-
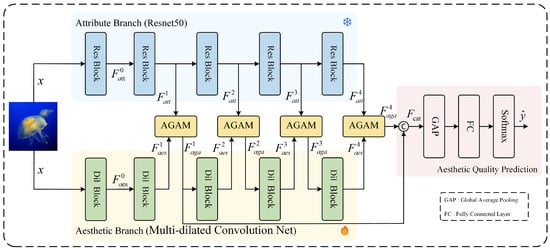
 Gated Attention-Augmented Double U-Net for White Blood Cell Segmentation
Gated Attention-Augmented Double U-Net for White Blood Cell Segmentation -
 Symbolic Regression for Interpretable Camera Calibration
Symbolic Regression for Interpretable Camera Calibration -
 GATF-PCQA: A Graph Attention Transformer Fusion Network for Point Cloud Quality Assessment
GATF-PCQA: A Graph Attention Transformer Fusion Network for Point Cloud Quality Assessment -
 Multi-Channel Spectro-Temporal Representations for Parkinson’s Detection
Multi-Channel Spectro-Temporal Representations for Parkinson’s Detection -
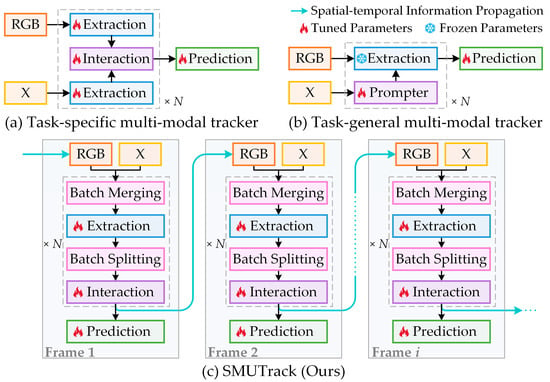
 Image Matching: Foundations, State of the Art, and Future Directions
Image Matching: Foundations, State of the Art, and Future Directions
Journal Description
Journal of Imaging
- Open Accessfree for readers, with article processing charges (APC) paid by authors or their institutions.
- High Visibility: indexed within Scopus, ESCI (Web of Science), PubMed, PMC, dblp, Inspec, Ei Compendex, and other databases.
- Journal Rank: JCR - Q2 (Imaging Science and Photographic Technology) / CiteScore - Q1 (Radiology, Nuclear Medicine and Imaging)
- Rapid Publication: manuscripts are peer-reviewed and a first decision is provided to authors approximately 15.3 days after submission; acceptance to publication is undertaken in 3.5 days (median values for papers published in this journal in the first half of 2025).
- Recognition of Reviewers: reviewers who provide timely, thorough peer-review reports receive vouchers entitling them to a discount on the APC of their next publication in any MDPI journal, in appreciation of the work done.
Latest Articles
E-Mail Alert
News
Topics
Deadline: 31 December 2025
Deadline: 31 March 2026
Deadline: 30 April 2026
Deadline: 31 May 2026
Conferences
Special Issues
Deadline: 30 November 2025
Deadline: 30 November 2025
Deadline: 30 November 2025
Deadline: 30 November 2025