Recent Approaches in Magnetic Nanoparticle-Based Biosensors of miRNA Detection
Abstract
:1. Introduction
2. MNPs
2.1. Synthesis
2.2. Coating/Stabilization and Functionalization Strategies
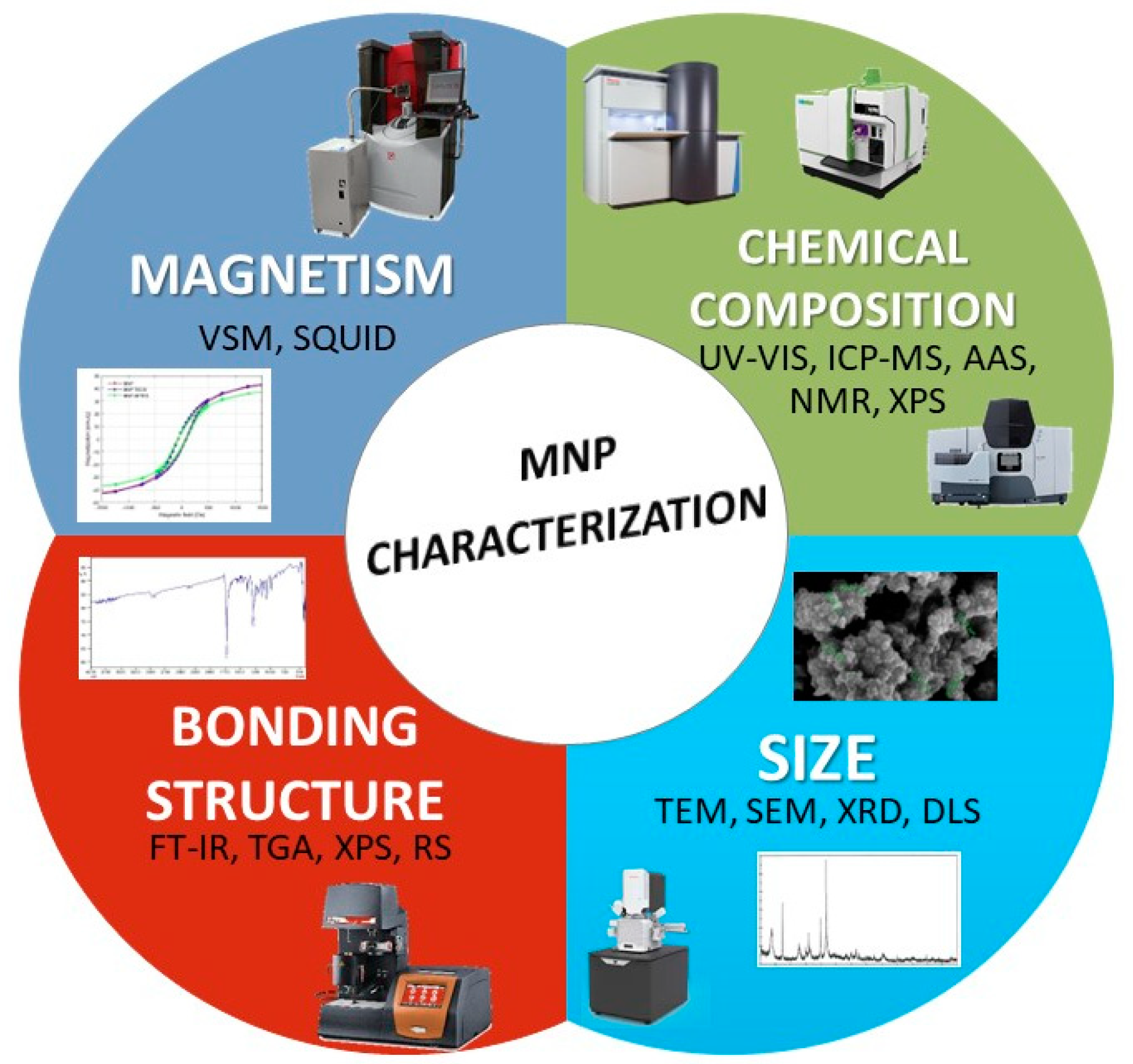
2.3. Characterization
2.4. Application Area
3. MNPs for Detection of miRNA
3.1. miRNA
3.2. Traditional Methods for miRNA Detection
3.2.1. Northern Blotting
3.2.2. RT-qPCR
3.2.3. Microarrays
3.3. MNP-Based Biosensors for miRNA Detection
3.3.1. Optical Biosensor Systems

3.3.2. Electrochemical Biosensor Systems
4. Conclusions and Future Perspective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MNPs | magnetic nanoparticles |
| miRNA | microRNA |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| PEG | polyethylene glycol |
| PVA | polyvinyl alcohol |
| PAA | polyacrylic acid |
| TEM | transmission electron microscopy |
| SEM | scanning electron microscopy |
| DLS | dynamic light scattering |
| XRF | X-ray fluorescence |
| XRD | X-ray diffraction |
| ICP-MS | inductively coupled plasma mass spectroscopy |
| AAS | atomic absorption spectrophotometry |
| XPS | X-ray photoelectron spectroscopy |
| FT-IR | Fourier transform infrared spectroscopy |
| XAS | X-ray absorption spectroscopy |
| TGA | thermogravimetric analysis |
| RS | Raman spectroscopy |
| VSM | magnetometer |
| SQUID | superconducting quantum interference device magnetometry |
| pri-miRNAs | primary transcript |
| pre-miRNA | precursor-miRNA |
| EDC | 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride |
| LNA | locked nucleic acid |
| cDNA | complementary DNA |
| PNA | peptide nucleic acids |
| SERS | surface-enhanced Raman spectroscopy |
| ssDNA | double-stranded DNA |
| LOD | limit of detection |
| SWV | square wave voltammetry |
| CV | cyclic voltammetry |
| DPV | differential pulse voltammetry |
References
- Tang, C.; He, Z.; Liu, H.; Xu, Y.; Huang, H.; Yang, G.; Xiao, Z.; Li, S.; Liu, H.; Deng, Y.; et al. Application of Magnetic Nanoparticles in Nucleic Acid Detection. J. Nanobiotechnol. 2020, 18, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akbarzadeh, A.; Samiei, M.; Davaran, S. Magnetic Nanoparticles: Preparation, Physical Properties, and Applications in Biomedicine. Nanoscale Res. Lett. 2012, 7, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gessner, I.; Fries, J.W.U.; Brune, V.; Mathur, S. Magnetic Nanoparticle-Based Amplification of MicroRNA Detection in Body Fluids for Early Disease Diagnosis. J. Mater. Chem. B 2021, 9, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Gloag, L.; Mehdipour, M.; Chen, D.; Tilley, R.D.; Gooding, J.J. Advances in the Application of Magnetic Nanoparticles for Sensing. Adv. Mater. 2019, 31, e1904385. [Google Scholar] [CrossRef]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, Biogenesis, and Their Evolving Role in Animal Development and Disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [Green Version]
- Harmanci, D.; Erkan, E.P.; Kocak, A.; Akdogan, G.G. Role of the MicroRNA-29 Family in Fibrotic Skin Diseases. Biomed. Rep. 2017, 6, 599–604. [Google Scholar] [CrossRef] [Green Version]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. MiRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [Green Version]
- Filipów, S.; Łaczmański, Ł. Blood Circulating MiRNAs as Cancer Biomarkers for Diagnosis and Surgical Treatment Response. Front. Genet. 2019, 10, 169. [Google Scholar] [CrossRef]
- Schade, A.; Delyagina, E.; Scharfenberg, D.; Skorska, A.; Lux, C.; David, R.; Steinhoff, G. Innovative Strategy for MicroRNA Delivery in Human Mesenchymal Stem Cells via Magnetic Nanoparticles. Int. J. Mol. Sci. 2013, 14, 10710–10726. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Xu, M.; Tian, X.; Cai, S.; Zeng, S. Research Advances in the Detection of MiRNA. J. Pharm. Anal. 2019, 9, 217–226. [Google Scholar] [CrossRef]
- Chaudhary, V.; Jangra, S.; Yadav, N.R. Nanotechnology Based Approaches for Detection and Delivery of MicroRNA in Healthcare and Crop Protection. J. Nanobiotechnol. 2018, 16, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Lin, Y.; Chi, Z.; Zhang, J.; Cai, F.; Zhu, Y.; Tang, D.; Lin, Q. Rolling Circle Amplification (RCA) -Based Biosensor System for the Fluorescent Detection of MiR-129-2-3p MiRNA. PeerJ 2022, 10, e14257. [Google Scholar] [CrossRef] [PubMed]
- Meng, T.; Zhao, D.; Ye, H.; Feng, Y.; Wang, H.; Zhang, Y. Construction of an Ultrasensitive Electrochemical Sensing Platform for MicroRNA-21 Based on Interface Impedance Spectroscopy. J. Colloid Interface Sci. 2020, 578, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Su, D.; Liu, J.; Saha, R.; Wang, J.-P. Magnetic Nanoparticles in Nanomedicine: A Review of Recent Advances. Nanotechnology 2019, 30, 502003. [Google Scholar] [CrossRef] [Green Version]
- Katz, E. Synthesis, Properties and Applications of Magnetic Nanoparticles and Nanowires—A Brief Introduction. Magnetochemistry 2019, 5, 61. [Google Scholar] [CrossRef] [Green Version]
- Rocha-Santos, T.A.P. Sensors and Biosensors Based on Magnetic Nanoparticles. TrAC Trends Anal. Chem. 2014, 62, 28–36. [Google Scholar] [CrossRef]
- Mohammed, L.; Gomaa, H.G.; Ragab, D.; Zhu, J. Magnetic Nanoparticles for Environmental and Biomedical Applications: A Review. Particuology 2017, 30, 1–14. [Google Scholar] [CrossRef]
- Mozharivskyj, Y. Magnetocaloric Effect and Magnetocaloric Materials. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Elsevier: Amsterdam, The Netherlands, 2016; ISBN 978-0-12-409547-2. [Google Scholar]
- Ali, A.; Shah, T.; Ullah, R.; Zhou, P.; Guo, M.; Ovais, M.; Tan, Z.; Rui, Y. Review on Recent Progress in Magnetic Nanoparticles: Synthesis, Characterization, and Diverse Applications. Front. Chem. 2021, 9, 629054. [Google Scholar] [CrossRef]
- Zhang, Z.; Wen, G. Synthesis and Characterization of Carbon-Encapsulated Magnetite, Martensite and Iron Nanoparticles by High-Energy Ball Milling Method. Mater. Charact. 2020, 167, 110502. [Google Scholar] [CrossRef]
- Zhao, Y.; Wen, G. Synthesis and Magnetic Properties of ε-Fe2O3 by Ball Milling and Post Annealing. J. Magn. Magn. Mater. 2020, 512, 167039. [Google Scholar] [CrossRef]
- Morone, M.V.; Dell’Annunziata, F.; Giugliano, R.; Chianese, A.; De Filippis, A.; Rinaldi, L.; Gambardella, U.; Franci, G.; Galdiero, M.; Morone, A. Pulsed Laser Ablation of Magnetic Nanoparticles as a Novel Antibacterial Strategy against Gram Positive Bacteria. Appl. Surf. Sci. Adv. 2022, 7, 100213. [Google Scholar] [CrossRef]
- Al-Alawy, A.F.; Al-Abodi, E.E.; Kadhim, R.M. Synthesis and Characterization of Magnetic Iron Oxide Nanoparticles by Co-Precipitation Method at Different Conditions. J. Eng. 2018, 24, 60–72. [Google Scholar] [CrossRef] [Green Version]
- De Mello, L.B.; Varanda, L.C.; Sigoli, F.A.; Mazali, I.O. Co-Precipitation Synthesis of (Zn-Mn)-Co-Doped Magnetite Nanoparticles and Their Application in Magnetic Hyperthermia. J. Alloys Compd. 2019, 779, 698–705. [Google Scholar] [CrossRef]
- Wang, G.; Cui, M.; Qiu, Y.; Miao, Y.; Ma, H.; Zhang, H.; Zhang, Y.; Liu, X.; Yi, J.; Peng, M.; et al. FeCO3 as a Novel Precursor for Controllable Synthesis of Monodisperse Iron Oxide Nanoparticles via Solution Thermal Decomposition. Micro Nano Lett. 2021, 16, 552–557. [Google Scholar] [CrossRef]
- Tomar, D.; Jeevanandam, P. Synthesis of Cobalt Ferrite Nanoparticles with Different Morphologies via Thermal Decomposition Approach and Studies on Their Magnetic Properties. J. Alloys Compd. 2020, 843, 155815. [Google Scholar] [CrossRef]
- Torres-Gómez, N.; Nava, O.; Argueta-Figueroa, L.; García-Contreras, R.; Baeza-Barrera, A.; Vilchis-Nestor, A.R. Shape Tuning of Magnetite Nanoparticles Obtained by Hydrothermal Synthesis: Effect of Temperature. J. Nanomater. 2019, 2019, e7921273. [Google Scholar] [CrossRef] [Green Version]
- Tovar, G.I.; Briceño, S.; Suarez, J.; Flores, S.; González, G. Biogenic Synthesis of Iron Oxide Nanoparticles Using Moringa oleifera and Chitosan and Its Evaluation on Corn Germination. Environ. Nanotechnol. Monit. Manag. 2020, 14, 100350. [Google Scholar] [CrossRef]
- Asab, G.; Zereffa, E.A.; Abdo Seghne, T. Synthesis of Silica-Coated Fe3O4 Nanoparticles by Microemulsion Method: Characterization and Evaluation of Antimicrobial Activity. Int. J. Biomater. 2020, 2020, e4783612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, P.; Chang, T.; Chen, W.-J.; Deng, J.; Li, S.-L.; Zuo, Y.-G.; Kang, L.; Yang, F.; Hostetter, M.; Volinsky, A.A. Temperature Effects on Magnetic Properties of Fe3O4 Nanoparticles Synthesized by the Sol-Gel Explosion-Assisted Method. J. Alloys Compd. 2019, 773, 605–611. [Google Scholar] [CrossRef]
- Moradnia, F.; Taghavi Fardood, S.; Ramazani, A.; Min, B.; Joo, S.W.; Varma, R.S. Magnetic Mg0.5Zn0.5FeMnO4 Nanoparticles: Green Sol-Gel Synthesis, Characterization, and Photocatalytic Applications. J. Clean. Prod. 2021, 288, 125632. [Google Scholar] [CrossRef]
- Benjamin, J.S. Dispersion Strengthened Superalloys by Mechanical Alloying. Metall. Trans. 1970, 1, 2943–2951. [Google Scholar] [CrossRef]
- Mohamed, A.E.-M.A.; Mohamed, M.A. Nanoparticles: Magnetism and Applications. In Magnetic Nanostructures; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–12. ISBN 978-3-030-16439-3. [Google Scholar]
- Niculescu, A.-G.; Chircov, C.; Grumezescu, A.M. Magnetite Nanoparticles: Synthesis Methods—A Comparative Review. Methods 2022, 199, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Svetlichnyi, V.A.; Shabalina, A.V.; Lapin, I.N.; Goncharova, D.A.; Kharlamova, T.S.; Stadnichenko, A.I. Comparative Study of Magnetite Nanoparticles Obtained by Pulsed Laser Ablation in Water and Air. Appl. Surf. Sci. 2019, 467–468, 402–410. [Google Scholar] [CrossRef]
- Verma, R.; Pathak, S.; Srivastava, A.K.; Prawer, S.; Tomljenovic-Hanic, S. ZnO Nanomaterials: Green Synthesis, Toxicity Evaluation and New Insights in Biomedical Applications. J. Alloys Compd. 2021, 876, 160175. [Google Scholar] [CrossRef]
- Duan, M.; Shapter, J.G.; Qi, W.; Yang, S.; Gao, G. Recent Progress in Magnetic Nanoparticles: Synthesis, Properties, and Applications. Nanotechnology 2018, 29, 452001. [Google Scholar] [CrossRef]
- Shabatina, T.I.; Vernaya, O.I.; Shabatin, V.P.; Melnikov, M.Y. Magnetic Nanoparticles for Biomedical Purposes: Modern Trends and Prospects. Magnetochemistry 2020, 6, 30. [Google Scholar] [CrossRef]
- Mascolo, M.C.; Pei, Y.; Ring, T.A. Room Temperature Co-Precipitation Synthesis of Magnetite Nanoparticles in a Large PH Window with Different Bases. Materials 2013, 6, 5549–5567. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Yang, H.C.; Yang, S.Y.; Horng, H.E.; Hung, J.C.; Chen, Y.C.; Hong, C.-Y. Preparation and Properties of Superparamagnetic Nanoparticles with Narrow Size Distribution and Biocompatible. J. Magn. Magn. Mater. 2004, 283, 210–214. [Google Scholar] [CrossRef]
- Dadfar, S.M.; Roemhild, K.; Drude, N.I.; von Stillfried, S.; Knüchel, R.; Kiessling, F.; Lammers, T. Iron Oxide Nanoparticles: Diagnostic, Therapeutic and Theranostic Applications. Adv. Drug Deliv. Rev. 2019, 138, 302–325. [Google Scholar] [CrossRef]
- Li, Z.; Sun, Q.; Gao, M. Preparation of Water-Soluble Magnetite Nanocrystals from Hydrated Ferric Salts in 2-Pyrrolidone: Mechanism Leading to Fe3O4. Angew. Chem. Int. Ed. 2005, 44, 123–126. [Google Scholar] [CrossRef]
- Farjadian, F.; Moradi, S.; Hosseini, M. Thin Chitosan Films Containing Super-Paramagnetic Nanoparticles with Contrasting Capability in Magnetic Resonance Imaging. J. Mater. Sci. Mater. Med. 2017, 28, 47. [Google Scholar] [CrossRef]
- Patsula, V.; Kosinová, L.; Lovrić, M.; Hamzić, L.F.; Rabyk, M.; Konefal, R.; Paruzel, A.; Šlouf, M.; Herynek, V.; Gajović, S.; et al. Superparamagnetic Fe3O4 Nanoparticles: Synthesis by Thermal Decomposition of Iron(III) Glucuronate and Application in Magnetic Resonance Imaging. ACS Appl. Mater. Interfaces 2016, 8, 7238–7247. [Google Scholar] [CrossRef] [PubMed]
- Reddy, L.H.; Arias, J.L.; Nicolas, J.; Couvreur, P. Magnetic Nanoparticles: Design and Characterization, Toxicity and Biocompatibility, Pharmaceutical and Biomedical Applications. Chem. Rev. 2012, 112, 5818–5878. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O.A.; Girgis, E.; Abdel-Mottaleb, M.M.S.A. Tailored Super Magnetic Nanoparticles Synthesized via Template Free Hydrothermal Technique. J. Magn. Magn. Mater. 2016, 397, 164–175. [Google Scholar] [CrossRef]
- Zhou, X.; Shi, Y.F.; Chen, Z.W. Facile Synthesis and Growth Mechanism of Uniform Fe3O4 Nanorod with Strong Magnetic Response. Adv. Mater. Res. 2013, 699, 616–619. [Google Scholar] [CrossRef]
- Sari, E.O.; Fadli, A.; Amri, A. The 3 Hours-Hydrothermal Synthesis of High Surface Area Superparamagnetic Fe3O4 Core-Shell Nanoparticles. J. Sains Mater. Indones. 2018, 19, 9–13. [Google Scholar] [CrossRef] [Green Version]
- Paul, B.K.; Moulik, S.P. Uses and Applications of Microemulsions. Curr. Sci. 2001, 80, 990–1001. [Google Scholar]
- Lu, A.-H.; Salabas, E.L.; Schüth, F. Magnetic Nanoparticles: Synthesis, Protection, Functionalization, and Application. Angew. Chem. Int. Ed. 2007, 46, 1222–1244. [Google Scholar] [CrossRef]
- Owens, G.J.; Singh, R.K.; Foroutan, F.; Alqaysi, M.; Han, C.-M.; Mahapatra, C.; Kim, H.-W.; Knowles, J.C. Sol–Gel Based Materials for Biomedical Applications. Prog. Mater. Sci. 2016, 77, 1–79. [Google Scholar] [CrossRef] [Green Version]
- Hasany, S.F.; Ahmed, I.; Jose, R.; Rehman, A. Systematic Review of the Preparation Techniques of Iron Oxide Magnetic Nanoparticles. Nanosci. Nanotechnol. 2012, 2, 148–158. [Google Scholar] [CrossRef] [Green Version]
- Itoh, H.; Sugimoto, T. Systematic Control of Size, Shape, Structure, and Magnetic Properties of Uniform Magnetite and Maghemite Particles. J. Colloid Interface Sci. 2003, 265, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Laibinis, P.E.; Hatton, T.A. Bilayer Surfactant Stabilized Magnetic Fluids: Synthesis and Interactions at Interfaces. Langmuir 1999, 15, 447–453. [Google Scholar] [CrossRef]
- Bohara, R.A.; Thorat, N.D.; Pawar, S.H. Role of Functionalization: Strategies to Explore Potential Nano-Bio Applications of Magnetic Nanoparticles. RSC Adv. 2016, 6, 43989–44012. [Google Scholar] [CrossRef]
- Farrell, D.; Majetich, S.A.; Wilcoxon, J.P. Preparation and Characterization of Monodisperse Fe Nanoparticles. J. Phys. Chem. B 2003, 107, 11022–11030. [Google Scholar] [CrossRef]
- Silva, S.M.; Tavallaie, R.; Sandiford, L.; Tilley, R.D.; Gooding, J.J. Gold Coated Magnetic Nanoparticles: From Preparation to Surface Modification for Analytical and Biomedical Applications. Chem. Commun. 2016, 52, 7528–7540. [Google Scholar] [CrossRef] [Green Version]
- Pham, X.-H.; Hahm, E.; Kim, H.-M.; Son, B.S.; Jo, A.; An, J.; Thi, T.A.T.; Nguyen, D.Q.; Jun, B.-H. Silica-Coated Magnetic Iron Oxide Nanoparticles Grafted onto Graphene Oxide for Protein Isolation. Nanomaterials 2020, 10, 117. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Hong, G.; Wang, H.; Welsher, K.; Tabakman, S.M.; Sherlock, S.P.; Robinson, J.T.; Liang, Y.; Dai, H. Graphite-Coated Magnetic Nanoparticle Microarray for Few-Cells Enrichment and Detection. ACS Nano 2012, 6, 1094–1101. [Google Scholar] [CrossRef]
- Monnier, C.A.; Burnand, D.; Rothen-Rutishauser, B.; Lattuada, M.; Petri-Fink, A. Magnetoliposomes: Opportunities and Challenges. Eur. J. Nanomed. 2014, 6, 201–215. [Google Scholar] [CrossRef]
- Arias, J.L.; López-Viota, M.; Ruiz, M.A.; López-Viota, J.; Delgado, A.V. Development of Carbonyl Iron/Ethylcellulose Core/Shell Nanoparticles for Biomedical Applications. Int. J. Pharm. 2007, 339, 237–245. [Google Scholar] [CrossRef]
- Goodhew, P.J.; Humphreys, J.; Beanland, R. Electron Microscopy and Analysis, 3rd ed.; CRC Press: Boca Raton, FL, USA, 2000; ISBN 978-0-7484-0968-6. [Google Scholar]
- Joy, D.C.; Pawley, J.B. High-Resolution Scanning Electron Microscopy. Ultramicroscopy 1992, 47, 80–100. [Google Scholar] [CrossRef]
- Kim, B.H.; Yang, J.; Lee, D.; Choi, B.K.; Hyeon, T.; Park, J. Liquid-Phase Transmission Electron Microscopy for Studying Colloidal Inorganic Nanoparticles. Adv. Mater. 2018, 30, 1703316. [Google Scholar] [CrossRef] [PubMed]
- Characterization of Magnetic Nanoparticle by Dynamic Light Scattering|SpringerLink. Available online: https://link.springer.com/article/10.1186/1556-276X-8-381 (accessed on 13 December 2022).
- Sandler, S.E.; Fellows, B.; Mefford, O.T. Best Practices for Characterization of Magnetic Nanoparticles for Biomedical Applications. Anal. Chem. 2019, 91, 14159–14169. [Google Scholar] [CrossRef] [Green Version]
- Mourdikoudis, S.; Pallares, R.M.; Thanh, N.T.K. Characterization Techniques for Nanoparticles: Comparison and Complementarity upon Studying Nanoparticle Properties. Nanoscale 2018, 10, 12871–12934. [Google Scholar] [CrossRef] [Green Version]
- Gunasekaran, D.; Gerchman, Y.; Vernick, S. Electrochemical Detection of Waterborne Bacteria Using Bi-Functional Magnetic Nanoparticle Conjugates. Biosensors 2022, 12, 36. [Google Scholar] [CrossRef]
- Kouhpanji, M.R.Z.; Stadler, B.J.H. A Guideline for Effectively Synthesizing and Characterizing Magnetic Nanoparticles for Advancing Nanobiotechnology: A Review. Sensors 2020, 20, 2554. [Google Scholar] [CrossRef]
- Kouhpanji, M.R.Z.; Stadler, B.J.H. Assessing the Reliability and Validity Ranges of Magnetic Characterization Methods. arXiv 2020, arXiv:2003.06911. [Google Scholar]
- Foner, S. Versatile and Sensitive Vibrating-Sample Magnetometer. Rev. Sci. Instrum. 1959, 30, 548–557. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Sun, J.; Liu, S.; Li, L.; Peng, X.; Ma, L.; Zhang, R. The Effect of Metal Ions on Endogenous Melanin Nanoparticles Used as Magnetic Resonance Imaging Contrast Agents. Biomater. Sci. 2020, 8, 379–390. [Google Scholar] [CrossRef]
- Salehipour, M.; Rezaei, S.; Mosafer, J.; Pakdin-Parizi, Z.; Motaharian, A.; Mogharabi-Manzari, M. Recent Advances in Polymer-Coated Iron Oxide Nanoparticles as Magnetic Resonance Imaging Contrast Agents. J. Nanopart. Res. 2021, 23, 48. [Google Scholar] [CrossRef]
- Gambhir, R.P.; Rohiwal, S.S.; Tiwari, A.P. Multifunctional Surface Functionalized Magnetic Iron Oxide Nanoparticles for Biomedical Applications: A Review. Appl. Surf. Sci. Adv. 2022, 11, 100303. [Google Scholar] [CrossRef]
- Ferreras, L.A.B.; Chan, S.Y.; Reina, S.V.; Dixon, J.E. Rapidly Transducing and Spatially Localized Magnetofection Using Peptide-Mediated Non-Viral Gene Delivery Based on Iron Oxide Nanoparticles. ACS Appl. Nano Mater. 2021, 4, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T. Cancer Hyperthermia Using Magnetic Nanoparticles. Biotechnol. J. 2011, 6, 1342–1347. [Google Scholar] [CrossRef] [PubMed]
- Jose, J.; Kumar, R.; Harilal, S.; Mathew, G.E.; Parambi, D.G.T.; Prabhu, A.; Uddin, S.; Aleya, L.; Kim, H.; Mathew, B. Magnetic Nanoparticles for Hyperthermia in Cancer Treatment: An Emerging Tool. Environ. Sci. Pollut. Res. 2020, 27, 19214–19225. [Google Scholar] [CrossRef] [PubMed]
- Fatima, H.; Charinpanitkul, T.; Kim, K.-S. Fundamentals to Apply Magnetic Nanoparticles for Hyperthermia Therapy. Nanomaterials 2021, 11, 1203. [Google Scholar] [CrossRef]
- Lee, E.A.; Yim, H.; Heo, J.; Kim, H.; Jung, G.; Hwang, N.S. Application of Magnetic Nanoparticle for Controlled Tissue Assembly and Tissue Engineering. Arch. Pharmacal. Res. 2014, 37, 120–128. [Google Scholar] [CrossRef]
- Van de Walle, A.; Perez, J.E.; Abou-Hassan, A.; Hémadi, M.; Luciani, N.; Wilhelm, C. Magnetic Nanoparticles in Regenerative Medicine: What of Their Fate and Impact in Stem Cells? Mater. Today Nano 2020, 11, 100084. [Google Scholar] [CrossRef]
- Jat, S.K.; Gandhi, H.A.; Bhattacharya, J.; Sharma, M.K. Magnetic Nanoparticles: An Emerging Nano-Based Tool to Fight against Viral Infections. Mater. Adv. 2021, 2, 4479–4496. [Google Scholar] [CrossRef]
- Weiss, C.; Carriere, M.; Fusco, L.; Capua, I.; Regla-Nava, J.A.; Pasquali, M.; Scott, J.A.; Vitale, F.; Unal, M.A.; Mattevi, C.; et al. Toward Nanotechnology-Enabled Approaches against the COVID-19 Pandemic. ACS Nano 2020, 14, 6383–6406. [Google Scholar] [CrossRef]
- Durmus, C.; Hanoglu, S.B.; Harmanci, D.; Moulahoum, H.; Tok, K.; Ghorbanizamani, F.; Sanli, S.; Zihnioglu, F.; Evran, S.; Cicek, C.; et al. Indiscriminate SARS-CoV-2 Multivariant Detection Using Magnetic Nanoparticle-Based Electrochemical Immunosensing. Talanta 2022, 243, 123356. [Google Scholar] [CrossRef]
- Hanoglu, S.B.; Man, E.; Harmanci, D.; Ruzgar, S.T.; Sanli, S.; Keles, N.A.; Ayden, A.; Tuna, B.G.; Duzgun, O.; Ozkan, O.F.; et al. Magnetic Nanoparticle-Based Electrochemical Sensing Platform Using Ferrocene-Labelled Peptide Nucleic Acid for the Early Diagnosis of Colorectal Cancer. Biosensors 2022, 12, 736. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Vaid, K.; Bansal, S.A.; Kim, K.-H. Nanomaterial-Based Immunosensors for Ultrasensitive Detection of Pesticides/Herbicides: Current Status and Perspectives. Biosens. Bioelectron. 2020, 165, 112382. [Google Scholar] [CrossRef] [PubMed]
- Nadar, S.S.; Kelkar, R.K.; Pise, P.V.; Patil, N.P.; Patil, S.P.; Chaubal-Durve, N.S.; Bhange, V.P.; Tiwari, M.S.; Patil, P.D. The Untapped Potential of Magnetic Nanoparticles for Forensic Investigations: A Comprehensive Review. Talanta 2021, 230, 122297. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Asano, Y.; Ui-Tei, K. Modulation of MicroRNA Processing by Dicer via Its Associated DsRNA Binding Proteins. Non-Coding RNA 2021, 7, 57. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Mishra, R.; Kumar, A.; Ingle, H.; Kumar, H. The Interplay Between Viral-Derived MiRNAs and Host Immunity During Infection. Front. Immunol. 2020, 10, 3079. [Google Scholar] [CrossRef] [Green Version]
- Wong-Bajracharya, J.; Singan, V.R.; Monti, R.; Plett, K.L.; Ng, V.; Grigoriev, I.V.; Martin, F.M.; Anderson, I.C.; Plett, J.M. The Ectomycorrhizal Fungus Pisolithus Microcarpus Encodes a MicroRNA Involved in Cross-Kingdom Gene Silencing during Symbiosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2103527119. [Google Scholar] [CrossRef]
- Millar, A.A. The Function of MiRNAs in Plants. Plants 2020, 9, 198. [Google Scholar] [CrossRef] [Green Version]
- The Roles of MicroRNAs in Mouse Development|Nature Reviews Genetics. Available online: https://www.nature.com/articles/s41576-020-00309-5 (accessed on 21 November 2022).
- Lee, L.W.; Zhang, S.; Etheridge, A.; Ma, L.; Martin, D.; Galas, D.; Wang, K. Complexity of the MicroRNA Repertoire Revealed by Next-Generation Sequencing. RNA 2010, 16, 2170–2180. [Google Scholar] [CrossRef] [Green Version]
- Galluzzo, A.; Gallo, S.; Pardini, B.; Birolo, G.; Fariselli, P.; Boretto, P.; Vitacolonna, A.; Peraldo-Neia, C.; Spilinga, M.; Volpe, A.; et al. Identification of Novel Circulating MicroRNAs in Advanced Heart Failure by Next-Generation Sequencing. ESC Heart Fail. 2021, 8, 2907–2919. [Google Scholar] [CrossRef]
- Raza, S.H.A.; Abdelnour, S.A.; Dhshan, A.I.M.; Hassanin, A.A.; Noreldin, A.E.; Albadrani, G.M.; Abdel-Daim, M.M.; Cheng, G.; Zan, L. Potential Role of Specific MicroRNAs in the Regulation of Thermal Stress Response in Livestock. J. Therm. Biol. 2021, 96, 102859. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, F.; Xia, H.; Yao, S. MicroRNA-155: Regulation of Immune Cells in Sepsis. Mediat. Inflamm. 2021, 2021, 8874854. [Google Scholar] [CrossRef] [PubMed]
- Tabaei, S.; Tabaee, S.S. Implications for MicroRNA Involvement in the Prognosis and Treatment of Atherosclerosis. Mol. Cell. Biochem. 2021, 476, 1327–1336. [Google Scholar] [CrossRef]
- Borbet, T.C.; Hines, M.J.; Koralov, S.B. MicroRNA Regulation of B Cell Receptor Signaling. Immunol. Rev. 2021, 304, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Yang, X.; Liu, M.; Zhang, Z.; Xing, E. Roles of MiRNA Dysregulation in the Pathogenesis of Multiple Myeloma. Cancer Gene Ther. 2021, 28, 1256–1268. [Google Scholar] [CrossRef]
- Siddika, T.; Heinemann, I.U. Bringing MicroRNAs to Light: Methods for MicroRNA Quantification and Visualization in Live Cells. Front. Bioeng. Biotechnol. 2021, 8, 619583. [Google Scholar] [CrossRef]
- Anelli, L.; Zagaria, A.; Specchia, G.; Musto, P.; Albano, F. Dysregulation of MiRNA in Leukemia: Exploiting MiRNA Expression Profiles as Biomarkers. Int. J. Mol. Sci. 2021, 22, 7156. [Google Scholar] [CrossRef]
- Nies, Y.H.; Najib, N.H.M.; Lim, W.L.; Kamaruzzaman, M.A.; Yahaya, M.F.; Teoh, S.L. MicroRNA Dysregulation in Parkinson’s Disease: A Narrative Review. Front. Neurosci. 2021, 15, 660379. [Google Scholar] [CrossRef]
- Ahmed, U.; Ashfaq, U.A.; Qasim, M.; Ahmad, I.; Ahmad, H.U.; Tariq, M.; Masoud, M.S.; Khaliq, S. Dysregulation of Circulating MiRNAs Promotes the Pathogenesis of Diabetes-Induced Cardiomyopathy. PLoS ONE 2021, 16, e0250773. [Google Scholar] [CrossRef]
- Liu, H.; Ma, L.; Wang, L.; Yang, Y. MicroRNA-937 Is Overexpressed and Predicts Poor Prognosis in Patients with Colon Cancer. Diagn. Pathol. 2019, 14, 136. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Y. Upregulation of MicroRNA-937 Predicts a Poor Prognosis and Promotes Hepatocellular Carcinoma Cell Proliferation, Migration, and Invasion. Mol. Biotechnol. 2022, 64, 33–41. [Google Scholar] [CrossRef]
- Shen, Q.; Xiong, P.; Yang, D.; Chen, L. Downregulated MicroRNA-149-3p Triggers Malignant Development and Predicts Worse Prognosis in Oral Squamous Cell Carcinoma. Arch. Oral Biol. 2022, 134, 105336. [Google Scholar] [CrossRef] [PubMed]
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA Biomarkers for Infectious Diseases: From Basic Research to Biosensing. Front. Microbiol. 2020, 11, 1197. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Zhu, X.; Jiang, C. MicroRNA as an Early Diagnostic Biomarker for Contrast-Induced Acute Kidney Injury. Drug Chem. Toxicol. 2022, 45, 1552–1557. [Google Scholar] [CrossRef] [PubMed]
- Vykoukal, J.; Fahrmann, J.F.; Patel, N.; Shimizu, M.; Ostrin, E.J.; Dennison, J.B.; Ivan, C.; Goodman, G.E.; Thornquist, M.D.; Barnett, M.J.; et al. Contributions of Circulating MicroRNAs for Early Detection of Lung Cancer. Cancers 2022, 14, 4221. [Google Scholar] [CrossRef] [PubMed]
- Iguchi, H.; Kosaka, N.; Ochiya, T. Secretory MicroRNAs as a Versatile Communication Tool. Commun. Integr. Biol. 2010, 3, 478–481. [Google Scholar] [CrossRef]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Groot, M.; Lee, H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020, 9, 1044. [Google Scholar] [CrossRef]
- Durur, D.Y.; Tastan, B.; Tufekci, K.U.; Olcum, M.; Uzuner, H.; Karakülah, G.; Yener, G.; Genc, S. Alteration of MiRNAs in Small Neuron-Derived Extracellular Vesicles of Alzheimer’s Disease Patients and the Effect of Extracellular Vesicles on Microglial Immune Responses. J. Mol. Neurosci. 2022, 72, 1182–1194. [Google Scholar] [CrossRef]
- Panvongsa, W.; Pegtel, D.M.; Voortman, J. More than a Bubble: Extracellular Vesicle MicroRNAs in Head and Neck Squamous Cell Carcinoma. Cancers 2022, 14, 1160. [Google Scholar] [CrossRef]
- Aryani, A.; Denecke, B. In Vitro Application of Ribonucleases: Comparison of the Effects on MRNA and MiRNA Stability. BMC Res. Notes 2015, 8, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Chen, D.; Wang, Q.; Tian, H.; Tan, M.; Peng, D.; Tan, Y.; Zhu, J.; Liang, W.; Zhang, L. MRNA and MicroRNA Stability Validation of Blood Samples under Different Environmental Conditions. Forensic Sci. Int. Genet. 2021, 55, 102567. [Google Scholar] [CrossRef] [PubMed]
- Shihana, F.; Wong, W.K.; Joglekar, M.V.; Mohamed, F.; Gawarammana, I.B.; Isbister, G.K.; Hardikar, A.A.; Seth, D.; Buckley, N.A. Urinary MicroRNAs as Non-Invasive Biomarkers for Toxic Acute Kidney Injury in Humans. Sci. Rep. 2021, 11, 9165. [Google Scholar] [CrossRef] [PubMed]
- Ban, E.; Kwon, H.; Seo, H.S.; Yoo, Y.S.; Song, E.J. Screening of MiRNAs in Plasma as a Diagnostic Biomarker for Cardiac Disease Based on Optimization of Extraction and QRT-PCR Condition Assay through Amplification Efficiency. BMC Biotechnol. 2021, 21, 50. [Google Scholar] [CrossRef]
- Demiray, A.; Sarı, T.; Çalışkan, A.; Nar, R.; Aksoy, L.; Akbubak, İ.H. Serum MicroRNA Signature Is Capable of Predictive and Prognostic Factor for SARS-COV-2 Virulence. Turk. J. Biochem. 2021, 46, 245–253. [Google Scholar] [CrossRef]
- Kopkova, A.; Sana, J.; Machackova, T.; Vecera, M.; Radova, L.; Trachtova, K.; Vybihal, V.; Smrcka, M.; Kazda, T.; Slaby, O.; et al. Cerebrospinal Fluid MicroRNA Signatures as Diagnostic Biomarkers in Brain Tumors. Cancers 2019, 11, 1546. [Google Scholar] [CrossRef] [Green Version]
- Bendifallah, S.; Suisse, S.; Puchar, A.; Delbos, L.; Poilblanc, M.; Descamps, P.; Golfier, F.; Jornea, L.; Bouteiller, D.; Touboul, C.; et al. Salivary MicroRNA Signature for Diagnosis of Endometriosis. J. Clin. Med. 2022, 11, 612. [Google Scholar] [CrossRef]
- Sabato, C.; Noviello, T.M.R.; Covre, A.; Coral, S.; Caruso, F.P.; Besharat, Z.M.; Splendiani, E.; Masuelli, L.; Battistelli, C.; Vacca, A.; et al. A Novel MicroRNA Signature for the Detection of Melanoma by Liquid Biopsy. J. Transl. Med. 2022, 20, 469. [Google Scholar] [CrossRef]
- Singh, P.; Singh, A.; Gupta, N.; Raja, K.D.; Singh, P.; Agarwal, S.; Sharma, A. Non-Invasive Diagnostic Potential of MicroRNA-203 in Liquid Biopsy of Urothelial Carcinoma of Bladder. Mol. Cell. Biochem. 2022, 477, 2173–2182. [Google Scholar] [CrossRef]
- Hussein, Z. Leading to Intention: The Role of Attitude in Relation to Technology Acceptance Model in E-Learning. Procedia Comput. Sci. 2017, 105, 159–164. [Google Scholar] [CrossRef]
- Huang, F.; Xiang, Y.; Li, T.; Huang, Y.; Wang, J.; Zhang, H.-M.; Li, H.-H.; Dai, Z.-T.; Li, J.-P.; Li, H.; et al. Metformin and MiR-365 Synergistically Promote the Apoptosis of Gastric Cancer Cells via MiR-365-PTEN-AMPK Axis. Pathol. Res. Pract. 2022, 230, 153740. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, Z. MiRNA-139-3p inhibits malignant progression in urothelial carcinoma of the bladder via targeting KIF18B and inactivating Wnt/beta-catenin pathway. Pharm. Genom. 2022, 33, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Han, B.; Zhu, X.; Wang, Y.; Ji, H.; Weng, J.; Liu, Y. Dysfunctional MicroRNA-144-3p/ZBTB20/ERK/CREB1 Signalling Pathway Is Associated with MK-801-Induced Schizophrenia-like Abnormalities. Brain Res. 2023, 1798, 148153. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xiao, C.; Sun, Q. MiRNA-375 Inhibits Retinoblastoma Progression through Targeting ERBB2 and Inhibiting MAPK1/MAPK3 Signalling Pathway. Cutan. Ocul. Toxicol. 2021, 41, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kehl, T.; Backes, C.; Kern, F.; Fehlmann, T.; Ludwig, N.; Meese, E.; Lenhof, H.-P.; Keller, A. About MiRNAs, MiRNA Seeds, Target Genes and Target Pathways. Oncotarget 2017, 8, 107167. [Google Scholar] [CrossRef] [Green Version]
- Špringer, T.; Krejčík, Z.; Homola, J. Detecting Attomolar Concentrations of MicroRNA Related to Myelodysplastic Syndromes in Blood Plasma Using a Novel Sandwich Assay with Nanoparticle Release. Biosens. Bioelectron. 2021, 194, 113613. [Google Scholar] [CrossRef]
- Ouyang, T.; Liu, Z.; Han, Z.; Ge, Q. MicroRNA Detection Specificity: Recent Advances and Future Perspective. Anal. Chem. 2019, 91, 3179–3186. [Google Scholar] [CrossRef] [Green Version]
- Streit, S.; Michalski, C.W.; Erkan, M.; Kleeff, J.; Friess, H. Northern Blot Analysis for Detection and Quantification of RNA in Pancreatic Cancer Cells and Tissues. Nat. Protoc. 2009, 4, 37–43. [Google Scholar] [CrossRef]
- He, S.L.; Green, R. Chapter Three—Northern Blotting. In Methods in Enzymology; Laboratory Methods in Enzymology: RNA; Lorsch, J., Ed.; Academic Press: Cambridge, MA, USA, 2013; Volume 530, pp. 75–87. [Google Scholar]
- Koscianska, E.; Starega-Roslan, J.; Sznajder, L.J.; Olejniczak, M.; Galka-Marciniak, P.; Krzyzosiak, W.J. Northern Blotting Analysis of MicroRNAs, Their Precursors and RNA Interference Triggers. BMC Mol. Biol. 2011, 12, 14. [Google Scholar] [CrossRef] [Green Version]
- Pall, G.S.; Codony-Servat, C.; Byrne, J.; Ritchie, L.; Hamilton, A. Carbodiimide-Mediated Cross-Linking of RNA to Nylon Membranes Improves the Detection of SiRNA, MiRNA and PiRNA by Northern Blot. Nucleic Acids Res. 2007, 35, e60. [Google Scholar] [CrossRef] [Green Version]
- Várallyay, E.; Burgyán, J.; Havelda, Z. MicroRNA Detection by Northern Blotting Using Locked Nucleic Acid Probes. Nat. Protoc. 2008, 3, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Li, Z.; Moore, P.S.; Monaghan, A.P.; Chang, Y.; Nichols, M.; John, B. A Sensitive Non-Radioactive Northern Blot Method to Detect Small RNAs. Nucleic Acids Res. 2010, 38, e98. [Google Scholar] [CrossRef] [PubMed]
- McClure, L.V.; Lin, Y.-T.; Sullivan, C.S. Detection of Viral MicroRNAs by Northern Blot Analysis. Methods Mol. Biol. 2011, 721, 153–171. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, W.; Gull, B.; Baby, J.; Mustafa, F. A Comprehensive Analysis of Northern versus Liquid Hybridization Assays for MRNAs, Small RNAs, and MiRNAs Using a Non-Radiolabeled Approach. Curr. Issues Mol. Biol. 2021, 43, 457–484. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tan, R.; Wong, L.; Fekete, R.; Halsey, J. Quantitation of MicroRNAs by Real-Time RT-QPCR. PCR Protoc. 2011, 687, 113–134. [Google Scholar] [CrossRef]
- Dellett, M.; Simpson, D.A. Considerations for Optimization of MicroRNA PCR Assays for Molecular Diagnosis. Expert Rev. Mol. Diagn. 2016, 16, 407–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotlarek, M.; Kubiak, A.; Jażdżewski, K.; Wójcicka, A. MicroRNA Analysis Using the Quantitative Real-Time PCR Reaction. Methods Mol. Biol. 2018, 1823, 69–85. [Google Scholar] [CrossRef]
- Busk, P.K. A Tool for Design of Primers for MicroRNA-Specific Quantitative RT-QPCR. BMC Bioinform. 2014, 15, 29. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.-T.; Hsieh, Y.-S.; Feng, C.-T.; Chen, Y.-T.; Yang, P.E.; Chen, W.-M. MiPrimer: An Empirical-Based QPCR Primer Design Method for Small Noncoding MicroRNA. RNA 2018, 24, 304–312. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-Time Quantification of MicroRNAs by Stem-Loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Yang, L.; Wang, S.; Tang, L.; Liu, B.; Ye, W.; Wang, L.; Wang, Z.; Zhou, M.; Chen, B. Universal Stem-Loop Primer Method for Screening and Quantification of MicroRNA. PLoS ONE 2014, 9, e115293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, F.; Ni, N.; Wang, H.; Zeng, Z.; Zhao, P.; Shi, D.; Xia, Y.; Chen, C.; Hu, D.A.; Qin, K.H.; et al. OUHP: An Optimized Universal Hairpin Primer System for Cost-Effective and High-Throughput RT-QPCR-Based Quantification of MicroRNA (MiRNA) Expression. Nucleic Acids Res. 2022, 50, e22. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Murashov, A.K.; Stellwag, E.J.; Zhang, B. Using Quantitative Real-Time PCR to Detect MicroRNA Expression Profile During Embryonic Stem Cell Differentiation. In RNAi and Small Regulatory RNAs in Stem Cells; Humana Press: New York, NY, USA, 2017; pp. 255–265. [Google Scholar]
- Lee, H.; He, X.; Le, T.; Carnino, J.M.; Jin, Y. Single-Step RT-QPCR for Detection of Extracellular Vesicle MicroRNAs in Vivo: A Time- and Cost-Effective Method. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 318, L742–L749. [Google Scholar] [CrossRef] [PubMed]
- Voss, G.; Edsjö, A.; Bjartell, A.; Ceder, Y. Quantification of MicroRNA Editing Using Two-Tailed RT-QPCR for Improved Biomarker Discovery. RNA 2021, 27, 1412–1424. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.; Follo, M.; Haenel, D.; Mauler, M.; Stallmann, D.; Tewari, M.; Duerschmied, D.; Peter, K.; Bode, C.; Ahrens, I.; et al. Droplet Digital PCR as a Novel Detection Method for Quantifying MicroRNAs in Acute Myocardial Infarction. Int. J. Cardiol. 2018, 257, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ding, Q.; Plant, P.; Basheer, M.; Yang, C.; Tawedrous, E.; Krizova, A.; Boulos, C.; Farag, M.; Cheng, Y.; et al. Droplet Digital PCR Improves Urinary Exosomal MiRNA Detection Compared to Real-Time PCR. Clin. Biochem. 2019, 67, 54–59. [Google Scholar] [CrossRef]
- Laprovitera, N.; Riefolo, M.; Porcellini, E.; Durante, G.; Garajova, I.; Vasuri, F.; Aigelsreiter, A.; Dandachi, N.; Benvenuto, G.; Agostinis, F.; et al. MicroRNA Expression Profiling with a Droplet Digital PCR Assay Enables Molecular Diagnosis and Prognosis of Cancers of Unknown Primary. Mol. Oncol. 2021, 15, 2732–2751. [Google Scholar] [CrossRef]
- Yan, N.; Lu, Y.; Sun, H.; Tao, D.; Zhang, S.; Liu, W.; Ma, Y. A Microarray for MicroRNA Profiling in Mouse Testis Tissues. Reproduction 2007, 134, 73–79. [Google Scholar] [CrossRef]
- Castoldi, M.; Schmidt, S.; Benes, V.; Hentze, M.W.; Muckenthaler, M.U. MiChip: An Array-Based Method for MicroRNA Expression Profiling Using Locked Nucleic Acid Capture Probes. Nat. Protoc. 2008, 3, 321–329. [Google Scholar] [CrossRef]
- Krichevsky, A.M.; King, K.S.; Donahue, C.P.; Khrapko, K.; Kosik, K.S. A MicroRNA Array Reveals Extensive Regulation of MicroRNAs during Brain Development. RNA 2003, 9, 1274–1281. [Google Scholar] [CrossRef] [Green Version]
- Liang, R.-Q.; Li, W.; Li, Y.; Tan, C.; Li, J.-X.; Jin, Y.-X.; Ruan, K.-C. An Oligonucleotide Microarray for MicroRNA Expression Analysis Based on Labeling RNA with Quantum Dot and Nanogold Probe. Nucleic Acids Res. 2005, 33, e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.-G.; Calin, G.A.; Volinia, S.; Croce, C.M. MicroRNA Expression Profiling Using Microarrays. Nat. Protoc. 2008, 3, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Forte, G.; Ventimiglia, G.; Pesaturo, M.; Petralia, S. A Highly Sensitive PNA-Microarray System for MiRNA122 Recognition. Biotechnol. J. 2022, 17, 2100587. [Google Scholar] [CrossRef]
- Mastriani, E.; Zhai, R.; Zhu, S. Microarray-Based MicroRNA Expression Data Analysis with Bioconductor. In Transcriptome Data Analysis: Methods and Protocols; Methods in Molecular, Biology; Wang, Y., Sun, M., Eds.; Springer: New York, NY, USA, 2018; pp. 127–138. ISBN 978-1-4939-7710-9. [Google Scholar]
- Witwer, K.W. Data Submission and Quality in Microarray-Based MicroRNA Profiling. Clin. Chem. 2013, 59, 392–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Lin, Y.; Jin, Y.; Zheng, C. RETRACTED: Microarray Analysis of MicroRNA Expression in Liver Cancer Tissues and Normal Control. Gene 2013, 523, 158–160. [Google Scholar] [CrossRef]
- Zhu, P.; Pan, J.; Cai, Q.Q.; Zhang, F.; Peng, M.; Fan, X.L.; Ji, H.; Dong, Y.W.; Wu, X.Z.; Wu, L.H. MicroRNA Profile as Potential Molecular Signature for Attention Deficit Hyperactivity Disorder in Children. Biomarkers 2022, 27, 230–239. [Google Scholar] [CrossRef]
- Prahm, K.P.; Høgdall, C.K.; Karlsen, M.A.; Christensen, I.J.; Novotny, G.W.; Høgdall, E. MicroRNA Characteristics in Epithelial Ovarian Cancer. PLoS ONE 2021, 16, e0252401. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.-F.; Zhang, Q.; Mo, X.-B.; Lin, J.; Wu, Y.-L.; Lu, X.; He, P.; Wu, J.; Guo, Y.-F.; Wang, M.-J.; et al. Identification of Novel Rheumatoid Arthritis-Associated MiRNA-204-5p from Plasma Exosomes. Exp. Mol. Med. 2022, 54, 334–345. [Google Scholar] [CrossRef]
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J. Optical Biosensors: An Exhaustive and Comprehensive Review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef]
- Kaur, B.; Kumar, S.; Kaushik, B.K. Recent Advancements in Optical Biosensors for Cancer Detection. Biosens. Bioelectron. 2022, 197, 113805. [Google Scholar] [CrossRef] [PubMed]
- McCracken, K.E.; Yoon, J.-Y. Recent Approaches for Optical Smartphone Sensing in Resource-Limited Settings: A Brief Review. Anal. Methods 2016, 8, 6591–6601. [Google Scholar] [CrossRef]
- Aydindogan, E.; Ceylan, A.E.; Timur, S. Paper-Based Colorimetric Spot Test Utilizing Smartphone Sensing for Detection of Biomarkers. Talanta 2020, 208, 120446. [Google Scholar] [CrossRef]
- Song, Y.; Wei, W.; Qu, X. Colorimetric Biosensing Using Smart Materials. Adv. Mater. 2011, 23, 4215–4236. [Google Scholar] [CrossRef] [PubMed]
- Hosseinzadeh, E.; Ravan, H.; Mohammadi, A.; Mohammad-rezaei, R.; Norouzi, A.; Hosseinzadeh, H. Target-Triggered Three-Way Junction in Conjugation with Catalytic Concatemers-Functionalized Nanocomposites Provides a Highly Sensitive Colorimetric Method for MiR-21 Detection. Biosens. Bioelectron. 2018, 117, 567–574. [Google Scholar] [CrossRef]
- Tang, S.; Li, Y.; Zhu, A.; Yao, Y.; Sun, J.; Zheng, F.; Lin, Z.; Shen, W. A Triple-Amplification Strategy Based on the Formation of Peroxidase-like Two-Dimensional DNA/Fe3O4 Networks Initiated by the Hybridization Chain Reaction for Highly Sensitive Detection of MicroRNA. Chem. Commun. 2019, 55, 8386–8389. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Z.-J.; Cao, H.-X.; Liang, G.-X. Ultrasensitive Colorimetric MiRNA Detection Based on Magnetic 3D DNA Walker and Unmodified AuNPs. Sens. Actuators B Chem. 2021, 337, 129813. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, C.; Tang, L.; Zhang, Y.; Zhang, G.-J. Magnetic-Enhanced Fluorescence Sensing of Tumor MiRNA by Combination of MNPs@PDA with Duplex Specific Nuclease. RSC Adv. 2021, 11, 2968–2975. [Google Scholar] [CrossRef]
- Tian, H.; Yuan, C.; Liu, Y.; Li, Z.; Xia, K.; Li, M.; Xie, F.; Chen, Q.; Chen, M.; Fu, W.; et al. A Novel Quantification Platform for Point-of-Care Testing of Circulating MicroRNAs Based on Allosteric Spherical Nanoprobe. J. Nanobiotechnol. 2020, 18, 158. [Google Scholar] [CrossRef]
- Fan, Y.; Liu, Y.; Zhou, Q.; Du, H.; Zhao, X.; Ye, F.; Zhao, H. Catalytic Hairpin Assembly Indirectly Covalent on Fe3O4@C Nanoparticles with Signal Amplification for Intracellular Detection of MiRNA. Talanta 2021, 223, 121675. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, J.; Zeng, J.; Yang, Z.; Ran, F.; Wu, L.; Yang, G.; Mei, Q.; Wang, X.; Chen, Q. Determination of MiRNA Derived from Exosomes of Prostate Cancer via Toehold-Aided Cyclic Amplification Combined with HRP Enzyme Catalysis and Magnetic Nanoparticles. Anal. Biochem. 2021, 630, 114336. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, J.; He, N.; Zhang, M.; Wu, L.; Chen, X.; Zhu, J.; Ran, F.; Chen, Q.; Zhang, H. CRISPR/Cas12a Coupling with Magnetic Nanoparticles and Cascaded Strand Displacement Reaction for Ultrasensitive Fluorescence Determination of Exosomal MiR-21. Molecules 2022, 27, 5338. [Google Scholar] [CrossRef] [PubMed]
- Shao, B.; Xiao, Z. Recent Achievements in Exosomal Biomarkers Detection by Nanomaterials-Based Optical Biosensors—A Review. Anal. Chim. Acta 2020, 1114, 74–84. [Google Scholar] [CrossRef]
- Wang, F.; Liu, Y.; Fu, C.; Li, N.; Du, M.; Zhang, L.; Ge, S.; Yu, J. Paper-Based Bipolar Electrode Electrochemiluminescence Platform for Detection of Multiple MiRNAs. Anal. Chem. 2021, 93, 1702–1708. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhang, Y.; Wang, P.; Nie, Y.; Ma, Q. Luminous MoS2 Nanosheet-Based Electrochemiluminescence Biosensor with Biomimetic Vesicle for MiRNA-210 Detection. Talanta 2022, 237, 122969. [Google Scholar] [CrossRef]
- Shao, H.; Lu, J.; Zhang, Q.; Hu, Y.; Wang, S.; Guo, Z. Ruthenium-Based Metal Organic Framework (Ru-MOF)-Derived Novel Faraday-Cage Electrochemiluminescence Biosensor for Ultrasensitive Detection of MiRNA-141. Sens. Actuators B Chem. 2018, 268, 39–46. [Google Scholar] [CrossRef]
- Üzek, R.; Sari, E.; Merkoçi, A. Optical-Based (Bio) Sensing Systems Using Magnetic Nanoparticles. Magnetochemistry 2019, 5, 59. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Zeng, Z.; Cao, Y.; Zhang, X.; Wu, C.; Luo, X. Ultrasenstive SERS Biosensor Based on Zn2+ from ZnO Nanoparticle Assisted DNA Enzyme Amplification for Detection of MiRNA. Anal. Chim. Acta 2022, 1228, 340340. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, X.; Li, P.; Lin, X.; Wang, J.; Hu, Z.; Zhang, P.; Chen, D.; Cai, H.; Niessner, R.; et al. Ultrasensitive and Simultaneous SERS Detection of Multiplex MicroRNA Using Fractal Gold Nanotags for Early Diagnosis and Prognosis of Hepatocellular Carcinoma. Anal. Chem. 2021, 93, 8799–8809. [Google Scholar] [CrossRef]
- Zhang, H.; Fu, C.; Wu, S.; Shen, Y.; Zhou, C.; Neng, J.; Yi, Y.; Jin, Y.; Zhu, Y. Magnetic-Capture-Based SERS Detection of Multiple Serum MicroRNA Biomarkers for Cancer Diagnosis. Anal. Methods 2019, 11, 783–793. [Google Scholar] [CrossRef]
- Zhang, H.; Fu, C.; Yi, Y.; Zhou, X.; Zhou, C.; Ying, G.; Shen, Y.; Zhu, Y. A Magnetic-Based SERS Approach for Highly Sensitive and Reproducible Detection of Cancer-Related Serum MicroRNAs. Anal. Methods 2018, 10, 624–633. [Google Scholar] [CrossRef]
- Xiao, X.; Yuan, C.; Li, T.; Fock, J.; Svedlindh, P.; Tian, B. Optomagnetic Biosensors: Volumetric Sensing Based on Magnetic Actuation-Induced Optical Modulations. Biosens. Bioelectron. 2022, 215, 114560. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Han, Y.; Wetterskog, E.; Donolato, M.; Hansen, M.F.; Svedlindh, P.; Strömberg, M. MicroRNA Detection through DNAzyme-Mediated Disintegration of Magnetic Nanoparticle Assemblies. ACS Sens. 2018, 3, 1884–1891. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Qiu, Z.; Ma, J.; Donolato, M.; Hansen, M.F.; Svedlindh, P.; Strömberg, M. On-Particle Rolling Circle Amplification-Based Core–Satellite Magnetic Superstructures for MicroRNA Detection. ACS Appl. Mater. Interfaces 2018, 10, 2957–2964. [Google Scholar] [CrossRef] [Green Version]
- Celikbas, E.; Balaban, S.; Evran, S.; Coskunol, H.; Timur, S. A Bottom-Up Approach for Developing Aptasensors for Abused Drugs: Biosensors in Forensics. Biosensors 2019, 9, 118. [Google Scholar] [CrossRef] [Green Version]
- Cui, F.; Zhou, Z.; Zhou, H.S. Review—Measurement and Analysis of Cancer Biomarkers Based on Electrochemical Biosensors. J. Electrochem. Soc. 2019, 167, 037525. [Google Scholar] [CrossRef]
- Islam, N.; Gorgannezhad, L.; Masud, M.K.; Tanaka, S.; Hossain, S.A.; Yamauchi, Y.; Nguyen, N.-T.; Shiddiky, M.J.A. Graphene-Oxide-Loaded Superparamagnetic Iron Oxide Nanoparticles for Ultrasensitive Electrocatalytic Detection of MicroRNA. ChemElectroChem 2018, 5, 2488–2495. [Google Scholar] [CrossRef] [Green Version]
- Daneshpour, M.; Karimi, B.; Omidfar, K. Simultaneous Detection of Gastric Cancer-Involved MiR-106a and Let-7a through a Dual-Signal-Marked Electrochemical Nanobiosensor. Biosens. Bioelectron. 2018, 109, 197–205. [Google Scholar] [CrossRef]
- Shen, Z.; He, L.; Wang, W.; Tan, L.; Gan, N. Highly Sensitive and Simultaneous Detection of MicroRNAs in Serum Using Stir-Bar Assisted Magnetic DNA Nanospheres-Encoded Probes. Biosens. Bioelectron. 2020, 148, 111831. [Google Scholar] [CrossRef]
- Tavallaie, R.; McCarroll, J.; Le Grand, M.; Ariotti, N.; Schuhmann, W.; Bakker, E.; Tilley, R.D.; Hibbert, D.B.; Kavallaris, M.; Gooding, J.J. Nucleic Acid Hybridization on an Electrically Reconfigurable Network of Gold-Coated Magnetic Nanoparticles Enables MicroRNA Detection in Blood. Nat. Nanotechnol. 2018, 13, 1066–1071. [Google Scholar] [CrossRef]
- Yu, L.; He, P.; Xu, Y.; Kou, X.; Yu, Z.; Xie, X.; Miao, P. Manipulations of DNA Four-Way Junction Architecture and DNA Modified Fe3O4@Au Nanomaterials for the Detection of MiRNA. Sens. Actuators B Chem. 2020, 313, 128015. [Google Scholar] [CrossRef]
- Liu, S.; Yang, Z.; Chang, Y.; Chai, Y.; Yuan, R. An Enzyme-Free Electrochemical Biosensor Combining Target Recycling with Fe3O4/CeO2@Au Nanocatalysts for MicroRNA-21 Detection. Biosens. Bioelectron. 2018, 119, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Jiang, J.; Wang, J.; Xia, J.; Wang, R.; Diao, G. Competitive Host-Guest Recognition Initiated by DNAzyme-Cleavage Cycling for Novel Ratiometric Electrochemical Assay of MiRNA-21. Sens. Actuators B Chem. 2021, 333, 129556. [Google Scholar] [CrossRef]
- Amoshahi, H.; Shafiee, M.R.M.; Kermani, S.; Mirmohammadi, M. A Biosensor for Detection of MiR-106 a by Using Duplex-Specific Nuclease, Assisted Target, Magnetic Nanoparticles, Gold Nanoparticles and Enzymatic Signal Amplification. ChemistrySelect 2022, 7, e202103115. [Google Scholar] [CrossRef]



| Target MNP | Method | Advantage | Disadvantage | Ref. |
|---|---|---|---|---|
| Carbon-encapsulated MNP | Ball milling |
|
| [20] |
| ε-Fe2O3 nanoparticles | Ball milling |
|
| [21] |
| MNP | Laser ablation |
|
| [22] |
| Fe3O4 nanoparticle | Co-precipitation |
|
| [23] |
| Zinc- and manganese-co-doped magnetic nanoparticles | Co-precipitation |
|
| [24] |
| FeCO3 | Thermal decomposition |
|
| [25] |
| CoFe2O4 | Thermal decomposition |
|
| [26] |
| Fe3O4 | Hydrothermal method |
|
| [27] |
| Iron oxide nanoparticle | Hydrothermal method and biological synthesis |
|
| [28] |
| Silica-Coated Fe3O4 Nanoparticles | Microemulsion |
|
| [29] |
| Fe3O4 | Sol-gel |
|
| [30] |
| Mg0.5Zn0.5FeMnO4 magnetic nanoparticles | Green Sol-gel |
|
| [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balaban Hanoglu, S.; Harmanci, D.; Ucar, N.; Evran, S.; Timur, S. Recent Approaches in Magnetic Nanoparticle-Based Biosensors of miRNA Detection. Magnetochemistry 2023, 9, 23. https://doi.org/10.3390/magnetochemistry9010023
Balaban Hanoglu S, Harmanci D, Ucar N, Evran S, Timur S. Recent Approaches in Magnetic Nanoparticle-Based Biosensors of miRNA Detection. Magnetochemistry. 2023; 9(1):23. https://doi.org/10.3390/magnetochemistry9010023
Chicago/Turabian StyleBalaban Hanoglu, Simge, Duygu Harmanci, Nursima Ucar, Serap Evran, and Suna Timur. 2023. "Recent Approaches in Magnetic Nanoparticle-Based Biosensors of miRNA Detection" Magnetochemistry 9, no. 1: 23. https://doi.org/10.3390/magnetochemistry9010023






