Comprehensive Transcriptome and Metabolome Characterization of Peony ‘Coral Sunset’ Petals Provides Insights into the Mechanism of Pigment Degradation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
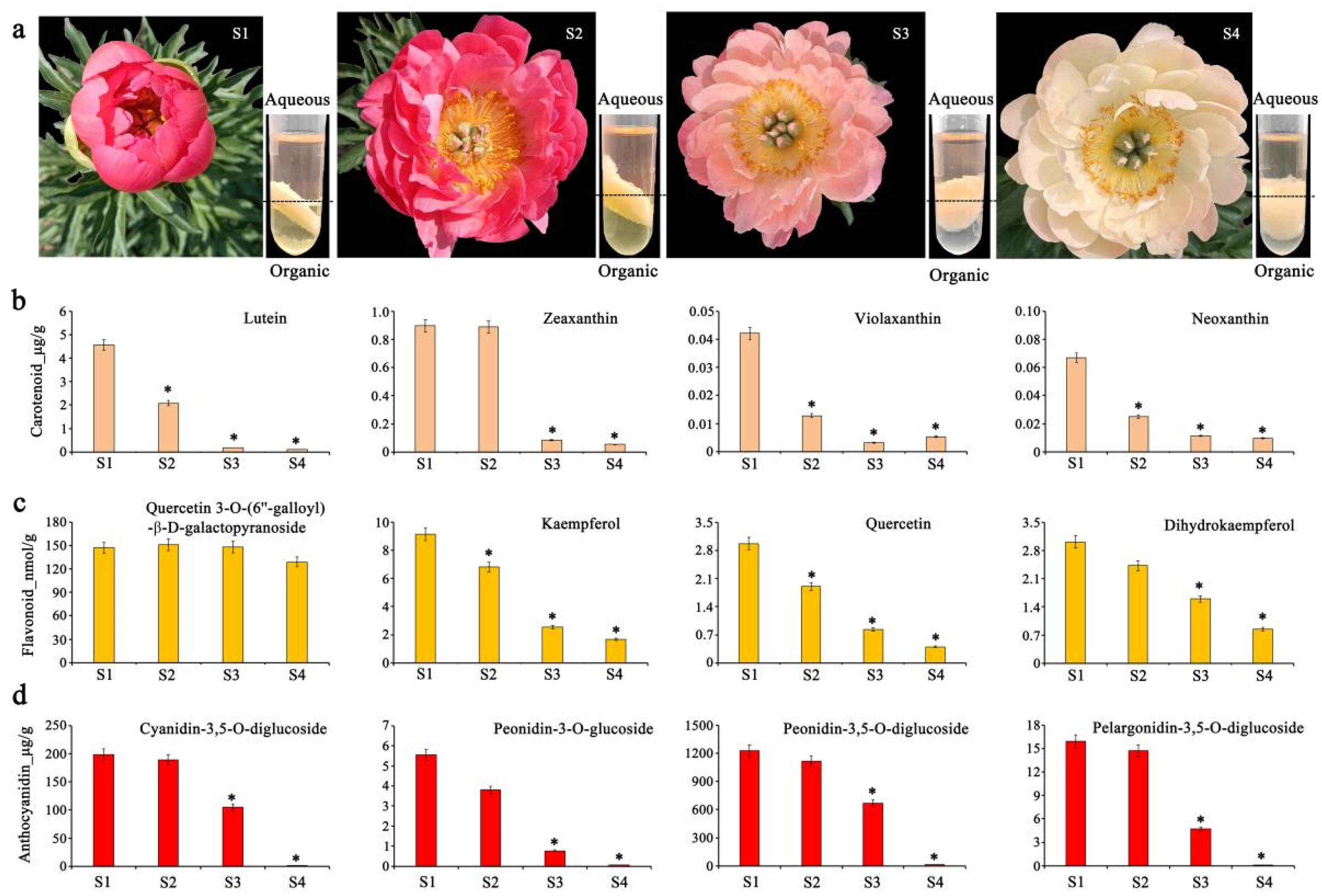
2.2. Identification of Petal Pigments
2.3. Illumina Sequencing
2.4. Library Construction and SMART Sequencing
2.5. Structural Analysis of the Transcriptome
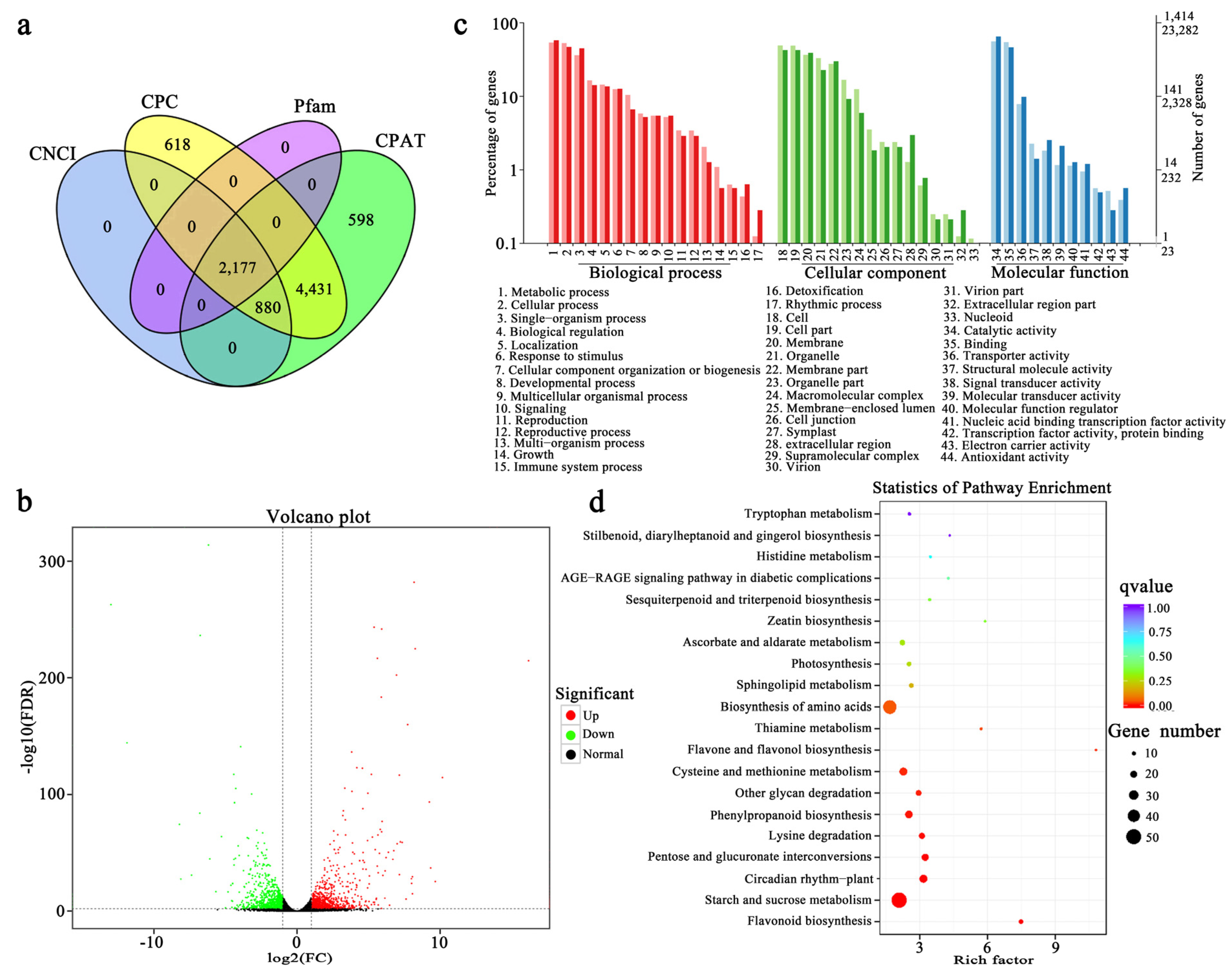
2.6. Coding RNA and Long Non-Coding RNA Analysis
2.7. Gene Functional Annotation
2.8. Quantification of Gene Expression Levels and Analysis of Differentially Expressed Genes
2.9. Quantitative Real-Time PCR Analysis
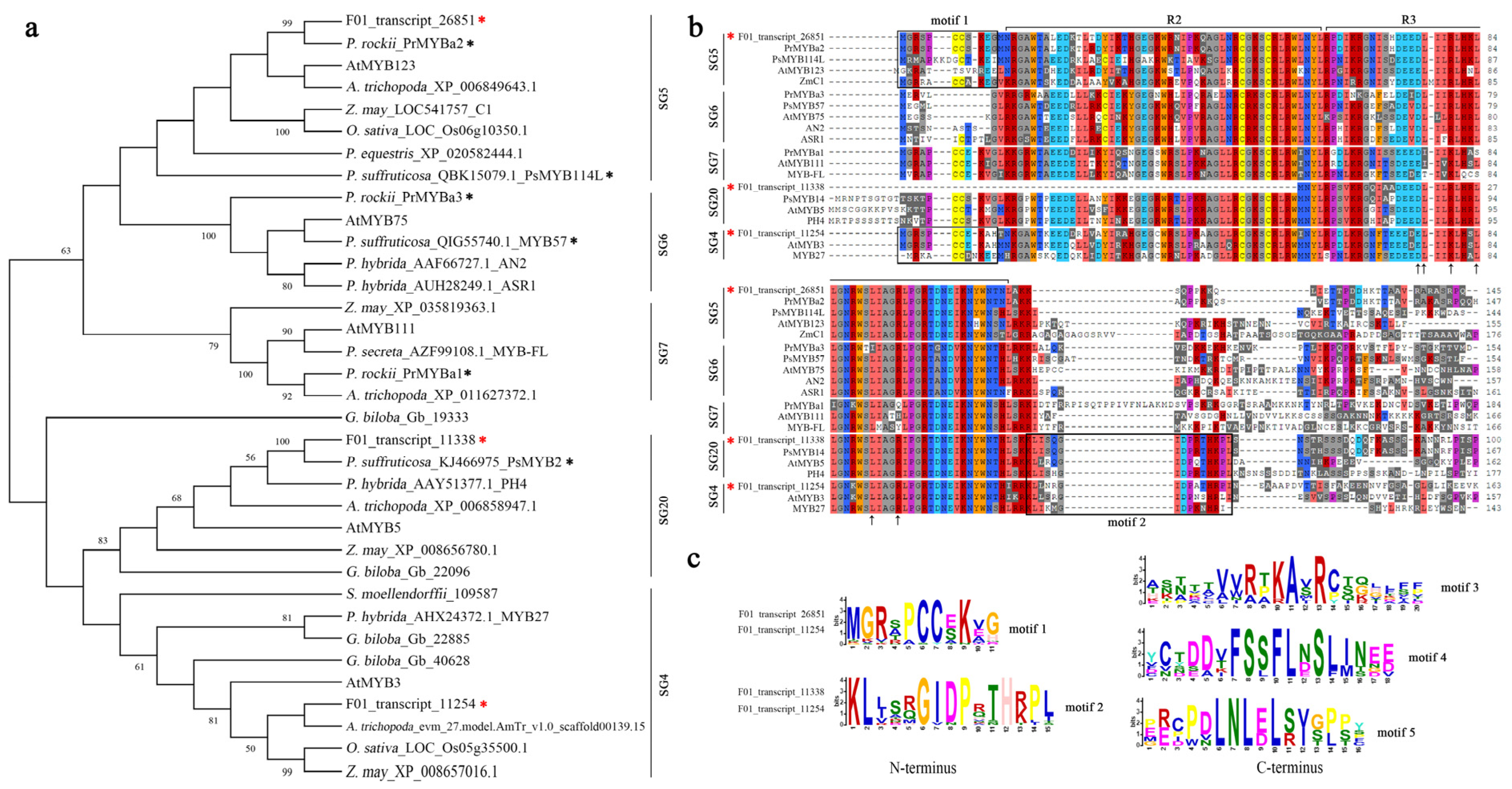
2.10. Phylogenetic and Amino Acid Analysis of the Flavonoid Regulatory R2R3-MYBs
2.11. Yeast Two-Hybrid Assay
2.12. Transient Transformation and Expression Pattern Analysis of Anthocyanin Synthesis Structural Genes in Petunia Corolla
2.13. Data Analysis
3. Results
3.1. Pigment Composition and Changes in Paeonia lactiflora ‘Coral Sunset’ Petals
3.2. RNA-Seq and Analysis of DEGs in Paeonia lactiflora ‘Coral Sunset’ Petals
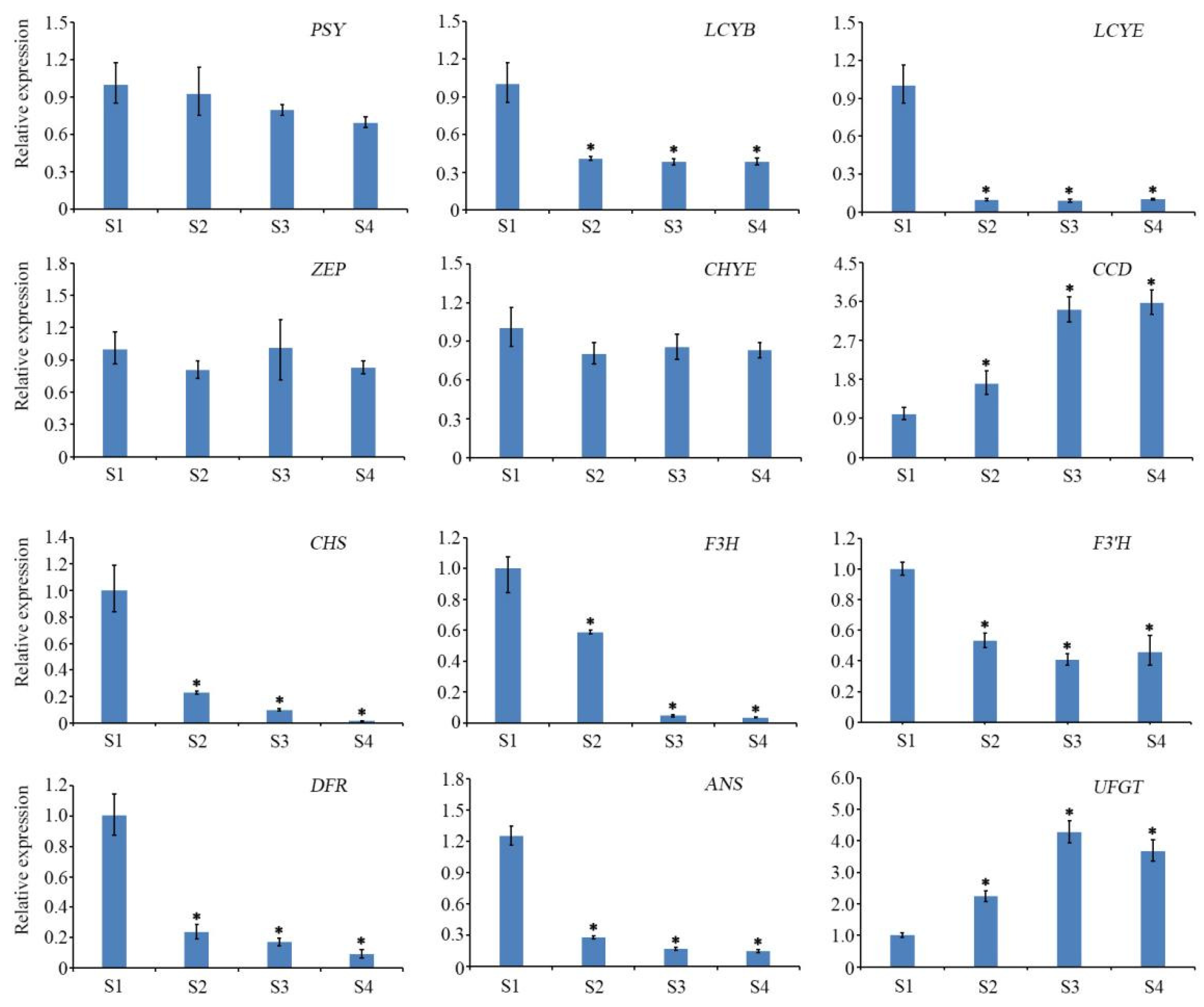
3.3. Pigment Synthesis Structural Genes Determined the Petal Coloration of Paeonia lactiflora ‘Coral Sunset’
3.4. Confirmation of Gene Expression Patterns by qRT-PCR
3.5. Characterization of R2R3-MYB Members in P. lactiflora ‘Coral Sunset’ Petals
3.6. Peony MYBs-SG4 Plays Roles in Anthocyanin Synthesis
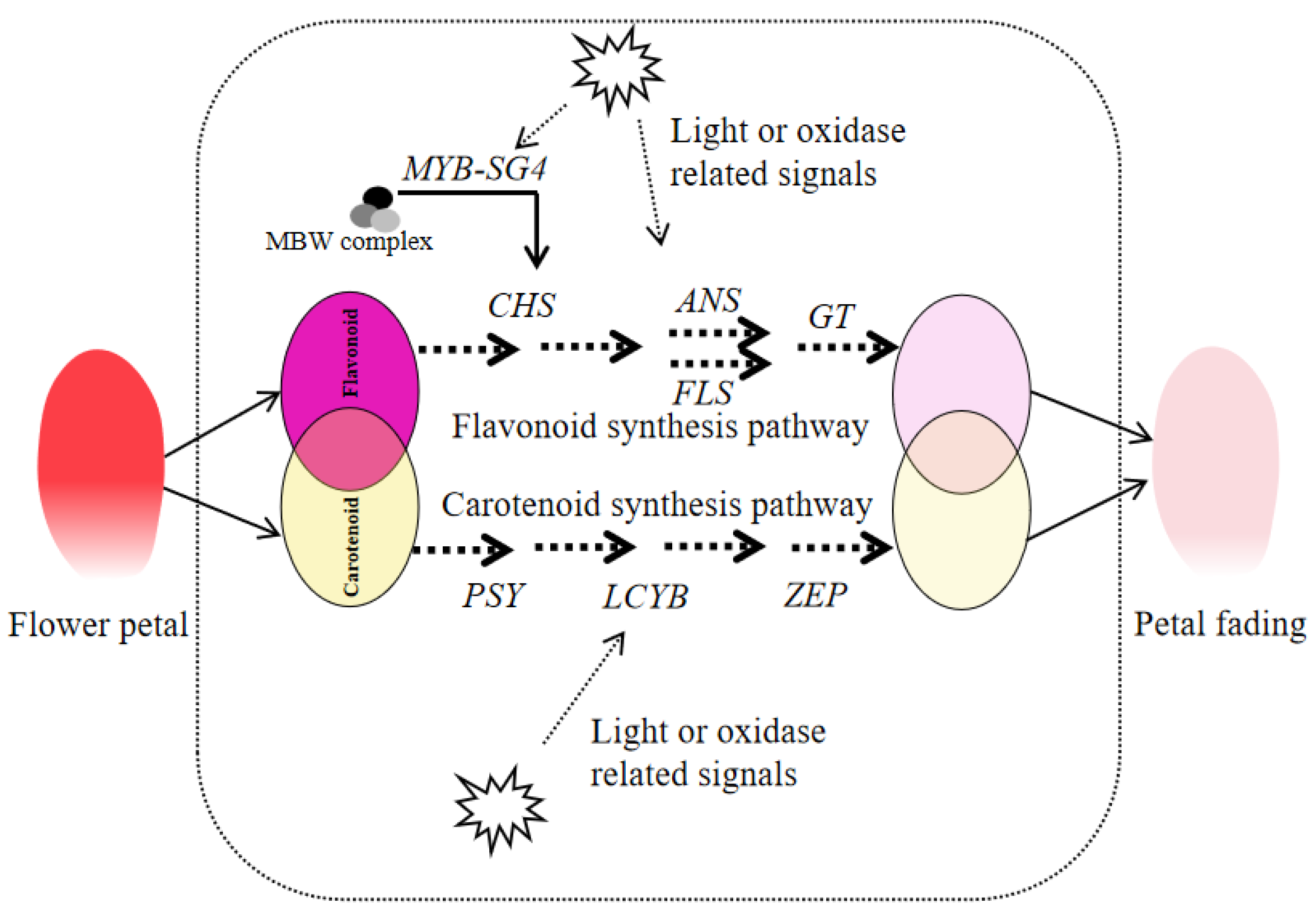
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Römer, S.; Fraser, P.D. Recent advances in carotenoid biosynthesis, regulation and manipulation. Planta 2005, 221, 305–308. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, M.; Nakatsuka, T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 2011, 33, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, R.T.; Glover, B.J. The evolution of flower colour. Curr. Biol. 2023, 33, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhang, Y.; Long, T.; Wang, S.; Yang, J. Regulation mechanism of plant pigments biosynthesis: Anthocyanins, carotenoids, and betalains. Metabolites 2022, 12, 871. [Google Scholar] [CrossRef] [PubMed]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef]
- Rosas-Saavedra, C.; Stange, C. Biosynthesis of carotenoids in plants: Enzymes and color. Subcell. Biochem. 2016, 79, 35–69. [Google Scholar]
- Gandía-Herrero, F.; García-Carmona, F. Biosynthesis of betalains: Yellow and violet plant pigments. Trends Plant Sci. 2013, 18, 334–343. [Google Scholar] [CrossRef]
- Liu, W.; Feng, Y.; Yu, S.; Fan, Z.; Li, X.; Li, J.; Yin, H. The flavonoid biosynthesis network in plants. Int. J. Mol. Sci. 2021, 22, 2824. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-mediated regulation of anthocyanin biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Zhang, H.; Koes, R.; Shang, H.; Fu, Z.; Wang, L.; Dong, X.; Zhang, J.; Passeri, V.; Li, Y.; Jiang, H.; et al. Identification and functional analysis of three new anthocyanin R2R3-MYB genes in Petunia. Plant Direct 2019, 3, e00114. [Google Scholar] [CrossRef] [PubMed]
- Albert, N.W.; Davies, K.M.; Lewis, D.H.; Zhang, H.; Montefiori, M.; Brendolise, C.; Boase, M.R.; Ngo, H.; Jameson, P.E.; Schwinn, K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 2014, 26, 962–980. [Google Scholar] [CrossRef] [PubMed]
- LaFountain, A.M.; Yuan, Y.W. Repressors of anthocyanin biosynthesis. New Phytol. 2021, 231, 933–949. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Oren-Shamir, M. Does anthocyanin degradation play a significant role in determining pigment concentration in plants? Plant Sci. 2009, 177, 310–316. [Google Scholar] [CrossRef]
- Passeri, V.; Koes, R.; Quattrocchio, F.M. New challenges for the design of high value plant products: Stabilization of anthocyanins in plant vacuoles. Front. Plant Sci. 2016, 7, 153. [Google Scholar] [CrossRef]
- Kader, F.; Haluk, J.; Nicolas, J.; Metche, M. Degradation of cyanidin 3-glucoside by blueberry polyphenol oxidase: Kinetic studies and mechanisms. J. Agric. Food Chem. 1998, 46, 3060–3065. [Google Scholar] [CrossRef]
- Sarni, P.; Fulcrand, H.; Souillol, V.; Souquet, J.-M.; Cheynier, V. Mechanisms of anthocyanin degradation in grape must-like model solutions. J. Sci. Food Agric. 1995, 69, 385–391. [Google Scholar] [CrossRef]
- Luo, H.; Deng, S.; Fu, W.; Zhang, X.; Zhang, X.; Zhang, Z.; Pang, X. Characterization of active anthocyanin degradation in the petals of Rosa chinensis and Brunfelsia calycina reveals the effect of gallated catechins on pigment maintenance. Int. J. Mol. Sci. 2017, 18, 699. [Google Scholar] [CrossRef] [PubMed]
- Zipor, G.; Duarte, P.; Carqueijeiro, I.; Shahar, L.; Ovadia, R.; Teper-Bamnolker, P.; Eshel, D.; Levin, Y.; Doron-Faigenboim, A.; Sottomayor, M.; et al. In planta anthocyanin degradation by a vacuolar class III peroxidase in Brunfelsia calycina flowers. New Phytol. 2015, 205, 653–665. [Google Scholar] [CrossRef] [PubMed]
- Quattrocchio, F.; Verweij, W.; Kroon, A.; Spelt, C.; Mol, J.; Koes, R. PH4 of Petunia is an R2R3-MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 2006, 18, 1274–1291. [Google Scholar] [CrossRef] [PubMed]
- Maoka, T. Carotenoids as natural functional pigments. J. Nat. Med. 2020, 74, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Moraga, A.; Rambla, J.L.; Fernández-De-Carmen, A.; Trapero-Mozos, A.; Ahrazem, O.; Orzáez, D.; Granell, A.; Gómez-Gómez, L. New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus. Plant Mol. Biol. 2014, 86, 555–569. [Google Scholar] [CrossRef] [PubMed]
- Dhar, M.K.; Mishra, S.; Bhat, A.; Chib, S.; Kaul, S. Plant carotenoid cleavage oxygenases: Structure-function relationships and role in development and metabolism. Brief. Funct. Genom. 2020, 19, 1–9. [Google Scholar] [CrossRef]
- Yoo, H.J.; Chung, M.Y.; Lee, H.A.; Lee, S.B.; Grandillo, S.; Giovannoni, J.J.; Lee, J.M. Natural overexpression of CAROTENOID CLEAVAGE DIOXYGENASE 4 in tomato alters carotenoid flux. Plant Physiol. 2023, 192, 1289–1306. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Rodríguez-Concepción, M. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 2010, 107, 11626–11631. [Google Scholar] [CrossRef]
- Quian-Ulloa, R.; Stange, C. Carotenoid biosynthesis and plastid development in plants: The role of light. Int. J. Mol. Sci. 2021, 22, 1184. [Google Scholar] [CrossRef]
- Sun, T.; Rao, S.; Zhou, X.; Li, L. Plant carotenoids: Recent advances and future perspectives. Mol. Hortic. 2022, 2, 3. [Google Scholar] [CrossRef]
- Ma, Y.; Ma, X.; Gao, X.; Wu, W.; Zhou, B. Light induced regulation pathway of anthocyanin biosynthesis in plants. Int. J. Mol. Sci. 2021, 22, 11116. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.; Liu, X.; Zhao, S.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, 345–349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Guo, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Wang, L.G.; Park, H.J.; Dasari, S.; Wang, S.Q.; Kocher, J.P.; Li, W. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identifcation using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Qin, X.; Liu, Y.; Zhang, Y.; Bao, H.; Hu, Y.; Shen, X. Analysis of flavonoid metabolism during the process of petal discoloration in three Malus Crabapple Cultivars. ACS Omega 2022, 7, 37304–37314. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Han, X.; Wang, G.; Qiu, J.; Zhou, L.J.; Chen, S.; Fang, W.; Chen, F.; Jiang, J. Transcriptome analysis reveals chrysanthemum flower discoloration under high-temperature stress. Front. Plant Sci. 2022, 13, 1003635. [Google Scholar] [CrossRef] [PubMed]
- Rong, H.; Han, X.; Xin, Y.; Ni, Z.; Zhang, W.; Xu, L. Small RNA and degradome sequencing reveal roles of miRNAs in the petal color fading of Malus Crabapple. Int. J. Mol. Sci. 2023, 24, 11384. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Yang, H.; Liu, H.; Yang, J.; Hu, Y. Extensive transcriptome changes underlying the flower color intensity variation in Paeonia ostii. Front. Plant Sci. 2016, 6, 1205. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Li, W.; Zhang, X.; Deng, S.; Xu, Q.; Hou, T.; Pang, X.; Zhang, Z.; Zhang, X. In planta high levels of hydrolysable tannins inhibit peroxidase mediated anthocyanin degradation and maintain abaxially red leaves of Excoecaria cochinchinensis. BMC Plant Biol. 2019, 19, 315. [Google Scholar] [CrossRef]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB repressors as regulators of phenylpropanoid metabolism in plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Z.; Yu, X.; Zhao, L.; Zhao, M.; Han, X.; Qi, S. Identification of two novel R2R3-MYB transcription factors, PsMYB114L and PsMYB12L, related to anthocyanin biosynthesis in Paeonia suffruticosa. Int. J. Mol. Sci. 2019, 20, 1055. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Cheng, Y.; Wang, J.; Wang, X.; Liu, R.; Han, J. Functional identification of PsMYB57 involved in anthocyanin regulation of tree peony. BMC Genet. 2020, 21, 124. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Ma, H.; Duan, X.; Gao, S.; Zhou, X.; Cheng, Y. The R2R3-MYB gene PsMYB58 positively regulates anthocyanin biosynthesis in tree peony flowers. Plant Physiol. Biochem. 2021, 164, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yan, L.; Zhang, C.; Kong, X.; Zheng, Y.; Dong, L. Glucose supply induces PsMYB2-mediated anthocyanin accumulation in Paeonia suffruticosa ‘Tai Yang’ cut flower. Front. Plant Sci. 2022, 13, 874526. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, Y.; Wang, Q.; Li, B.; Wang, X.; Zhou, X.; Zhang, H.; Xu, W.; Li, S.; Wang, L. The combination of DNA methylation and positive regulation of anthocyanin biosynthesis by MYB and bHLH transcription factors contributes to the petal blotch formation in Xibei tree peony. Hortic. Res. 2023, 10, uhad100. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Jeong, C.Y.; Kang, G.H.; Yoo, S.D.; Hong, S.W.; Lee, H. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 2015, 84, 1192–1205. [Google Scholar] [CrossRef]
- Jiang, Z.H.; Chen, C.; Wang, J.; Xie, W.Y.; Wang, M.; Li, X.; Zhang, X.Y. Purple potato (Solanum tuberosum L.) anthocyanins attenuate alcohol-induced hepatic injury by enhancing antioxidant defense. J. Nat. Med. 2016, 70, 45–53. [Google Scholar] [CrossRef]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef]


| Database | Number of Isoforms | Proportion (%) |
|---|---|---|
| COG | 14,040 | 42.03 |
| GO | 23,282 | 69.70 |
| KEGG | 14,960 | 44.79 |
| KOG | 22,574 | 67.58 |
| Pfam | 26,084 | 78.09 |
| SwissProt | 24,900 | 74.54 |
| eggNOG | 32,634 | 97.70 |
| Nr | 33,268 | 99.60 |
| All | 33,403 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Yuan, X.; Wang, R.; Wang, L.; Gao, J.; Wang, H.; Li, Y.; Fu, Z. Comprehensive Transcriptome and Metabolome Characterization of Peony ‘Coral Sunset’ Petals Provides Insights into the Mechanism of Pigment Degradation. Horticulturae 2023, 9, 1295. https://doi.org/10.3390/horticulturae9121295
Zhang H, Yuan X, Wang R, Wang L, Gao J, Wang H, Li Y, Fu Z. Comprehensive Transcriptome and Metabolome Characterization of Peony ‘Coral Sunset’ Petals Provides Insights into the Mechanism of Pigment Degradation. Horticulturae. 2023; 9(12):1295. https://doi.org/10.3390/horticulturae9121295
Chicago/Turabian StyleZhang, Hechen, Xin Yuan, Rui Wang, Limin Wang, Jie Gao, Huijuan Wang, Yanmin Li, and Zhenzhu Fu. 2023. "Comprehensive Transcriptome and Metabolome Characterization of Peony ‘Coral Sunset’ Petals Provides Insights into the Mechanism of Pigment Degradation" Horticulturae 9, no. 12: 1295. https://doi.org/10.3390/horticulturae9121295
APA StyleZhang, H., Yuan, X., Wang, R., Wang, L., Gao, J., Wang, H., Li, Y., & Fu, Z. (2023). Comprehensive Transcriptome and Metabolome Characterization of Peony ‘Coral Sunset’ Petals Provides Insights into the Mechanism of Pigment Degradation. Horticulturae, 9(12), 1295. https://doi.org/10.3390/horticulturae9121295






