Decoding Breast Cancer: Using Radiomics to Non-Invasively Unveil Molecular Subtypes Directly from Mammographic Images
Abstract
:1. Introduction
2. Materials and Methods
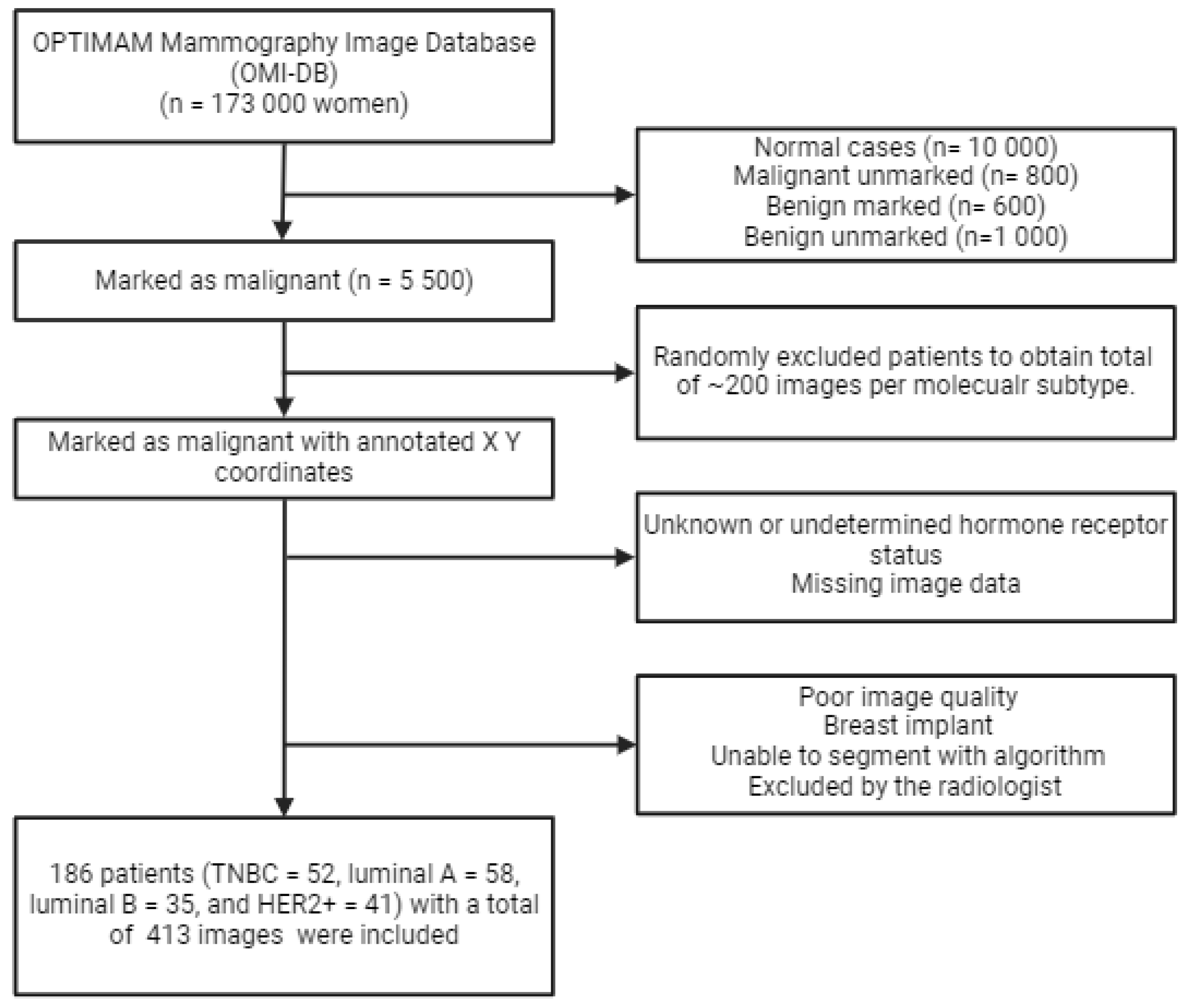
2.1. Database
2.2. In- and Exclusion Criteria
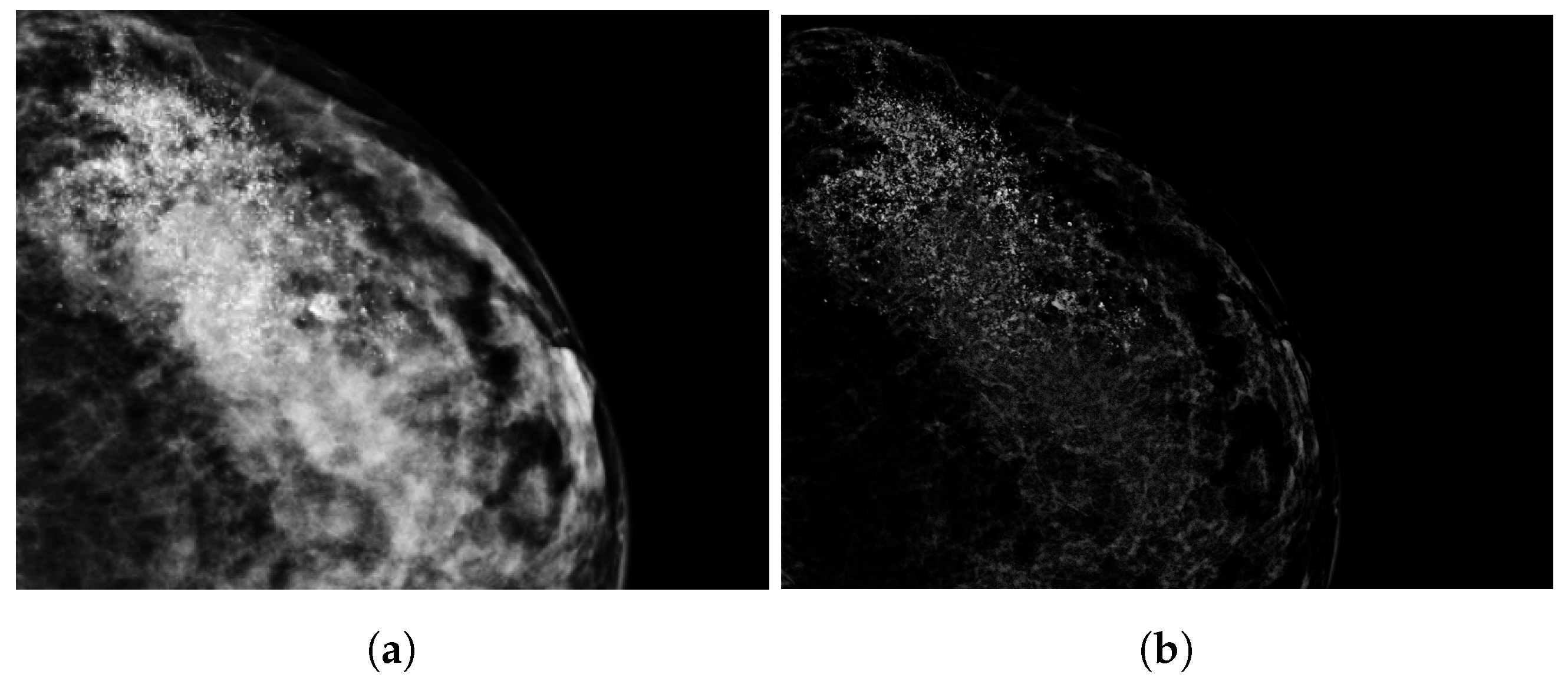
2.3. Tumor Segmentation
2.4. Radiomics Features
2.5. Statistical Analysis
3. Results
3.1. Radiomic Features
3.2. Classification Performance
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AUC | Area Under the Curve |
| CESM | Contrast-enhanced spectral mammography |
| CC | Cranio caudal |
| CNB | Core needle biopsy |
| DBT | Digital Breast Tomosynthesis |
| DM | Digital mammography |
| ER | Estrogen receptor |
| HER | Human epidermal growth factor receptor |
| IHC | Immunohistochemical |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| ML | Machine learning |
| MLO | Medio-lateral oblique |
| MRI | Magnetic Resonance Imaging |
| NB | Naive Bayes |
| OMI-DB | OPTIMAM Mammography Image Database |
| PR | Progesteron receptor |
| ROC | Receiver Operating Curve |
| ROI | Region of interest |
| SMOTE | Synthetic Minority Oversampling Technique |
| SVM | Support vector machine |
| TNBC | Triple-negative breast cancer |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA A Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- WHO. A Short Guide to Cancer Screening: Increase Effectiveness, Maximize Benefits and Minimize Harm; World Health Organization, Regional Office for Europe: Geneva, Switzerland, 2022; pp. 1–45. [Google Scholar]
- Harbeck, N.; Penault-Llorca, F.; Cortés, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast cancer. Nat. Rev. Dis. Primers 2019, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Orrantia-Borunda, E.; Anchondo-Nuñez, P.; Acuña-Aguilar, L.E.; Gómez-Valles, F.O.; Ramírez-Valdespino, C.A. Subtypes of Breast Cancer. In Breast Cancer; National Library of Medicine: Bethesda, ML, USA, 2022; pp. 31–42. [Google Scholar] [CrossRef]
- Phipps, A.I.; Li, C.I. Breast Cancer Biology and Clinical Characteristics. In Breast Cancer Epidemiology; Li, C., Ed.; Springer New York: New York, NY, USA, 2010; pp. 21–46. [Google Scholar] [CrossRef]
- Bilous, M. Breast core needle biopsy: Issues and controversies. Mod. Pathol. 2010, 23, S36–S45. [Google Scholar] [CrossRef]
- Panet, F.; Papakonstantinou, A.; Borrell, M.; Vivancos, J.; Vivancos, A.; Oliveira, M. Use of ctDNA in early breast cancer: Analytical validity and clinical potential. NPJ Breast Cancer 2024, 10, 50. [Google Scholar] [CrossRef]
- Sant, M.; Bernat-Peguera, A.; Felip, E.; Margelí, M. Role of ctDNA in Breast Cancer. Cancers 2022, 14, 310. [Google Scholar] [CrossRef] [PubMed]
- Mayerhoefer, M.E.; Materka, A.; Langs, G.; Häggström, I.; Szczypiński, P.; Gibbs, P.; Cook, G. Introduction to Radiomics. J. Nucl. Med. 2020, 61, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Van Timmeren, J.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baeßler, B. Radiomics in medical imaging—“How-to” guide and critical reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Panico, A.; Gatta, G.; Salvia, A.; Grezia, G.D.; Fico, N.; Cuccurullo, V. Radiomics in Breast Imaging: Future Development. J. Pers. Med. 2023, 13, 862. [Google Scholar] [CrossRef]
- Son, J.; Lee, S.E.; Kim, E.K.; Kim, S. Prediction of breast cancer molecular subtypes using radiomics signatures of synthetic mammography from Digital Breast Tomosynthesis. Sci. Rep. 2020, 10, 21566. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, F.; Zhang, H.; Ma, H.; Sun, J.; Zhang, R.; Song, L.; Shi, H. Diagnostic value of radiomics analysis in contrast-enhanced spectral mammography for identifying triple-negative breast cancer. Front. Oncol. 2021, 11, 773196. [Google Scholar] [CrossRef]
- Petrillo, A.; Fusco, R.; Bernardo, E.; Petrosino, T.; Barretta, M.; Porto, A.; Granata, V.; Bonito, M.; Fanizzi, A.; Massafra, R.; et al. Prediction of Breast Cancer Histological Outcome by Radiomics and Artificial Intelligence Analysis in Contrast-Enhanced Mammography. Cancers 2022, 14, 2132. [Google Scholar] [CrossRef] [PubMed]
- Forgia, D.; Fanizzi, A.; Campobasso, F.; Bellotti, R.; Didonna, V.; Lorusso, V.; Moschetta, M.; Massafra, R.; Tamborra, P.; Tangaro, S.; et al. Radiomic Analysis in Contrast-Enhanced Spectral Mammography for Predicting Breast Cancer Histological Outcome. Diagnostics 2020, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Wang, S.; Guo, S.; Wu, R.; Zhang, J.; Kong, M.; Pan, L.; Gu, Y.; Yu, S. Contrast-enhanced mammography radiomics analysis for preoperative prediction of breast cancer molecular subtypes. Acad. Radiol. 2023, 31, 2228–2238. [Google Scholar] [CrossRef]
- Ma, W.; Zhao, Y.; Ji, Y.; Guo, X.; Jian, X.; Liu, P.; Wu, S. Breast Cancer Molecular Subtype Prediction by Mammographic Radiomic Features. Acad. Radiol. 2019, 26, 196–201. [Google Scholar] [CrossRef]
- Zhou, J.; Tan, H.; Bai, Y.; Li, J.; Lu, Q.; Chen, R.; Zhang, M.; Feng, Q.; Wang, M. Evaluating the HER-2 status of breast cancer using mammography radiomics features. Eur. J. Radiol. 2019, 121, 108718. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Lu, Y.; Li, X.; Zhu, Y.; Zhao, Y.; Ruan, Z.; Mei, N.; Yin, B.; Liu, L. Prediction of human epidermal growth factor receptor 2 (HER2) status in breast cancer by mammographic radiomics features and clinical characteristics: A multicenter study. Eur. Radiol. 2024, 34, 5464–5476. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, W.; Xie, X.; Liu, W.; Wang, H.; Shen, J.; Ding, Y.; Zhang, B.; Song, B. Application of digital mammography-based radiomics in the differentiation of benign and malignant round-like breast tumors and the prediction of molecular subtypes. Gland. Surg. 2020, 9, 2005–2016. [Google Scholar] [CrossRef]
- Ge, S.; Yixing, Y.; Jia, D.; Ling, Y. Application of mammography-based radiomics signature for preoperative prediction of triple-negative breast cancer. BMC Med. Imaging 2022, 22, 166. [Google Scholar] [CrossRef]
- Niu, S.; Jiang, W.; Zhao, N.; Jiang, T.; Dong, Y.; Luo, Y.; Yu, T.; Jiang, X. Intra- and peritumoral radiomics on assessment of breast cancer molecular subtypes based on mammography and MRI. J. Cancer Res. Clin. Oncol. 2022, 148, 1–10. [Google Scholar] [CrossRef]
- Halling-Brown, M.D.; Warren, L.M.; Ward, D.; Lewis, E.; Mackenzie, A.; Wallis, M.G.; Wilkinson, L.S.; Given-Wilson, R.M.; McAvinchey, R.; Young, K.C. Optimam Mammography Image Database: A large-scale resource of mammography images and Clinical Data. Radiol. Artif. Intell. 2021, 3, e200103. [Google Scholar] [CrossRef]
- Wisselink, H.J. RegGrow. 2024. Available online: https://github.com/thrynae/RegGrow/releases/tag/v1.3.0 (accessed on 30 April 2024).
- Taneja, S.; Evans, A.J.; Rakha, E.A.; Green, G.; Ellis, I.O. The mammographic correlations of a new immunohistochemical classification of invasive breast cancer. Clin. Radiol. 2008, 63, 1228–1235. [Google Scholar] [CrossRef]
- Boisserie-Lacroix, M.; Hurtevent-Labrot, G.; Ferron, S.; Lippa, N.; Bonnefoi, H.; Mac Grogan, G. Correlation between imaging and molecular classification of breast cancers. Diagn. Interv. Imaging 2013, 94, 1069–1080. [Google Scholar] [CrossRef]
- Nicosia, L.; Bozzini, A.; Ballerini, D.; Palma, S.; Pesapane, F.; Raimondi, S.; Gaeta, A.; Bellerba, F.; Origgi, D.; De Marco, P.; et al. Radiomic Features Applied to Contrast Enhancement Spectral Mammography: Possibility to Predict Breast Cancer Molecular Subtypes in a Non-Invasive Manner. Int. J. Mol. Sci. 2022, 23, 15322. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, Y.; Burnside, E.; Huang, E.; Drukker, K.; Hoadley, K.; Fan, C.; Conzen, S.; Zuley, M.; Net, J.; et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. npj Breast Cancer 2016, 2, 16012. [Google Scholar] [CrossRef]
- Jalloul, R.; Chethan, H.; Alkhatib, R. A Review of Machine Learning Techniques for the Classification and Detection of Breast Cancer from Medical Images. Diagnostics 2023, 13, 2460. [Google Scholar] [CrossRef] [PubMed]
- Berrar, D. Bayes’ Theorem and Naive Bayes Classifier. Ref. Modul. Life Sci. 2018, 1, 403–412. [Google Scholar] [CrossRef]
- Mao, N.; Yin, P.; Wang, Q.; Liu, M.; Dong, J.; Zhang, X.; Xie, H.; Hong, N. Added Value of Radiomics on Mammography for Breast Cancer Diagnosis: A Feasibility Study. J. Am. Coll. Radiol. 2019, 16, 485–491. [Google Scholar] [CrossRef]
- Scapicchio, C.; Gabelloni, M.; Barucci, A.; Cioni, D.; Saba, L.; Neri, E. A deep look into radiomics. La Radiol. Medica. 2021, 126, 1296–1311. [Google Scholar] [CrossRef]
- Haarburger, C.; Müller-Franzes, G.; Weninger, L.; Kuhl, C.; Truhn, D.; Merhof, D. Radiomics feature reproducibility under inter-rater variability in segmentations of CT images. Sci. Rep. 2020, 10, 12688. [Google Scholar] [CrossRef]
- Conti, A.; Duggento, A.; Indovina, I.; Guerrisi, M.; Toschi, N. Radiomics in breast cancer classification and prediction. Semin. Cancer Biol. 2021, 72, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Lowry, K.; Coley, R.; Miglioretti, D.; Kerlikowske, K.; Henderson, L.; Onega, T.; Sprague, B.; Lee, J.; Herschorn, S.; Tosteson, A.; et al. Screening Performance of Digital Breast Tomosynthesis vs. Digital Mammography in Community Practice by Patient Age, Screening Round, and Breast Density. JAMA Netw. Open 2020, 3, e2011792. [Google Scholar] [CrossRef] [PubMed]
- Niu, S.; Wang, X.; Zhao, N.; Liu, G.; Kan, Y.; Dong, Y.; Cui, E.N.; Luo, Y.; Yu, T.; Jiang, X. Radiomic Evaluations of the Diagnostic Performance of DM, DBT, DCE MRI, DWI, and Their Combination for the Diagnosisof Breast Cancer. Front. Oncol. 2021, 11, 725922. [Google Scholar] [CrossRef] [PubMed]
- Mota, A.; Mendes, J.; Matela, N. Breast Cancer Molecular Subtype Prediction: A Mammography-Based AI Approach. Biomedicines 2024, 12, 1371. [Google Scholar] [CrossRef]




| Technique | Purpose | Findings | |
|---|---|---|---|
| W. Ma [18] | DM | Luminal vs. non-luminal TNBC vs. non-TNBC HER2 vs. non-HER2 | TNBC was differentiated from non-TNBC with an AUC/accuracy of 0.865/0.796. HER2 could be distinguished with an AUC/accuracy of 0.784/0.748 and the luminal type 0.752/0.788 |
| J. Son [13] | Synthetic DM | Luminal vs. non-luminal TNBC vs. non-TNBC HER2 vs. non-HER2 | The AUC, accuracy, sensitivity, and specificity for the TNBC model were 0.838, 0.803, 0.833, and 0.797. For HER2, this resulted in values of 0.556, 0.704, 0.111, and 0.790, respectively. When distinguishing the luminal subtype, AUC, accuracy, sensitivity, and specificity values of 0.645, 0.507, 0.440, and 0.667 were obtained. |
| J. Zhou [19] | DM | HER2 vs. non-HER2 | The SVM classifier resulted in AUC, accuracy, sensitivity, and specificity values of 0.740, 0.730, 0.688, and 0.609. The logistic regression model resulted in AUC/ACC/SENS/SPEC of 0.787/0.770/0.688/0.739. |
| Y. Deng [20] | DM | HER2 vs. non-HER2 | The AUC and accuracy of distinguishing HER2 vs. non-HER2 was 0.776 and 0.712 during testing. In the external validation set, the AUC and accuracy was 0.702 and 0.700. |
| L. Wang [21] | DM | TNBC vs. non-TNBC | Accuracy, sensitivity, and specificity values of 0.84, 0.81, and 0.78, respectively, were obtained. |
| Y. Zhang [14] | CESM | TNBC vs. non-TNBC | Resulted in AUC, sensitivity, and specificity values of 0.90, 0.97, and 0.69. |
| A. Petrillo [15] | CESM | HER2 vs. non-HER2 | Tested accuracies, sensitivities, and specificities for the logistic regression, CART, and Random Forest models. A combination of features from CC and MLO showed the highest accuracies of > 90% using a classification tree algorithm. For HER2 classification, the best accuracies were obtained with an RF algorithm. |
| D. La Forgia [16] | CESM | Histological outcome | Resulted in AUC values of ER+/ER−: 0.838, PR+/PR−: 0.755, Ki67+/Ki67−: 0.848, high-grade/low-grade: 0.799, TNBC/NTNBC: 0.768, and HER2/HER2−: 0.909. |
| S. Zhu [17] | CESM | Luminal vs. non-luminal TNBC vs. non-TNBC HER2 vs. non-HER2 | Showed AUC values during combined low energy and recombined images during testing for luminal, HER2, and TNBC values of 0.93, 0.89, and 0.87, respectively. For the external dataset, this resulted in AUC values of 0.82, 0.83, and 0.68 for luminal, HER2, and TNBC, respectively. |
| S. Niu [23] | DM, DBT, and MRI | Intra- and peritumoral regions | AUC values for distinguishing luminal A, luminal B, HER2, and TNBC of 0.762, 0.757, 0.756, and 0.771 were obtained for DM images. |
| S. GE [22] | DM | TNBC vs. non-TNBC | Distinguishing TNBC vs. non-TNBC resulted in AUC, accuracy, sensitivity, and specificity values of 0.809, 0.806, 0.720, and 0.801. |
| ER | PR | HER2 | |
|---|---|---|---|
| Luminal A | + | + | − |
| Luminal B | + | +/− | − |
| TNBC | − | − | − |
| HER2 | − | − | + |
| SVM | NB | |||
|---|---|---|---|---|
| Accuracy | AUC | Accuracy | AUC | |
| (95%-CI) | (95%-CI) | |||
| Luminal A | 0.815 | 0.855 | 0.726 | 0.714 |
| (0.779–0.930) | (0.616–0.812) | |||
| Luminal B | 0.734 | 0.812 | 0.750 | 0.746 |
| (0.736–0.889) | (0.655–0.837) | |||
| TNBC | 0.581 | 0.789 | 0.484 | 0.593 |
| (0.701–0.878) | (0.482–0.704) | |||
| HER2 | 0.637 | 0.755 | 0.718 | 0.714 |
| (0.644–0.867) | (0.608–0.819) | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bakker, M.A.G.; Ovalho, M.d.L.; Matela, N.; Mota, A.M. Decoding Breast Cancer: Using Radiomics to Non-Invasively Unveil Molecular Subtypes Directly from Mammographic Images. J. Imaging 2024, 10, 218. https://doi.org/10.3390/jimaging10090218
Bakker MAG, Ovalho MdL, Matela N, Mota AM. Decoding Breast Cancer: Using Radiomics to Non-Invasively Unveil Molecular Subtypes Directly from Mammographic Images. Journal of Imaging. 2024; 10(9):218. https://doi.org/10.3390/jimaging10090218
Chicago/Turabian StyleBakker, Manon A. G., Maria de Lurdes Ovalho, Nuno Matela, and Ana M. Mota. 2024. "Decoding Breast Cancer: Using Radiomics to Non-Invasively Unveil Molecular Subtypes Directly from Mammographic Images" Journal of Imaging 10, no. 9: 218. https://doi.org/10.3390/jimaging10090218
APA StyleBakker, M. A. G., Ovalho, M. d. L., Matela, N., & Mota, A. M. (2024). Decoding Breast Cancer: Using Radiomics to Non-Invasively Unveil Molecular Subtypes Directly from Mammographic Images. Journal of Imaging, 10(9), 218. https://doi.org/10.3390/jimaging10090218













