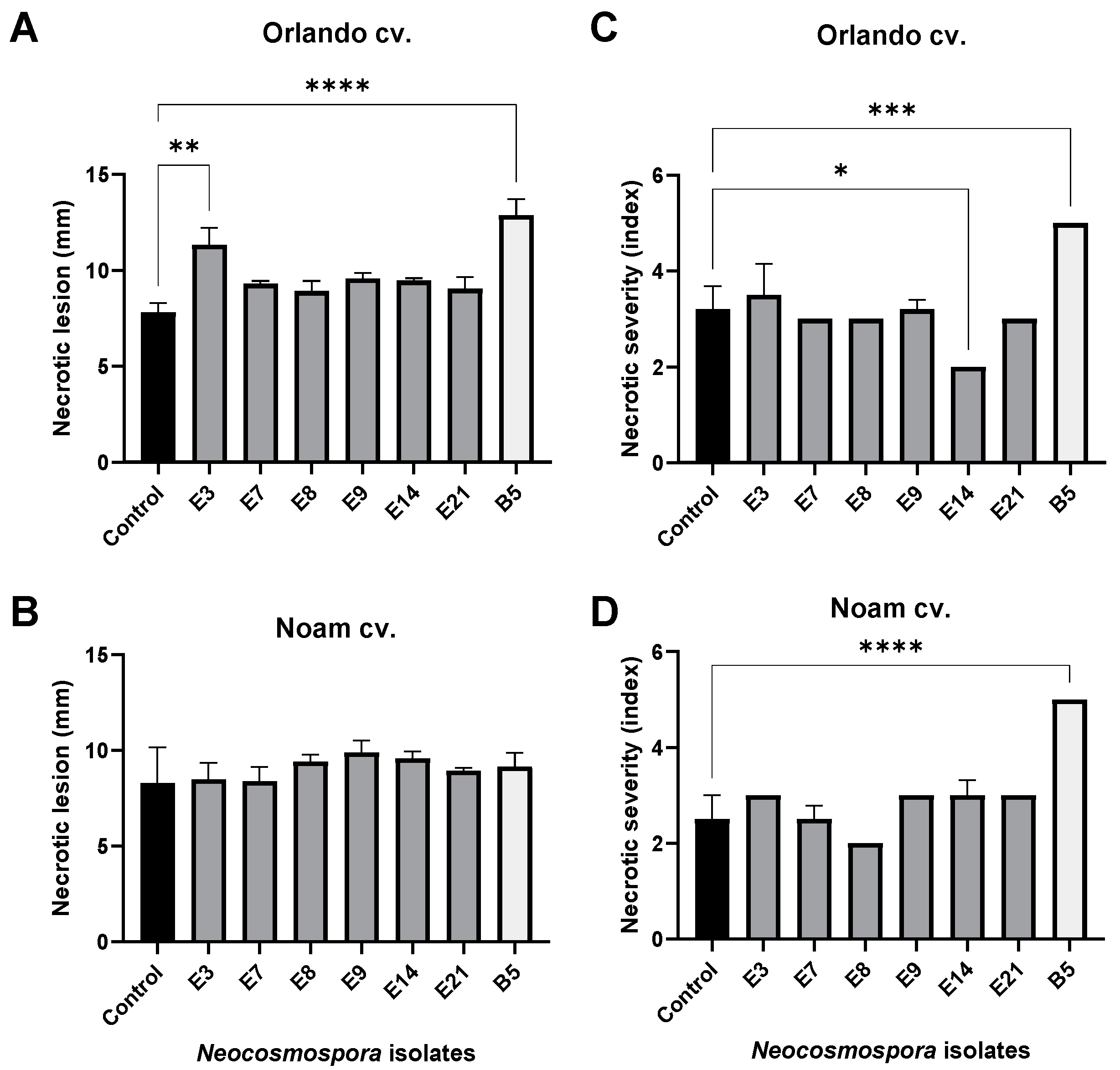
The bulb infection assay results (Figure 12 and Figure 13) were similar to the seedlings’ assay outcome. The intact onion bulb virulence assay rapidly assesses the Fusarium isolates’ ability to invade and thrive in host tissues [21]. In this assay, onion bulbs of the Orlando and Noam varieties were inoculated with six selected Neocosmospora isolates, resulting in the appearance of early symptoms two weeks post-infection. The bulbs’ basal plate exterior and interior tissue decay (up to 45% and 20% necrotic lesion dimensions and severity) and mycelial growth emergence on the bulb surface all indicated infection. The symptoms observed in this assay closely resembled those surveyed in onion fields naturally infected with the pathogen (Figure 2). Here, also (as in the seedling assay), the Neocosmospora isolates were less aggressive (41–45% less necrotic) than F. acutatum (B5 isolate). Also, the Orlando cv. was more susceptible to FBR than the Noam cv. Among the Neocosmospora SC isolates tested, E3 (N. falciformis isolate) was the most virulent strain in the bulbs’ pathogenicity assay, with a significantly larger necrotic lesion than the control in the Orlando cv. At the end of the experiment, Neocosmospora species were re-isolated from the infected bulbs and identified to satisfy Koch’s postulates.
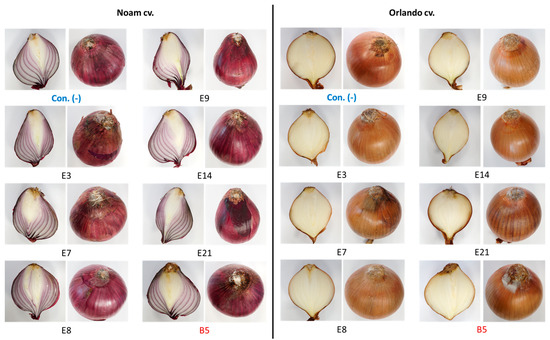
Figure 12.
Onion bulb inoculation assay. The assay evaluated the pathogenicity of selected Neocosmospora isolates (listed in Table 3). The procedure involved injecting a conidial suspension into the basal plate of the Orlando and Noam cultivar bulbs and then incubating them in moisture bags for two weeks at 28 ± 1 °C in the dark. The F. acutatum (isolate B5 [21], highlighted in red) was used as a positive control. The control group (Con. [-], highlighted in blue) was injected with DDW instead of fungal mycelia and spores. The external symptoms on the onion’s basal plate and a cross-section of the bulbs were examined, which revealed the development of white hyphae on the outer surface, accompanied by onion tissue decay.
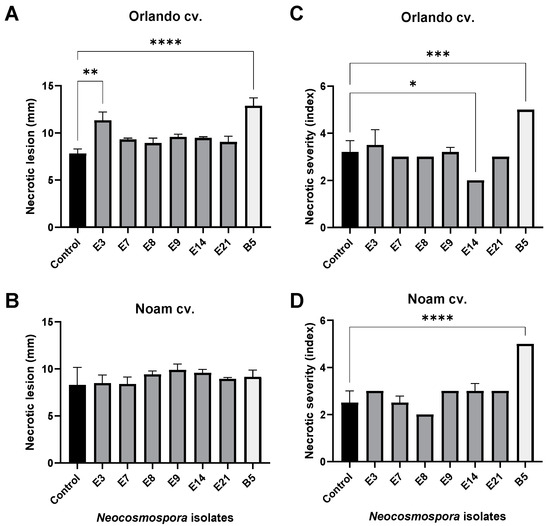
Figure 13.
Quantitative assessment of the disease symptoms in the onion bulb inoculation assay (described in Figure 12). Each onion bulb’s necrotic lesion dimensions (A,B) and severity (C,D) were evaluated. The necrotic lesion dimensions were measured as the length from an onion’s lower (root) tip to the scales (fleshy leaves). The necrotic lesion severity was assessed using five categories, of which 5 indicated severe rotting and 1 indicated healthy tissue. The standard error of the mean of 4–5 replications is shown by the vertical upper bars, with asterisks above the error bars indicating a significant difference (* p < 0.05, ** p <0.005, *** p < 0.0005, **** p < 0.00005) between the groups in ANOVA test.
4. Discussion
Fusarium basal rot (FBR) places a significant limitation on Allium production worldwide [2]. Damage by the disease can be observed throughout the entire crop cycle. Here, we report a new species complex of the Fusarium genus, Neocosmospora (previously F. solani), which also contributes to the FBR disease epidemic in onions along with F. oxysporum SC and F. acutatum in the northeastern region of Israel.
This species was already registered as one of the species involved in Allium FBR worldwide [2]. Such reports on onion plants include those from Serbia, Sri Lanka, Iran, and Vietnam [3,4,5,6]. In addition to uncovering Neocosmospora SC as an FBR causal agent in northeastern Israel, the composition and prevalence of Fusarium species were analyzed in two commercial fields and three onion cultivars. This study revealed an interesting pattern. Neocosmospora SC appeared to be a generalist pathogen group with a weak host specialization and lesser virulence capability than the other more aggressive specialist Fusarium species (adapted to a narrow host range). Those included F. oxysporum f. sp. cepae, dominating red onions (Ha4/Ha2 cv.), and F. acutatum, abundant in yellow onions (Ha1 cv.) [21]. This Fusarium species composition was characteristic of the Golan Heights field sampled. At the same time, all three Fusarium species were found together in the Galilee (Hula Valley) field (planted with a different yellow onion cultivar), about 40 km to the north. Thus, it seems a high host specialization degree is characteristic of some Fusarium species, as reported previously [21]. Still, a different pathobiome pattern (i.e., Fusarium species composition and incidence) exists depending on the host plant cultivar or location, as demonstrated here.
A previous study in one of the regions studied in this work (the Golan Heights) with the same onion cultivars and the same soil [9] revealed that F. oxysporum f. sp. cepae exhibited greater aggression towards the red onion Noam cv. At the same time, F. acutatum was more virulent towards the yellow Orlando cv. This observation could be attributed to the origin of these pathogens, as F. oxysporum f. sp. cepae was isolated from the red onion variety (Ha4/Ha2 cv.). In contrast, F. acutatum originated from the yellow Orlando cv. ([21] and the current work). In addition, co-inoculation of both pathogens resulted in severe disease in the red Noam cv., similar to the F. oxysporum f. sp. cepae single infection, but with reduced disease symptoms in the yellow Orlando cv. [9]. This result suggests that antagonistic interactions among certain onion genotypes may exist within the Fusarium population.
How does the presence of Neocosmospora species affect the onion basal rot disease outbreak? Does it comprise harmful endophyte species or high-virulence disease agents? Does this species complex balance or restrain the more aggressive Fusarium species? These are excellent questions to follow up on. The different combinations of Fusarium species in diverse onion species result from the host plant and the environment. They raise fascinating questions about the nature of intraspecies relationships in Fusarium populations and their interactions with the host plant. For example, are these populations fixed or altered according to plant developmental stage and season-related climatic conditions?
Previous studies provide some clues to the answers to these questions. For instance, it was observed that discernible divergences in pathogenicity across and within Fusarium species resulted in contrasting disease pathogenesis outcomes [2,30,31]. Such variability appears to be more related to the host plant than the geographical origin or climatic variables, which have a lesser impact in some instances [32]. The mechanism behind the FBR pathogenesis is now gradually being revealed. The disease severity results from pathogen metabolites and virulence factors interacting with the plant defense system. One such metabolite is fumonisin B1, secreted by F. proliferatum [1]. This toxin’s expression can vary depending on the infected host organ and phenological development stage. In response, the plant’s defense-related genes are expressed differentially during the seedling and bulb infection. These plant metabolites include lipoxygenase (LOX2), phenylalanine ammonia-lyase (PAL1, PAL2), anthocyanidin synthase (ANS), chalcone synthase (CHS), and pectin methyl esterase (PME) [1]. Thus, the plant’s defense variations are primarily linked to pathogen specialization towards specific host species, modulated by genetic mechanisms that repress host defense responses.
The current study’s findings are economically significant since each Fusarium species may react differently to control treatments [9]. Thus, knowing the exact population structure may assist in tailoring FBR protection to maximize its efficiency. The strategies currently employed in Israel to manage FBR disease are limited and consist of a four-year crop rotation cycle and soil disinfection using metam sodium [21]. However, despite these measures, the disease persists and is spreading to new areas where contaminated equipment and agricultural tools, such as harrows and plows, and the workforce unintentionally contribute to its propagation [21]. The problem is not unique to Israel but crosses borders. According to a review by Le et al. [2], Allium producers worldwide continue to face a significant disease problem despite implementing numerous control measures.
A recent study [9] explored the potential of chemical control methods in mitigating FBR disease damage in Israel. Initially, novel substances effective against the pathogens involved were identified using a plate screening technique. Subsequently, selected formulations from earlier trials in seedlings were evaluated for an entire growing season. One of the preparations, prochloraz, added to the irrigation, displayed efficacy against the principal causal agent of onion FBR disease, F. oxysporum f. sp. cepae (B14 isolate). However, it was relatively less effective against F. acutatum. Another compound based on fludioxonil + sedaxen (Fl-Se) applied in a seed coating could protect both onion cultivars against the two Fusarium species tested. Thus, a combined treatment that relies on both prochloraz and Fl-Se could be preferable.
Other important conclusions could be drawn from the results presented here. For instance, it was shown for the first time that the level of contamination in some fields is significantly higher than previously assumed and reaches 8% in certain varieties. While such a high incidence is alarming, it may be affected by the level of inbreeding. Since onion is an outcrossing species, it may suffer inbreeding depression [33]. The different lines used in onion hybrid cultivar development are often inbred to a certain extent to ensure uniformity within the hybrid cultivar. Inbred lines are frequently weaker in their growth than hybrid or open-pollinated cultivars. Thus, the survey presented here must be followed by a more comprehensive and dedicated study to evaluate the disease severity in different commercial onion cultivars in various geographic regions, considering the pollination method and other cultivation aspects.
Moreover, field losses are only a partial picture of the disease impact. The disease spread in onions is enhanced during storage, particularly in open sheds or packing houses. In this scenario, the disease can spread to other onions, and there is also a concern that infected bulbs that do not show visible symptoms could make their way to markets throughout the country. This concern increases significantly in light of the presence of toxins known to be produced by these pathogens [34].
According to Cramer [28], losses resulting from FBR can vary depending on growth stages and regions. Among the damping-off pathogens, Fusarium spp. can cause up to 70% of damage in nurseries [35]. Fusarium spp. can also lead to significant losses in bulbs, with reported losses of up to 50% in the field and 30–40% in storage in Asia [35,36]. Dauda and colleagues [8] observed FBR affecting 50% of seedlings in African growth areas. In southern New Mexico (USA), Cramer [28] reported a 40% and 29% disease incidence for fall-planted and spring-planted cultivars, respectively. Meanwhile, in Zambia, even when cultivated in virgin soil, the FBR prevalence was found to be high, with 80–90% of transplants infected by F. oxysporum f. sp. cepae, leading to significant losses in post-transplanting seedlings (44%) and potential yield (69%) [37]. Organic farms typically experience higher losses than conventional farms [38].
So, what future directions are needed to create efficient strategies for managing FBR? One (so far poorly explored) option is integrated pest management [2]. Integrating disease-resistant crops and biological control measures can provide proactive prevention against FBR disease damage. This approach should also involve using disease-free planting materials and regularly maintaining field hygiene to limit the spread of infections.
5. Conclusions
Fusarium basal rot disease (FBR) in onion (Allium cepa) is common worldwide, causing severe damage typified by an infection that spreads from the roots to the onion stem base and leaves. Significant knowledge gaps exist today regarding Israel’s FBR, the pathogen population involved, and the damage they cause. The current study analyzed the composition and prevalence of Fusarium species in two commercial fields in northeastern Israel, one in the Golan Heights and the other in Galilee (Hula Valley). The results revealed for the first time that Neocosmospora (previously F. solani) SC is part of the Fusarium population in onion FBR in northeastern Israel, and we found it to be the most common Fusarium in bulbs sampled from both areas. Furthermore, while in yellow onions of the Orlando cv. grown in the Galilee field, this species was found with two other species, F. oxysporum f. sp. cepae and F. acutatum, the Golan Heights field’s composition of Fusarium species was divided between onion cultivars. The red Ha2 cv. onions were populated by F. oxysporum f. sp. cepae, while the yellow Ha1 cv. onions were infected by F. acutatum. Meanwhile, Neocosmospora SC was found in both onion varieties. An in vitro seed and bulb pathogenicity assay showed that Neocosmospora species are moderately aggressive disease agents. Yet, the impact of these species’ combinations and interspecies relationships on host plant health is yet to be explored. The results of this and other global studies indicate that the Fusarium pathobiome composition and structure require specifically adapted pest control solutions since each Fusarium species may react differently with fungicide treatments.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae10040373/s1, Figure S1: Sequence alignment of the TEF1 gene of the Fusarium isolates (presented in Table 3); Figure S2: Full-length gels for the DNA fingerprinting of the Fusarium isolates shown in Figure 7A,B; Table S1: NCBI BLASTN identification of the Fusarium isolates from this study; Table S2: Seed germination in the seedling pathogenicity assay for Neocosmospora (F. solani) isolates.
Author Contributions
Conceptualization, O.D., E.D. and E.M.; data curation, O.D. and E.D.; formal analysis, O.D. and E.D.; funding acquisition, O.D. and E.M.; investigation, O.D. and E.D.; methodology, O.D. and E.D.; project administration, O.D.; resources, O.D. and E.M.; supervision, O.D.; validation, O.D. and E.D.; visualization, O.D. and E.D.; writing (original draft), O.D.; writing (review and editing), O.D., E.D. and E.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a one-year research grant (2022) from the Israel Plants Production and Marketing Board, the Ministry of Agriculture and Rural Development.
Data Availability Statement
Data generated by DNA sequencing were deposited in the NCBI repository. GenBank accession numbers for the nucleotide sequences are in Appendix A. All other data generated or analyzed during this study are included in this published article (and its Supplementary Information Files).
Acknowledgments
We would like to thank the owners of the lands, Kibbutz Yiron (Hula Valley, Upper Galilee, northern Israel) and Givat Yoav (Golan Heights), for their permission to conduct the study on these sites. We would also like to acknowledge Galia Shofman, Asaf Gordani, and Shaul Graph (Migal—Galilee Research Institute) for their helpful advice.
Conflicts of Interest
The authors declare no conflicts of interest. All authors have read and agreed to the published version of the manuscript.
Appendix A. GenBank Accession Numbers for the Nucleotide Sequences (https://www.ncbi.nlm.nih.gov/, Accessed on 1 April 2024)

References
- Le, D.; Ameye, M.; Landschoot, S.; Audenaert, K.; Haesaert, G. Phenology-regulated defence mechanisms as drivers for Fusarium basal rot in onion (Allium cepa). Plant Pathol. 2022, 71, 1440–1453. [Google Scholar] [CrossRef]
- Le, D.; Audenaert, K.; Haesaert, G. Fusarium basal rot: Profile of an increasingly important disease in Allium spp. Trop. Plant Pathol. 2021, 46, 241–253. [Google Scholar] [CrossRef]
- Klokočar-Šmit, Z.; Lević, J.; Maširević, S.; Gvozdanović-Varga, J.; Vasić, M.; Aleksić, S. Fusarium rot of onion and possible use of bioproduct. Zb. Matice Srp. Prir. Nauke. 2008, 144, 135–148. [Google Scholar] [CrossRef]
- Gunaratna, L.; Deshappriya, N.; Jayaratne, D.; Rajapaksha, R. Damping-off disease of big onion (Allium cepa L.) in Sri Lanka and evaluation of Trichoderma asperellum and Trichoderma virens for its control. Trop. Plant Res. 2019, 6, 2349–9265. [Google Scholar] [CrossRef]
- Le, D.; Ameye, M.; De Boevre, M.; De Saeger, S.; Audenaert, K.; Haesaert, G. Population, virulence, and mycotoxin profile of Fusarium spp. associated with basal rot of Allium spp. in Vietnam. Plant Dis. 2021, 105, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Ghanbarzadeh, B.; Mohammadi Goltapeh, E.; Safaie, N. Identification of Fusarium species causing basal rot of onion in east Azerbaijan province, Iran and evaluation of their virulence on onion bulbs and seedlings. Arch. Phytopathol. Plant Prot. 2014, 47, 1050–1062. [Google Scholar] [CrossRef]
- Özer, N.; Köycü, N.; Chilosi, G.; Magro, P. Resistance to Fusarium basal rot of onion in greenhouse and field and associated expression of antifungal compounds. Phytoparasitica 2004, 32, 388–394. [Google Scholar] [CrossRef]
- Dauda, W.; Alao, S.; Zarafi, A.; Alabi, O. First report of die-back disease of onion (Allium cepa L.) induced by Fusarium equiseti (mart) sacc in Nigeria. Int. J. Plant Soil Sci. 2018, 21, 2320–7035. [Google Scholar] [CrossRef]
- Degani, O.; Dimant, E.; Gordani, A.; Graph, S.; Margalit, E. Prevention and control of Fusarium spp., the causal agents of onion (Allium cepa) basal rot. Horticulturae 2022, 8, 1071. [Google Scholar] [CrossRef]
- Galván, G.A.; Koning-Boucoiran, C.F.; Koopman, W.J.; Burger-Meijer, K.; González, P.H.; Waalwijk, C.; Kik, C.; Scholten, O.E. Genetic variation among Fusarium isolates from onion, and resistance to Fusarium basal rot in related Allium species. Eur. J. Plant Pathol. 2008, 121, 499–512. [Google Scholar] [CrossRef]
- Boehnke, B.; Karlovsky, P.; Pfohl, K.; Gamliel, A.; Isack, Y.; Dehne, H. Identification of different Fusarium spp. in Allium spp. in Germany. Commun. Agric. Appl. Biol. Sci. 2015, 80, 453–463. [Google Scholar] [PubMed]
- Gebremichael, D.E.; Haile, Z.M.; Negrini, F.; Sabbadini, S.; Capriotti, L.; Mezzetti, B.; Baraldi, E. RNA interference strategies for future management of plant pathogenic fungi: Prospects and challenges. Plants 2021, 10, 650. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.; Rajasekaran, K.; Cary, J.W. RNA interference (RNAi) as a potential tool for control of mycotoxin contamination in crop plants: Concepts and considerations. Front. Plant Sci. 2017, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef] [PubMed]
- McDonald, T.; Brown, D.; Keller, N.P.; Hammond, T.M. RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol. Plant. Microbe. Interact. 2005, 18, 539–545. [Google Scholar] [CrossRef]
- Abdel-Hadi, A.M.; Caley, D.P.; Carter, D.R.; Magan, N. Control of aflatoxin production of Aspergillus flavus and Aspergillus parasiticus using RNA silencing technology by targeting AFLD (nor-1) gene. Toxins 2011, 3, 647–659. [Google Scholar] [CrossRef]
- Ghag, S.B.; Shekhawat, U.K.; Ganapathi, T.R. Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 2014, 12, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Lebiush-Mordechai, S.; Erlich, O.; Maymon, M.; Freeman, S.; Ben-David, T.; Ofek, T.; Palevsky, E.; Tsror Lahkin, L. Bulb and root rot in lily (Lilium longiflorum) and onion (Allium cepa) in Israel. J Phytopathol. 2014, 162, 466–471. [Google Scholar] [CrossRef]
- Gamliel, A.; Gillett, D.; Minkovsky, N.; Benikhis, M.; Dobrynin, S.; Margalit, E. Fusarium proliferatum Disease Outburst in White Onions from Different Fields in the Southern Israel Arava Area; 30 October 2012. Available online: https://aravard.org.il/wp-content/uploads/2013/10/12VegOniFusTissueCult.pdf (accessed on 1 April 2024). (In Hebrew).
- Degani, O.; Kalman, B. Assessment of commercial fungicides against onion (Allium cepa) basal rot disease caused by Fusarium oxysporum f. sp. cepae and Fusarium acutatum. J. Fungi 2021, 7, 235. [Google Scholar] [CrossRef]
- Kalman, B.; Abraham, D.; Graph, S.; Perl-Treves, R.; Meller Harel, Y.; Degani, O. Isolation and identification of Fusarium spp., the causal agents of onion (Allium cepa) basal rot in northeastern Israel. Biology 2020, 9, 69. [Google Scholar] [CrossRef]
- Dimant, E.; Degani, O. Molecular real-time PCR monitoring of onion Fusarium basal rot chemical control. J. Fungi 2023, 9, 809. [Google Scholar] [CrossRef]
- Reddy, M.P.; Sarla, N.; Siddiq, E.A. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Hong, S.-Y.; Kang, M.-R.; Cho, E.-J.; Kim, H.-K.; Yun, S.-H. Specific PCR detection of four quarantine Fusarium species in Korea. Plant Pathol. J. 2010, 26, 409–416. [Google Scholar] [CrossRef]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Sarver, B.A.; Balajee, S.A.; Schroers, H.-J.; Summerbell, R.C.; Robert, V.A.; Crous, P.W.; Zhang, N. Internet-accessible DNA sequence database for identifying Fusaria from human and animal infections. J. Clin. Microbiol. 2010, 48, 3708–3718. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. Seaview version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Cramer, C.S. Breeding and genetics of Fusarium basal rot resistance in onion. Euphytica 2000, 115, 159–166. [Google Scholar] [CrossRef]
- Bayraktar, H. Genetic diversity and population structure of Fusarium oxysporum f. sp. cepae, the causal agent of Fusarium basal plate rot on onion using RAPD markers. J. Agric. Sci. 2010, 16, 3. [Google Scholar]
- Caligiore Gei, P.F.; Valdez, J.G.; Piccolo, R.J.; Galmarini, C.R. Influence of Fusarium spp. Isolate and inoculum density on resistance screening tests in onion. Trop. Plant Pathol. 2014, 39, 19–27. [Google Scholar] [CrossRef]
- Nasr Esfahani, M. Genetic and virulence variation in Fusarium oxysporum f. sp. cepae causing root and basal rot of common onion in Iran. J. Phytopathol. 2018, 166, 572–580. [Google Scholar] [CrossRef]
- Brizuela, A.M.; Lalak-Kańczugowska, J.; Koczyk, G.; Stępień, Ł.; Kawaliło, M.; Palmero, D. Geographical origin does not modulate pathogenicity or response to climatic variables of Fusarium oxysporum associated with vascular wilt on asparagus. J. Fungi 2021, 7, 1056. [Google Scholar] [CrossRef] [PubMed]
- Khar, A.; Singh, H. Rapid methods for onion breeding. In Accelerated Plant Breeding, Volume 2: Vegetable Crops; Springer: Berlin/Heidelberg, Germany, 2020; pp. 77–99. [Google Scholar]
- Stankovic, S.; Levic, J.; Petrovic, T.; Logrieco, A.; Moretti, A. Pathogenicity and mycotoxin production by Fusarium proliferatum isolated from onion and garlic in Serbia. Eur. J. Plant Pathol. 2007, 118, 165–172. [Google Scholar] [CrossRef]
- Mishra, R.; Jaiswal, R.; Kumar, D.; Saabale, P.; Singh, A. Management of major diseases and insect pests of onion and garlic: A comprehensive review. Plant Breed. Crop Sci. 2014, 6, 160–170. [Google Scholar]
- Gupta, R.; Gupta, R. Effect of integrated disease management packages on diseases incidence and bulb yield of onion (Allium cepa L.). SAARC J. Agric. 2013, 11, 49–59. [Google Scholar] [CrossRef]
- Naik, D.; Burden, O. Chemical control of basal rot of onion in Zambia. Int. J. Pest Manag. 1981, 27, 455–460. [Google Scholar] [CrossRef]
- Haapalainen, M.; Latvala, S.; Kuivainen, E.; Qiu, Y.; Segerstedt, M.; Hannukkala, A. Fusarium oxysporum, F. proliferatum and F. redolens associated with basal rot of onion in Finland. Plant Pathol. 2016, 65, 1310–1320. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).