Epithelium and Stroma Identification in Histopathological Images Using Unsupervised and Semi-Supervised Superpixel-Based Segmentation †
Abstract
:1. Introduction
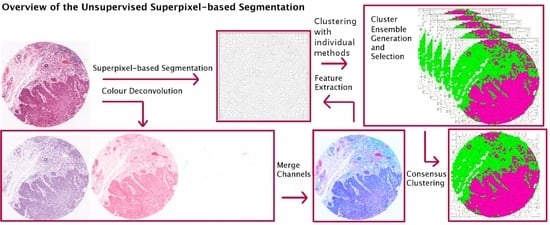
- Propose an extended version of our work in [4], in which we investigate CC in the context of superpixel-based segmentation of haematoxylin and eosin (H&E) stained histopathological images and suggest a multi-stage segmentation process.First, the recently proposed Simple Linear Iterative Clustering (SLIC) superpixel framework [1,5] is used to segment the image into compact regions. Colour features from each dye of H & E staining are extracted from the superpixels and used as input to multiple base clustering algorithms with various parameter initializations. The generated results (denoted here as ‘partitions’) pass through a selection scheme, which generates an ‘ensemble’ based on partitions diversity. Two consensus functions are considered here: the Evidence Accumulation Clustering (EAC) [6] and the voting-based consensus function (e.g., [7,8]).
- Suggest a new implementation for the voting-based CC method, based on image processing operations, to solve the label mismatching problem occurring among the base clustering outcomes.Unlike supervised methods, labels resulting from unsupervised techniques are symbolic (i.e., labels do not represent a meaningful class), and, consequently, an individual partition in the ensemble will likely include clusters that do not necessarily correspond to the labels of other clusters in different partitions of the ensemble. In the voting-based consensus function, this label mismatch is defined as the problem of finding the optimal re-labelling of a given partition with respect to a reference partition. This problem is commonly formulated as a weighted bipartite matching formulation [7,8] and it is solved by inspecting whether data patterns in two partitions share labels more frequently than with other clusters. In this paper, we present an alternative simple, yet robust, implementation for generating a consistent labelling scheme among the different partitions of the ensemble. Our approach considers the space occupied by each individual cluster in an image and exploits the fact that pairs of individual clusters from different partitions would match when their pixels overlap in a segmented image.
- Introduce a Semi-Supervised Classification (SSC) framework based on the CC method for the epithelium-stroma identification in histopathological images.Current supervised classification methods have reported promising results (e.g., [9,10]); however, they require large volumes of manually segmented training sets (i.e., labelled images) that are time-consuming to obtain. By contrast, our proposed unsupervised epithelium-stroma segmentation CC techniques do not require labelled data during training, but can result in a relatively lower segmentation accuracy than the supervised results. In such situations, semi-supervised learning techniques [11,12] can be of practical value as they combine the two learning strategies (supervised and unsupervised). Such approaches rely on the presence of very few labelled training instances as well as large volumes of unlabelled training samples and additionally exploit the use of unlabelled data during the learning course. Here, we propose an SSC framework based on the CC method for the epithelium-stroma identification in microscopy images. Our SSC model is based on a simple and effective semi-supervised learning methodology named the self-training method [13,14]. In this approach, a classifier repeatedly labels unlabelled training examples and retrains itself on an enlarged labelled training set. In particular, the proposed classifier is initially trained with few labelled samples and then takes advantage of the obtained CC clustering results to perform the self-training procedure. Unlike other supervised methods for epithelium and stroma segmentation, the proposed SSC approach offers an effective segmentation while relying on a small number of labelled segments. This is done by taking advantage of the knowledge acquired from clustering the unlabelled data (clustering distribution) to build a classifier that predicts the classes of unseen data (class distribution).
2. Related Work
3. Unsupervised Superpixel-Based Segmentation with Consensus Clustering
3.1. Dataset and Preprocessing
3.2. Superpixel-Based Segmentation
3.3. Feature Extraction
3.4. Consensus Clustering (CC) Frameworks
3.4.1. Ensemble Generation and Selection
3.4.2. Evidence Accumulation Consensus (EAC) Function
3.4.3. Voting-Based Consensus Function
| Algorithm 1: Label Matching Algorithm for the Vote-CC Method |
| Input: P, , S, n, K |
| Output: Labels matched for P with respect to |
| for (s = 1 to n) do |
| Assign label of to superpixel and save in image |
| Assign label of to superpixel and save in image |
| end for |
| for ( = 1 to K) do |
| for ( = 1 to K) do |
| Threshold in , convert to mask and save in |
| Threshold in , convert to mask and save in |
| Compute using Equation (4) |
| end for |
| if () |
| SwappedLabels |
| JISwappedLabels |
| if () ∉ SwappedLabels |
| Swap and and save result in |
| else if (() ∈ SwappedLabels) and (JISwappedLabels JISwappedLabels) |
| Swap and and save result in |
| end If |
| end If |
| end for |
| Assign the new labels in to partition P |
4. Self-Training Semi-Supervised Classification Based on Consensus Clustering
| Algorithm 2: Semi-Supervised Self-training Classification Based on Consensus Clustering |
| Input: h, L, U, T, |
| Output: Predicted labels of T |
| Create variables P, , , Exit = False |
| Apply the Vote-CC (described in Section 3.4.3) on U and save the obtained labels in P |
| while (not (Exit)) do |
| Train h with L to predict the labels of U and save predicted labels in |
| Match P with (reference partition) using Algorithm 1 |
| Compute the LabelSimilarity between P and using Equation (6) |
| if (LabelSimilarity(P, ) ≤ ) and (LabelSimilarity(P, ) ≠ 0) |
| for () do |
| if() select the instances with confident predictions |
| add to the reliable instance i to |
| end if |
| end for |
| else |
| Exit = True |
| end if |
| end while |
| Train h with L to predict the labels of T |
5. Experiments and Evaluation
5.1. Clustering Evaluation Methods
- The Rand Index (RI) was used to compare the final consensus clustering solution given in image with their corresponding reference partition given in the gold-standard image R and it is estimated as(see Equation (2)), where , , , or were calculated by considering the overlapping superpixels of and R (as explained before).
- F1-score is defined as:whereand
- Jaccard Index (JI) is defined as:
5.2. Comparing the Proposed CC with Individual Clustering Methods
5.3. Comparing the Proposed ST-CC with Supervised Methods
6. Discussion
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CC | Consensus Clustering |
| EAC | Evidence Accumulation Clustering |
| SSC | Semi-Supervised Classification |
| SLIC | Simple Linear Iterative Clustering |
| jSLIC | Java Simple Linear Iterative Clustering |
| H&E | Haematoxylin and Eosin |
| ST-CC | Self-Training CC-based method |
| EM | Expectation-Maximisation |
| LVQ | Learning Vector Quantization |
| MDB | Make Density Based |
| AH | Agglomerative Hierarchical Clustering |
| TMA | Tissue Micro Arrays |
| RI | Rand Index |
| TP | True Positive |
| FP | False Positive |
| TN | True Negative |
| FN | False Negative |
| Vote-CC | Voting-Based Consensus Function |
| EAC-CC | EAC Consensus Function |
| JI | Jaccard Index |
References
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Susstrunk, S. SLIC Superpixels; Technical Report; École Polytechnique Fédérale de Lausanne (EPFL): Lausanne, Switzerland, 2010. [Google Scholar]
- Borovec, J. Fully Automatic Segmentation of Stained Histological Cuts. In Proceedings of the 17th International Student Conference on Electrical Engineering, Prague, Czech Republic, 16 May 2013; pp. 1–7. [Google Scholar]
- Vega-Pons, S.; Ruiz-Shulcloper, J. A Survey of Clustering Ensemble Algorithms. Int. J. Pattern Recognit. Artif. Intell. 2011, 25, 337–372. [Google Scholar] [CrossRef]
- Fouad, S.; Randell, D.; Galton, A.; Mehanna, H.; Landini, G. Unsupervised Superpixel-Based Segmentation of Histopathological Images with Consensus Clustering. In Medical Image Understanding and Analysis; Valdés Hernández, M., González-Castro, V., Eds.; Springer: Edinburgh, UK, 2017; Volume 723, pp. 767–779. [Google Scholar]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Susstrunk, S. SLIC Superpixels Compared to State-of-the-art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Fred, A.L.; Jain, A.K. Combining Multiple Clusterings Using Evidence Accumulation. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Topchy, A.P.; Law, M.H.C.; Jain, A.K.; Fred, A.L. Analysis of Consensus Partition in Cluster Ensemble. In Proceedings of the 4th IEEE International Conference on Data Mining, Brighton, UK, 1–4 November 2004; pp. 225–232. [Google Scholar]
- Dudoit, S.; Fridlyand, J. Bagging to Improve the Accuracy of a Clustering Procedure. Bioinformatics 2003, 19, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Bianconi, F.; Álvarez-Larrán, A.; Fernández, A. Discrimination Between Tumour Epithelium and Stroma Via Perception-based Features. Neurocomputing 2015, 154, 119–126. [Google Scholar] [CrossRef]
- Linder, N.; Konsti, J.; Turkki, R.; Rahtu, E.; Lundin, M.; Nordling, S.; Haglund, C.; Ahonen, T.; Pietikäinen, M.; Lundin, J. Identification of Tumor Epithelium and Stroma in Tissue Microarrays using Texture Analysis. Diagn. Pathol. 2012, 7, 22. [Google Scholar] [CrossRef] [PubMed]
- Chapelle, O.; Scholkopf, B.; Zien, A. Semi-supervised Learning (chapelle, o. et al., eds.; 2006) [book reviews]. IEEE Trans. Neural Netw. 2009, 20, 542. [Google Scholar] [CrossRef]
- Zhu, X. Semi-Supervised Learning Literature Survey; University of Wisconsin-Madison: Madison, WI, USA, 2006. [Google Scholar]
- Rosenberg, C.; Hebert, M.; Schneiderman, H. Semi-supervised Self-training of Object Detection Models. In Proceedings of the 7th IEEE Workshop on Applications of Computer Vision, Breckenridge, CO, USA, 9–11 January 2005; pp. 29–36. [Google Scholar]
- Azmi, R.; Norozi, N.; Anbiaee, R.; Salehi, L.; Amirzadi, A. IMPST: A New Interactive Self-training Approach to Segmentation Suspicious Lesions in Breast MRI. J. Med. Signals Sens. 2011, 1, 138. [Google Scholar] [PubMed]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum Likelihood From Incomplete Data Via the EM Algorithm. J. R. Stat. Soc. B. 1977, 39, 1–38. [Google Scholar]
- Nguyen, B.P.; Heemskerk, H.; So, P.T.; Tucker-Kellogg, L. Superpixel-based Segmentation of Muscle Fibers in Multi-channel Microscopy. BMC Syst. Biol. 2016, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Nguyen, B.P.; Chui, C.K.; Ong, S.H. Reworking Multilabel Brain Tumor Segmentation: An Automated Framework Using Structured Kernel Sparse Representation. IEEE Syst. Man Cybern. Mag. 2017, 3, 18–22. [Google Scholar] [CrossRef]
- Simsek, A.C.; Tosun, A.B.; Aykanat, C.; Sokmensuer, C.; Gunduz-Demir, C. Multilevel Segmentation of Histopathological Images Using Cooccurrence of Tissue Objects. IEEE Trans. Biomed. Eng. 2012, 59, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.M.; El-Daly, H.; Rajpoot, N. RanPEC: Random Projections with Ensemble Clustering for Segmentation of Tumor Areas in Breast Histology Images. 2012. Available online: http://miua2012.swansea.ac.uk/uploads/Site/Programme/CS01.pdf (accessed on 9 December 2017).
- Agrawala, A. Learning With a Probabilistic Teacher. IEEE Trans. Inf. Theory 1970, 16, 373–379. [Google Scholar] [CrossRef]
- Ruifrok, A.C.; Johnston, D.A. Quantification of Histochemical Staining by Color Deconvolution. Anal. Quant. Cytol. Histol. 2001, 23, 291–299. [Google Scholar] [PubMed]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-means Clustering Algorithm. J. R. Stat. Soc. Ser. C 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Borovec, J.; Kybic, J. jSLIC: Superpixels in ImageJ. Computer Vision Winter Workshop. 2014. Available online: http://hdl.handle.net/10467/60967 (accessed on 15 January 2016).
- Borovec, J. The jSLIC-Superpixels Plugin. Available online: http://imagej.net/CMP-BIA_tools (accessed on 15 January 2016).
- Landini, G. Advanced Shape Analysis with ImageJ. In Proceedings of the Second ImageJ User and Developer Conference, Luxembourg, 6–7 November 2008; pp. 116–121. Available online: http://www.mecourse.com/landinig/software/software.html (accessed on 1 September 2015).
- Hadjitodorov, S.T.; Kuncheva, L.I.; Todorova, L.P. Moderate Diversity for Better Cluster Ensembles. Inf. Fusion 2006, 7, 264–275. [Google Scholar] [CrossRef]
- Hubert, L.; Arabie, P. Comparing Partitions. J. Classif. 1985, 2, 193–218. [Google Scholar] [CrossRef]
- Defays, D. An Efficient Algorithm for a Complete Link Method. Comput. J. 1977, 20, 364–366. [Google Scholar] [CrossRef]
- Rasband, W.S. ImageJ.; US National Institutes of Health: Bethesda, MD, USA, 1997. Available online: http://imagej.nih.gov/ij/ (accessed on 1 September 2015).
- Frank, E.; Hall, M.; Witten, I.H. The WEKA Workbench, 4th ed.; Morgan Kaufmann: Burlington, MA, USA, 2016. [Google Scholar]
- Kohonen, T. Learning Vector Quantization. In The Handbook of Brain Theory and Neural Networks, 2nd ed.; MIT Press: Cambridge, MA, USA, 2003; pp. 631–634. [Google Scholar]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A Density-based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996; pp. 226–231. [Google Scholar]
- Keerthi, S.S.; Shevade, S.K.; Bhattacharyya, C.; Murthy, K.R.K. Improvements to Platt’s SMO Algorithm for SVM Classifier Design. Neural Comput. 2001, 13, 637–649. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Quinlan, J.R. C4.5: Programs for Machine Learning; Elsevier: San Mateo, CA, USA, 2014. [Google Scholar]
- Beucher, S.; Lantuéjoul, C. Use of Watersheds in Contour Detection. In International Workshop on Image Processing: Real-time Edge and Motion Detection/Estimation; BibSonomy: Rennes, France, 1979. [Google Scholar]
- Beucher, S. Watershed, Hierarchical Segmentation and Waterfall Algorithm. In Proceedings of the Mathematical Morphology and Its Applications to Image Processing, Fontainebleau, France, 5–9 September 1994; pp. 69–76. [Google Scholar]
- Xu, J.; Luo, X.; Wang, G.; Gilmore, H.; Madabhushi, A. A Deep Convolutional Neural Network for Segmenting and Classifying Epithelial and Stromal Regions in Histopathological Images. Neurocomputing 2016, 191, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhou, Z.H. SETRED: Self-training with Editing. In Proceedings of the PAKDD: Pacific-Asia Conference on Knowledge Discovery and Data Mining, Hanoi, Vietnam, 18–20 May 2005; pp. 611–621. [Google Scholar]
- Leistner, C.; Saffari, A.; Santner, J.; Bischof, H. Semi-supervised random forests. In Proceedings of the 2009 IEEE 12th International Conference Computer Vision, Kyoto, Japan, 29 September–2 October 2009; pp. 506–513. [Google Scholar]
Sample Availability: The full data set with original images (tissue micro-arrays (TMA)) cannot be publicly shared due to ethical restrictions (REC Ethics Reference, 10/h1210/9). The full results of the methods described in the paper are available from the authors, School of Dentistry, University of Birmingham, Birmingham, UK. |



| Measure | EAC-CC | Vote-CC | k-Means | LVQ | EM | MDB | AH |
|---|---|---|---|---|---|---|---|
| RI (±) | 0.81 ±(0.05) | 0.82 ±(0.05) | 0.78 ±(0.15) | 0.77 ±(0.11) | 0.76 ±(0.16) | 0.79 ±(0.07) | 0.72 ±(0.12) |
| F1-score (±) | 0.74 ±(0.08) | 0.75 ±(0.09) | 0.71 ±(0.15) | 0.72 ±(0.12) | 0.69 ±(0.14) | 0.73 ±(0.09) | 0.68 ±(0.15) |
| JI (±) | 0.71 ±(0.10) | 0.72 ±(0.10) | 0.69 ±(0.18) | 0.61 ±(0.12) | 0.66 ±(0.20) | 0.68 ±(0.13) | 0.60 ±(0.17) |
| Time(millisecond) | 748.95 | 31.02 |
| Learning Approach | Precision | Recall | F1-Score |
|---|---|---|---|
| (self-training semi-supervised) ST-CC | 0.91 ± (0.06) | 0.89 ± (0.07) | 0.90 ± (0.07) |
| (Supervised) Random Forest | 0.88 ± (0.08) | 0.83 ± (0.10) | 0.85 ± (0.10) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fouad, S.; Randell, D.; Galton, A.; Mehanna, H.; Landini, G. Epithelium and Stroma Identification in Histopathological Images Using Unsupervised and Semi-Supervised Superpixel-Based Segmentation. J. Imaging 2017, 3, 61. https://doi.org/10.3390/jimaging3040061
Fouad S, Randell D, Galton A, Mehanna H, Landini G. Epithelium and Stroma Identification in Histopathological Images Using Unsupervised and Semi-Supervised Superpixel-Based Segmentation. Journal of Imaging. 2017; 3(4):61. https://doi.org/10.3390/jimaging3040061
Chicago/Turabian StyleFouad, Shereen, David Randell, Antony Galton, Hisham Mehanna, and Gabriel Landini. 2017. "Epithelium and Stroma Identification in Histopathological Images Using Unsupervised and Semi-Supervised Superpixel-Based Segmentation" Journal of Imaging 3, no. 4: 61. https://doi.org/10.3390/jimaging3040061
APA StyleFouad, S., Randell, D., Galton, A., Mehanna, H., & Landini, G. (2017). Epithelium and Stroma Identification in Histopathological Images Using Unsupervised and Semi-Supervised Superpixel-Based Segmentation. Journal of Imaging, 3(4), 61. https://doi.org/10.3390/jimaging3040061