Transcriptome Analyses Reveal the Key Regulators of Tomato Compound Leaf Development
Abstract
:1. Introduction
2. Results
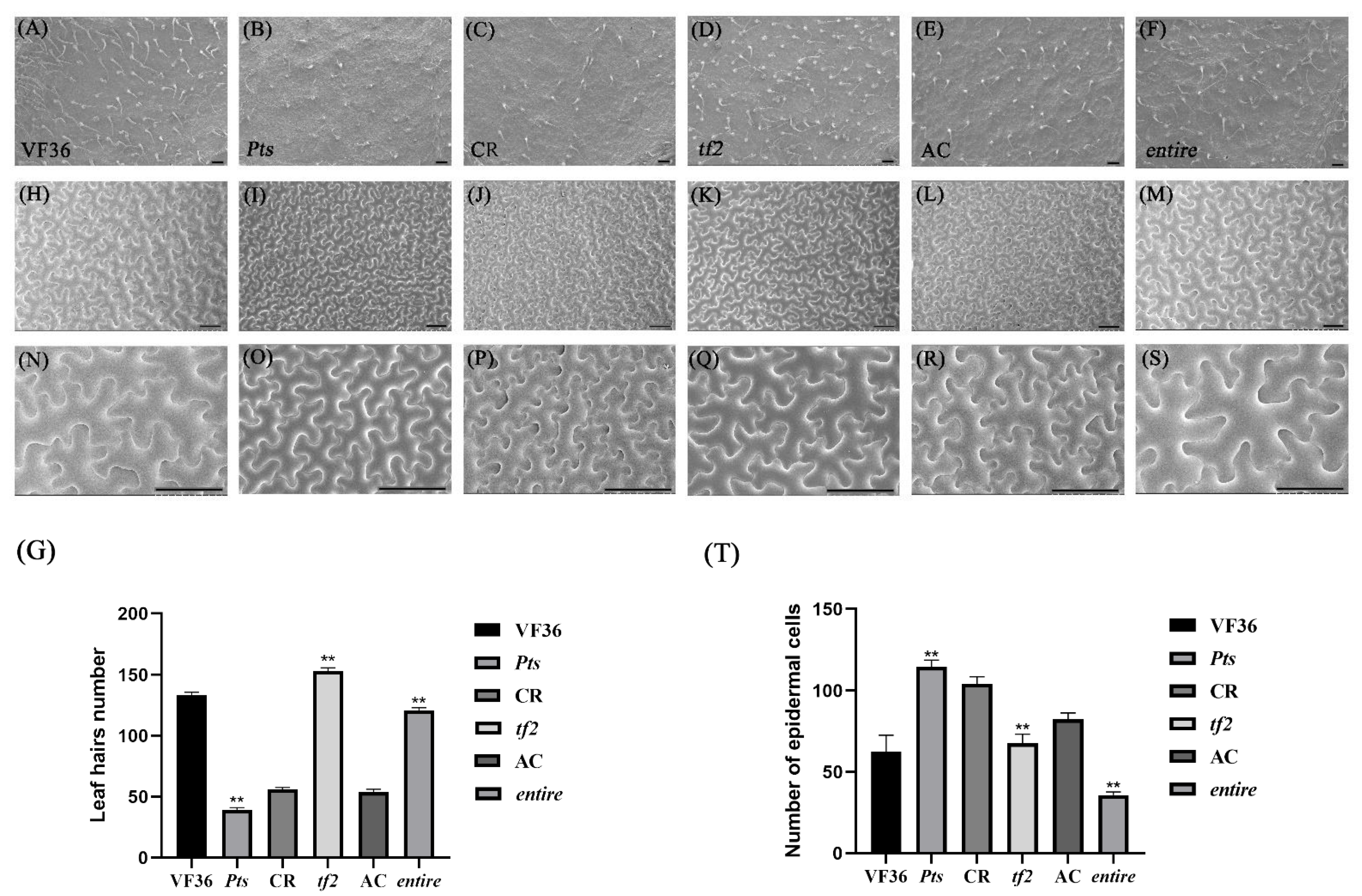
2.1. Leaf Shape Characteristics of the Mutant Plants
2.2. The Simplification and Complexity of Leaves Can Affect Leaf Hair Development and Cell Division
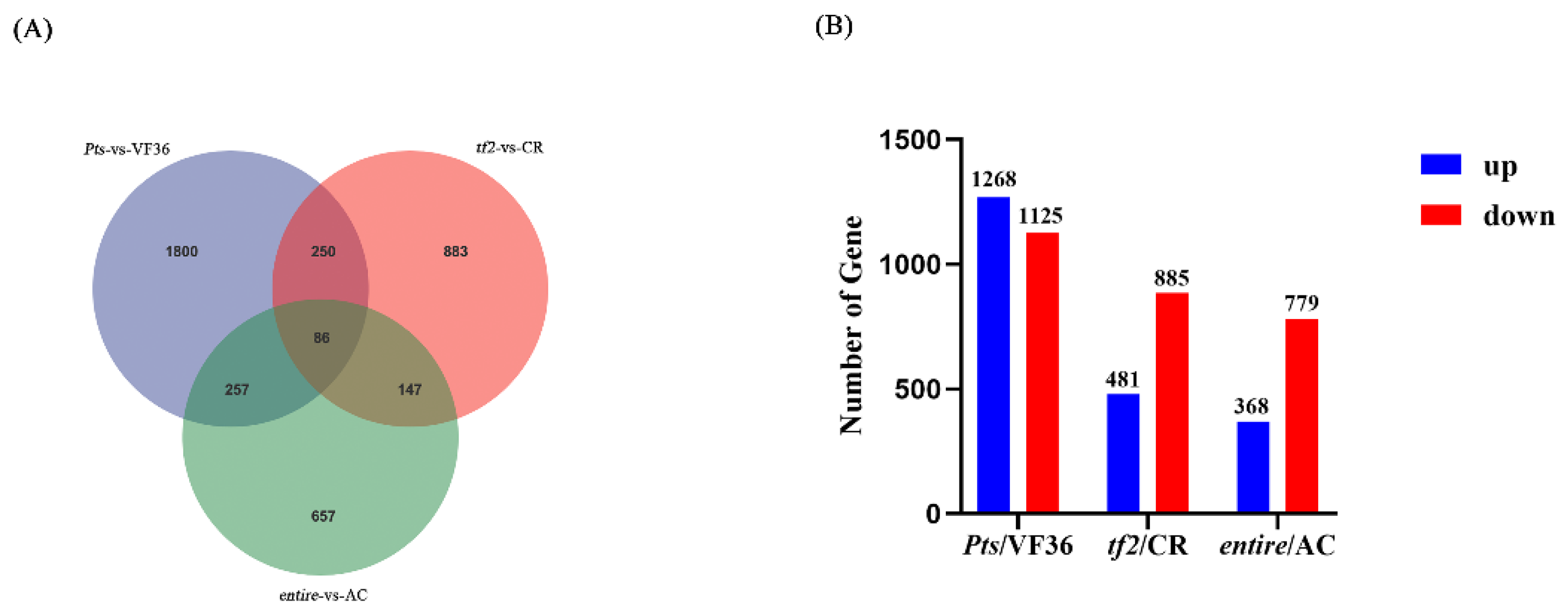
2.3. Transcriptome Sequencing among Pts/VF36, tf2/CR, and e/AC Groups
2.4. Transpiration Rate, Stomatal Conductance, and Epidermal Cell Permeability Are Closely Related to GDSL Esterase/Lipase Expression
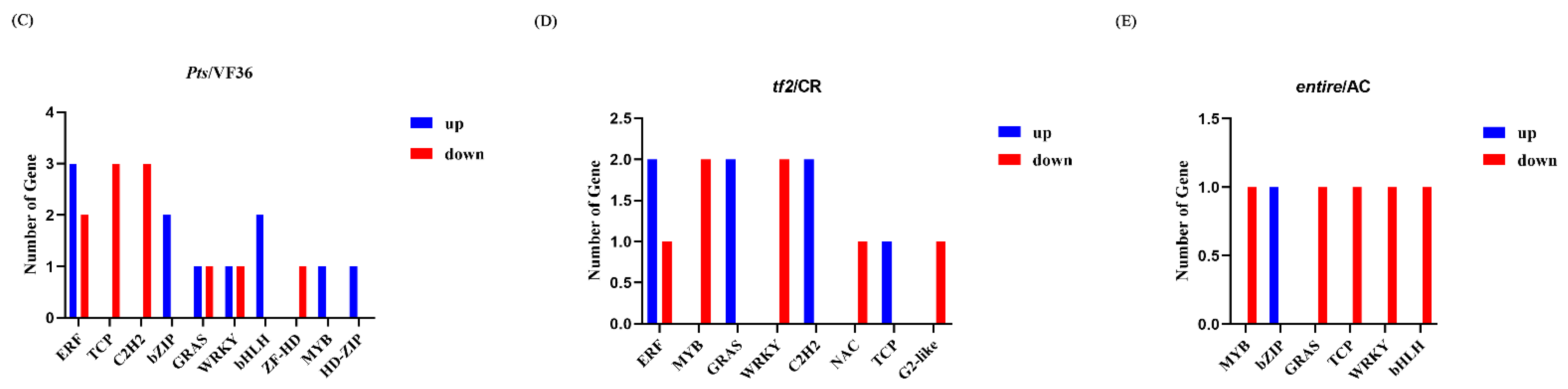
2.5. Differentially Expressed Genes in Pts/VF36 and tf2/CR, Pts/VF36 and e/AC, and tf2/CR and e/AC
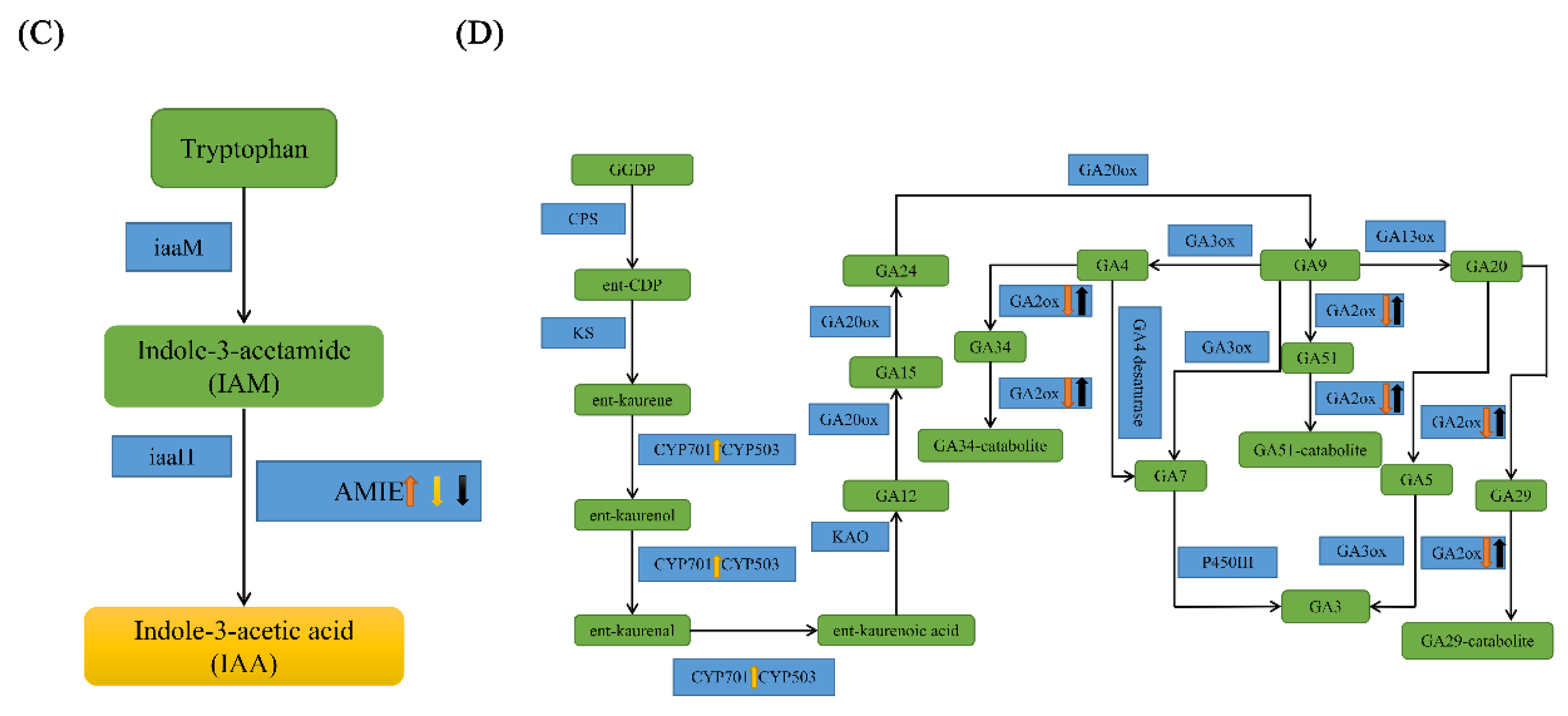
2.6. IAA and GA May Regulate Compound Leaf Shape Formation in Tomatoes
2.7. Identification of the Expression of Candidate Genes at Different Leaf Development Stages in Tomatoes
2.8. Morphological Changes in Plants with VIGS
3. Discussion
3.1. Different Compound Leaf Shapes Influenced GDSL Expression in Tomatoes
3.2. The Roles of the Plant Hormones IAA and GA in Tomato Compound Leaf Development
3.3. Key Regulators of Tomato Compound Leaf Development
4. Materials and Methods
4.1. Plant Materials and Culture Conditions
4.2. RNA Isolation
4.3. Library Preparation
4.4. Sequencing and Data Analysis
4.5. Reverse Transcription Quantitative PCR (RT-qPCR) Analysis
4.6. Scanning Electron Microscopy
4.7. Transpiration Rate Measurements
4.8. Tissue Collection
4.9. Statistical Analysis
4.10. UFLC-ESI-MS/MS Assay
4.11. VIGS Assays
4.12. Chlorophyll Leaching Assay
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| AC | Ailsa Craig |
| ARF | Auxin response factor |
| bHLH | Basic helix-loop-helix |
| BLAST | Basic local alignment search tool |
| BR | Brassinosteroids |
| CKs | Cytokinins |
| CKX | Cytokinin oxidase genes |
| CR | Condine Red |
| CPS | Ent-copalyl diphosphate synthase |
| DEGs | Differentially expressed genes |
| dpy | Defective mutant dumpy |
| e | Entire |
| ET | Ethylene |
| FPKM | Fragments per kilobase of exon model per million mapped reads |
| GA | Gibberellic acid |
| GA2ox7 | Gibberellin 2-beta-dioxygenase 7 |
| GA20ox | Gibberellin 20-oxidase-like |
| GDSL | GDSL esterases/lipases |
| GO | Gene ontology |
| GhFP1 | Cotton bHLH protein |
| HD | Homeodomain |
| IAA | Indole-3-acetic acid |
| IAA-aah | IAA-amino acid hydrolase |
| IBH1 | ILI1-binding BHLH Protein 1 |
| IPT7 | Iso-pentenyl transferase 7 |
| JA | Jasmonic acid |
| KAO | Ent-kaurenoic acid oxidase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KNOX | Knotted1-like homeobox |
| KO | Ent-kaurene oxidase |
| KS | Ent-kaurene synthase |
| LA | LANCEOLATE |
| LP1 | Leaf-associated protein 1 |
| LP2 | Leaf-associated protein 2 |
| MYC | Myelocytomatosis |
| OCL | Outer cuticular ledge |
| OSP1 | Occlusion of Stomatal Pore 1 |
| PIFs | Phytochrome-interacting factors |
| PM | Primary morphogenesis |
| Pts | Petroselinum |
| SAGs | Senescence-related gene |
| SAM | Shoot apical meristem |
| SM | Secondary morphogenesis |
| tf2 | Trifoliate |
| TFs | Transcription factors |
| VIGS | Virus-induced gene silencing |
| wdl1 | Wilted dwarf and lethal 1 |
| WOX | WUSCHEL-related homeobox |
References
- Berger, Y.; Harpaz-Saad, S.; Brand, A.; Melnik, H.; Sirding, N.; Alvarez, J.P.; Zinder, M.; Samach, A.; Eshed, Y.; Ori, N. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 2009, 136, 823–832. [Google Scholar] [CrossRef] [Green Version]
- Tsiantis, M. Control of shoot cell fate: Beyond homeoboxes. Plant Cell 2001, 13, 733–738. [Google Scholar] [CrossRef] [PubMed]
- Champagne, C.; Sinha, N. Compound leaves: Equal to the sum of their parts? Development 2004, 131, 4401–4412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef] [Green Version]
- Ferris, K.G.; Rushton, T.; Greenlee, A.B.; Toll, K.; Blackman, B.K.; Willis, J.H. Leaf shape evolution has a similar genetic architecture in three edaphic specialists within the Mimulus guttatus species complex. Ann. Bot. 2015, 116, 213–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogel, S. Leaves in the lowest and highest winds: Temperature, force and shape. New Phytol. 2009, 183, 13–26. [Google Scholar] [CrossRef]
- Semchenko, M.; Zobel, K. The role of leaf lobation in elongation responses to shade in the rosette-forming forb Serratula tinctoria (Asteraceae). Ann. Bot. 2007, 100, 83–90. [Google Scholar] [CrossRef] [Green Version]
- Burglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.Q.; Zhao, B.L.; He, L.L.; Zhou, S.L.; Liu, Y.; Zhao, W.Y.; Guo, S.Q.; Wang, R.R.; Bai, Q.Z.; Li, Y.H.; et al. The WOX family transcriptional regulator SlLAM1 controls compound leaf and floral organ development in Solanum lycopersicum. J. Exp. Bot. 2021, 72, 1822–1835. [Google Scholar] [CrossRef]
- Romani, F.; Reinheimer, R.; Florent, S.N.; Bowman, J.L.; Moreno, J.E. Evolutionary history of HOMEODOMAIN LEUCINE ZIPPER transcription factors during plant transition to land. New Phytol. 2018, 219, 408–421. [Google Scholar] [CrossRef] [Green Version]
- Bar, M.; Ori, N. Compound leaf development in model plant species. Curr. Opin. Plant Biol. 2015, 23, 61–69. [Google Scholar] [CrossRef]
- Tsuda, K.; Hake, S. Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 2015, 27, 91–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hareven, D.; Gutfinger, T.; Parnis, A.; Eshed, Y.; Lifschitz, E. The making of a compound leaf: Genetic manipulation of leaf architecture in tomato. Cell 1996, 84, 735–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ori, N.; Cohen, A.R.; Etzioni, A.; Brand, A.; Yanai, O.; Shleizer, S.; Menda, N.; Amsellem, Z.; Efroni, I.; Pekker, I.; et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 2007, 39, 787–791. [Google Scholar] [CrossRef]
- Shleizer-Burko, S.; Burko, Y.; Ben-Herzel, O.; Ori, N. Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 2011, 138, 695–704. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Zhang, J.; Wu, Y.W.; Wang, Y.; Zhang, J.; Zheng, Y.; Li, Y.; Li, X.B. bHLH transcription factors LP1 and LP2 regulate longitudinal cell elongation. Plant Physiol. 2021, 187, 2577–2591. [Google Scholar] [CrossRef]
- Pekker, I.; Alvarez, J.P.; Eshed, Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 2005, 17, 2899–2910. [Google Scholar] [CrossRef] [Green Version]
- Chitwood, D.H.; Headland, L.R.; Ranjan, A.; Martinez, C.C.; Braybrook, S.A.; Koenig, D.P.; Kuhlemeier, C.; Smith, R.S.; Sinha, N.R. Leaf Asymmetry as a Developmental Constraint Imposed by Auxin-Dependent Phyllotactic Patterning. Plant Cell 2012, 24, 2318–2327. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jones, B.; Li, Z.; Frasse, P.; Delalande, C.; Regad, F.; Chaabouni, S.; Latche, A.; Pech, J.-C.; Bouzayen, M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 2005, 17, 2676–2692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Gera, H.; Dafna, A.; Alvarez, J.P.; Bar, M.; Mauerer, M.; Ori, N. Auxin-mediated lamina growth in tomato leaves is restricted by two parallel mechanisms. Plant J. 2016, 86, 443–457. [Google Scholar] [CrossRef]
- Ben-Gera, H.; Shwartz, I.; Shao, M.R.; Shani, E.; Estelle, M.; Ori, N. ENTIRE and GOBLET promote leaflet development in tomato by modulating auxin response. Plant J. 2012, 70, 903–915. [Google Scholar] [CrossRef]
- Hendelman, A.; Buxdorf, K.; Stav, R.; Kravchik, M.; Arazi, T. Inhibition of lamina outgrowth following Solanum lycopersicum AUXIN RESPONSE FACTOR 10 (SlARF10) derepression. Plant Mol. Biol. 2012, 78, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Koenig, D.; Bayer, E.; Kang, J.; Kuhlemeier, C.; Sinha, N. Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 2009, 136, 2997–3006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassel, G.W.; Mullen, R.T.; Bewley, J.D. Procera is a putative DELLA mutant in tomato (Solanum lycopersicum): Effects on the seed and vegetative plant. J. Exp. Bot. 2008, 59, 585–593. [Google Scholar] [CrossRef] [Green Version]
- Fleishon, S.; Shani, E.; Ori, N.; Weiss, D. Negative reciprocal interactions between gibberellin and cytokinin in tomato. New Phytol. 2011, 190, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Kaur, H.; Phillips, A.; Hedden, P.; Hake, S.; Tsiantis, M. The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. CB 2002, 12, 1557–1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jasinski, S.; Tattersall, A.; Piazza, P.; Hay, A.; Martinez-Garcia, J.F.; Schmitz, G.; Theres, K.; McCormick, S.; Tsiantis, M. PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato. Plant J. 2009, 60, 397-397. [Google Scholar]
- Livne, S.; Lor, V.S.; Nir, I.; Eliaz, N.; Aharoni, A.; Olszewski, N.E.; Eshed, Y.; Weiss, D. Uncovering DELLA-Independent Gibberellin Responses by Characterizing New Tomato procera Mutants. Plant Cell 2015, 27, 1579–1594. [Google Scholar] [CrossRef] [Green Version]
- Gordon, S.P.; Chickarmane, V.S.; Ohno, C.; Meyerowitz, E.M. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 2009, 106, 16529–16534. [Google Scholar] [CrossRef] [Green Version]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef]
- Chen, J.J.; Janssen, B.J.; Williams, A.; Sinha, N. A gene fusion at a homeobox locus: Alterations in leaf shape and implications for morphological evolution. Plant Cell 1997, 9, 1289–1304. [Google Scholar] [PubMed] [Green Version]
- Shalit, A.; Rozman, A.; Goldshmidt, A.; Alvarez, J.P.; Bowman, J.L.; Eshed, Y.; Lifschitz, E. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc. Natl. Acad. Sci. USA 2009, 106, 8392–8397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shani, E.; Ben-Gera, H.; Shleizer-Burko, S.; Burko, Y.; Weiss, D.; Ori, N. Cytokinin Regulates Compound Leaf Development in Tomato. Plant Cell 2010, 22, 3206–3217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saini, S.; Sharma, I.; Pati, P.K. Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front. Plant Sci. 2015, 6, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.P.; Savaldi-Goldstein, S. Growth control: Brassinosteroid activity gets context. J. Exp. Bot. 2015, 66, 1123–1132. [Google Scholar] [CrossRef] [Green Version]
- Koka, C.V.; Cerny, R.E.; Gardner, R.G.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Clouse, S.D. A putative role for the tomato genes DUMPY and CURL-3 in brassinosteroid biosynthesis and response. Plant Physiol. 2000, 122, 85–98. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zhao, Y.; McCaig, B.C.; Wingerd, B.A.; Wang, J.; Whalon, M.E.; Pichersky, E.; Howe, G.A. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 2004, 16, 126–143. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.K.; Li, Y.L.; Chen, G.P.; Ren, L.J.; Xie, Q.L.; Zhao, Z.P.; Hu, Z.L. Silencing SlELP2L, a tomato Elongator complex protein 2-like gene, inhibits leaf growth, accelerates leaf, sepal senescence, and produces dark-green fruit. Sci. Rep. 2015, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Ecker, J.R. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell 2003, 115, 667–677. [Google Scholar] [CrossRef] [Green Version]
- Tieman, D.M.; Ciardi, J.A.; Taylor, M.G.; Klee, H.J. Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J. Cell Mol. Biol. 2001, 26, 47–58. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Yang, X.P.; Xiao, C.L.; Li, J.Y.; Chen, Y.Q.; Li, R.Y.; Li, S.P.; Lu, S.Y.; Hu, H.H. GDSL lipase occluded stomatal pore 1 is required for wax biosynthesis and stomatal cuticular ledge formation. New Phytol. 2020, 228, 1880–1896. [Google Scholar] [CrossRef] [PubMed]
- Shwartz, I.; Levy, M.; Ori, N.; Bar, M. Hormones in tomato leaf development. Dev. Biol. 2016, 419, 132–142. [Google Scholar] [CrossRef] [PubMed]
- Shinjiro; Yamaguchi, Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 2008, 59, 225–251. [CrossRef] [PubMed]
- Ren, X.J.; Huang, W.D.; Li, W.Z.; Yu, D.Q. Tobacco transcription factor WRKY4 is a modulator of leaf development and disease resistance. Biol. Plant. 2010, 54, 684–690. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Poole, A.; Peacock, W.J.; Dennis, E.S. Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol. 1999, 119, 507–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helliwell, C.A.; Sullivan, J.A.; Mould, R.M.; Gray, J.C.; Peacock, W.J.; Dennis, E.S. A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. Cell Mol. Biol. 2001, 28, 201–208. [Google Scholar] [CrossRef]
- Park, J.J.; Jin, P.; Yoon, J.; Yang, J.I.; Jeong, H.J.; Ranathunge, K.; Schreiber, L.; Franke, R.; Lee, I.J.; An, G. Mutation in Wilted Dwarf and Lethal 1 (WDL1) causes abnormal cuticle formation and rapid water loss in rice. Plant Mol.Biol. 2010, 74, 91–103. [Google Scholar] [CrossRef]
- Bilsborough, G.D.; Runions, A.; Barkoulas, M.; Jenkins, H.W.; Hasson, A.; Galinha, C.; Laufs, P.; Hay, A.; Prusinkiewicz, P.; Tsiantis, M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. USA 2011, 108, 3424–3429. [Google Scholar] [CrossRef] [Green Version]
- Hay, A.; Tsiantis, M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 2006, 38, 942–947. [Google Scholar] [CrossRef]
- Bartel, B. AUXIN BIOSYNTHESIS. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 51–66. [Google Scholar] [CrossRef]
- Normanly, J.; Grisafi, P.; Bartel, F.B. Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 1997, 9, 1781–1790. [Google Scholar] [PubMed]
- Davies, R.T.; Goetz, D.H.; Lasswell, J.; Anderson, M.N.; Bartel, B. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 1999, 11, 365–376. [Google Scholar] [CrossRef] [Green Version]
- D’Ippolito, S.; Vankova, R.; Joosten, M.; Casalongue, C.A.; Fiol, D.F. Knocking down expression of the auxin-amidohydrolase IAR3 alters defense responses in Solanaceae family plants. Plant Sci. 2016, 253, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Yanai, O.; Shani, E.; Russ, D.; Ori, N. Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 2011, 68, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Shohat, H.; Cheriker, H.; Kilambi, H.V.; Eliaz, N.I.; Blum, S.; Amsellem, Z.; Tarkowska, D.; Aharoni, A.; Eshed, Y.; Weiss, D. Inhibition of gibberellin accumulation by water deficiency promotes fast and long-term ‘drought avoidance’ responses in tomato. New Phytol. 2021, 232, 1985–1998. [Google Scholar] [CrossRef]
- Liu, Z.H.; Chen, Y.; Wang, N.N.; Chen, Y.H.; Wei, N.; Lu, R.; Li, Y.; Li, X.B. A basic helix-loop-helix protein (GhFP1) promotes fibre elongation of cotton (Gossypium hirsutum) by modulating brassinosteroid biosynthesis and signalling. New Phytol. 2020, 225, 2439–2452. [Google Scholar] [CrossRef]
- Li, J.; Gong, J.; Zhang, L.C.; Shen, H.; Chen, G.P.; Xie, Q.L.; Hu, Z.L. Overexpression of SlPRE5, an atypical bHLH transcription factor, affects plant morphology and chlorophyll accumulation in tomato. J. Plant Physiol. 2022, 273, 11. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Bai, M.Y.; Wu, J.X.; Zhu, J.Y.; Wang, H.; Zhang, Z.G.; Wang, W.F.; Sun, Y.; Zhao, J.; Sun, X.H.; et al. Antagonistic HLH/bHLH Transcription Factors Mediate Brassinosteroid Regulation of Cell Elongation and Plant Development in Rice and Arabidopsis. Plant Cell 2009, 21, 3767–3780. [Google Scholar] [CrossRef] [Green Version]
- Bai, M.Y.; Fan, M.; Oh, E.; Wang, Z.Y. A Triple Helix-Loop-Helix/Basic Helix-Loop-Helix Cascade Controls Cell Elongation Downstream of Multiple Hormonal and Environmental Signaling Pathways in Arabidopsis. Plant Cell 2012, 24, 4917–4929. [Google Scholar] [CrossRef] [Green Version]
- Sakulsingharoj, C.; Inta, P.; Sukkasem, R.; Pongjaroenkit, S.; Chowpongpang, S.; Sangtong, V. Overexpression of OSB2 gene in transgenic rice up-regulated expression of structural genes in anthocyanin biosynthesis pathway. Thai J. Genet. 2014, 7. [Google Scholar]
- Zheng, K.; Wang, Y.; Na, Z.; Jia, Q.; Wang, S. Involvement of PACLOBUTRAZOL RESISTANCE6/KIDARI, an atypical bHLH transcription factor, in auxin responses in arabidopsis. Front. Plant Sci. 2017, 8, 1813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, K.J.; Wang, Y.T.; Wang, S.C. The non-DNA binding bHLH transcription factor Paclobutrazol Resistances are involved in the regulation of ABA and salt responses in Arabidopsis. Plant Physiol. Biochem. 2019, 139, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Yang, C.W.; Gao, S.; Zhang, W.; Li, L.; Kuai, B.K. Age-Triggered and Dark-Induced Leaf Senescence Require the bHLH Transcription Factors PIF3, 4, and 5. Mol. Plant. 2014, 7, 1776–1787. [Google Scholar] [CrossRef] [Green Version]
- Schuster, C.; Gaillochet, C.; Lohmann, J.U. Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 2015, 142, 3343–3350. [Google Scholar] [CrossRef] [Green Version]
- Feng, S.; Martinez, C.; Gusmaroli, G.; Yu, W.; Zhou, J.; Wang, F.; Chen, L.; Lu, Y.; Iglesias-Pedraz, J.M.; Kircher, S. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 2008, 451, 475–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 Interacts with DELLA Proteins in Regulating Sesquiterpene Synthase Gene Expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.H.; Israeli, A.; Ori, N.; Sun, T.P. The Interaction between DELLA and ARF/IAA Mediates Crosstalk between Gibberellin and Auxin Signaling to Control Fruit Initiation in Tomato. Plant Cell 2018, 30, 1710–1728. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.J.; Dou, L.L.; Guo, Y.N.; Wang, H.T.; Li, L.B.; Wang, C.C.; Ma, L.; Wei, H.L.; Yu, S.X. The WRKY transcription factor GhWRKY27 coordinates the senescence regulatory pathway in upland cotton (Gossypium hirsutum L.). BMC Plant Biol. 2019, 19, 14. [Google Scholar] [CrossRef] [Green Version]
- Niu, F.F.; Cui, X.; Zhao, P.Y.; Sun, M.T.; Yang, B.; Deyholos, M.K.; Li, Y.; Zhao, X.J.; Jiang, Y.Q. WRKY42 transcription factor positively regulates leaf senescence through modulating SA and ROS synthesis in Arabidopsis thaliana. Plant J. 2020, 104, 171–184. [Google Scholar] [CrossRef]
- Zentgraf, U.; Laun, T.; Miao, Y. The complex regulation of WRKY53 during leaf senescence of Arabidopsis thaliana. Eur. J. Cell Biol. 2010, 89, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.G.; Xiang, S.Y.; Chen, Y.L.; Li, D.B.; Yu, D.Q. Arabidopsis WRKY45 Interacts with the DELLA Protein RGL1 to Positively Regulate Age-Triggered Leaf Senescence. Mol. Plant. 2017, 10, 1174–1189. [Google Scholar] [CrossRef] [Green Version]
- Gan, S.; Amasino, R.M. Making Sense of Senescence (Molecular Genetic Regulation and Manipulation of Leaf Senescence). Plant Physiol. 1997, 113, 313–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Fukushige, H.; Hildebrand, D.F.; Gan, S. Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 2002, 128, 876–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, K.; MacKerness, S.A.; Page, T.; John, C.F.; Murphy, A.M.; Carr, J.P.; Buchanan-Wollaston, V. Salicylic acid has a role in regulating gene expression during leaf senescence. Plant J. Cell Mol. Biol. 2000, 23, 677–685. [Google Scholar] [CrossRef]
- Naz, A.A.; Raman, S.; Martinez, C.C.; Sinha, N.R.; Schmitz, G.; Theres, K. Trifoliate encodes an MYB transcription factor that modulates leaf and shoot architecture in tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 2401–2406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Liu, H.; Li, X.; Xiao, J.; Wang, S. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: Application in study of rice-bacterium interaction. Plant Methods 2012, 8, 2. [Google Scholar] [CrossRef] [Green Version]









| Gene ID | Pts/VF36 | Log2 (FC) | tf2/CR | Log2 (FC) | entire/AC | Log2 (FC) | Description |
|---|---|---|---|---|---|---|---|
| Solyc06g064820.3 | Downregulation | −2.30 | Downregulation | −3.52 | Downregulation | −5.58 | GDSL esterase/lipase At1g71691 |
| Solyc11g031960.2 | Downregulation | −1.36 | Downregulation | −1.79 | Downregulation | −1.80 | GDSL esterase/lipase At1g33811 |
| Solyc11g006250.2 | Downregulation | −1.71 | Downregulation | −3.50 | Downregulation | −3.63 | GDSL esterase/lipase At5g33370 |
| Solyc01g095450.4 | Downregulation | −1.94 | Downregulation | −3.42 | Downregulation | −5.72 | GDSL esterase/lipase At4g01130 |
| Solyc03g006830.3 | Downregulation | −1.89 | Downregulation | −2.37 | Downregulation | −2.50 | MADS-box transcription factor 2 |
| Solyc05g009270.4 | Downregulation | −3.40 | Downregulation | −3.84 | Downregulation | −4.03 | Fatty acid elongase 3-ketoacyl-CoA synthase |
| Solyc09g090500.3 | Downregulation | −1.42 | Downregulation | −1.91 | Downregulation | −2.10 | Cyclopropane-fatty-acyl-phospholipid synthase |
| Solyc06g050500.2 | Downregulation | −1.26 | Upregulation | 1.66 | Upregulation | 1.82 | Abscisic acid receptor PYL4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, G.; Ma, C.; Yu, S.; Zhang, X.; Jiang, J.; Liu, X. Transcriptome Analyses Reveal the Key Regulators of Tomato Compound Leaf Development. Horticulturae 2023, 9, 363. https://doi.org/10.3390/horticulturae9030363
Zhu G, Ma C, Yu S, Zhang X, Jiang J, Liu X. Transcriptome Analyses Reveal the Key Regulators of Tomato Compound Leaf Development. Horticulturae. 2023; 9(3):363. https://doi.org/10.3390/horticulturae9030363
Chicago/Turabian StyleZhu, Guangwu, Chongtian Ma, Shuimiao Yu, Xueying Zhang, Jing Jiang, and Xin Liu. 2023. "Transcriptome Analyses Reveal the Key Regulators of Tomato Compound Leaf Development" Horticulturae 9, no. 3: 363. https://doi.org/10.3390/horticulturae9030363
APA StyleZhu, G., Ma, C., Yu, S., Zhang, X., Jiang, J., & Liu, X. (2023). Transcriptome Analyses Reveal the Key Regulators of Tomato Compound Leaf Development. Horticulturae, 9(3), 363. https://doi.org/10.3390/horticulturae9030363





