Considerations on Baseline Generation for Imaging AI Studies Illustrated on the CT-Based Prediction of Empyema and Outcome Assessment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.1.1. Eligibility Criteria
2.1.2. Intended Sample Size
2.2. CT and Acquisition
2.3. Pleural CT Features
2.3.1. Radiological Report-Based CT Feature Extraction
2.3.2. Prespecified CT Based CT Feature Extraction
2.4. Reference Standard
2.5. Possible Applications
2.6. Analysis
3. Results
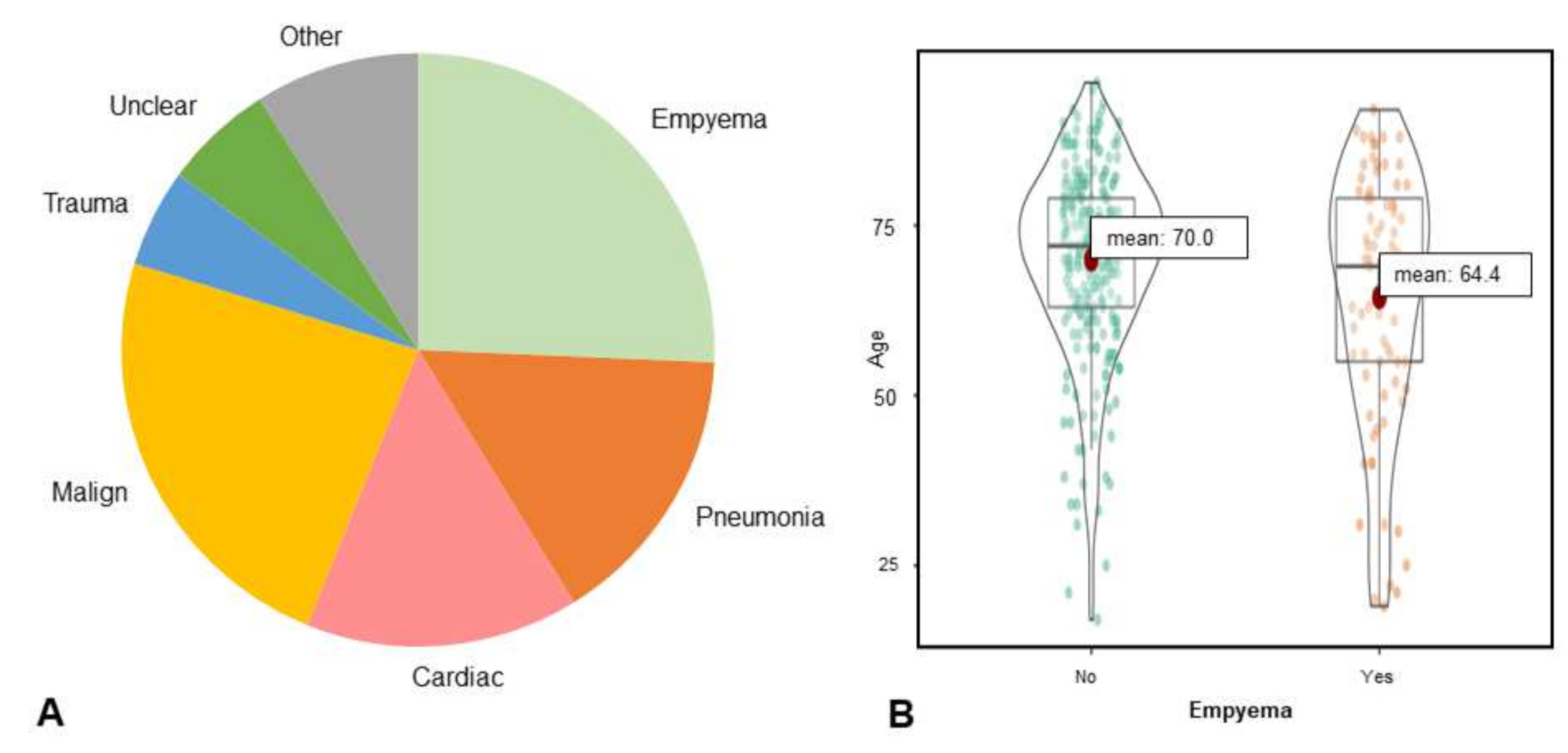
3.1. Study Population
3.2. Estimates of Diagnostic Accuracy
3.2.1. Diagnostic Accuracy Based on Radiology Report
3.2.2. Interrater Agreement
3.2.3. Diagnostic Accuracy to Differentiate Pleural Empyema from Other Effusions
3.3. Outcome Measures
3.4. Possible Applications
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Anastasopoulos, C.; Weikert, T.; Yang, S.; Abdulkadir, A.; Schmülling, L.; Bühler, C.; Paciolla, F.; Sexauer, R.; Cyriac, J.; Nesic, I.; et al. Development and clinical implementation of tailored image analysis tools for COVID-19 in the midst of the pandemic: The synergetic effect of an open, clinically embedded software development platform and machine learning. Eur. J. Radiol. 2020, 131, 109233. [Google Scholar] [CrossRef]
- Harmon, S.A.; Sanford, T.H.; Xu, S.; Turkbey, E.B.; Roth, H.; Xu, Z.; Yang, D.; Myronenko, A.; Anderson, V.; Amalou, A.; et al. Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets. Nat. Commun. 2020, 11, 4080. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529. [Google Scholar] [CrossRef] [PubMed]
- Khan, J.; Lehtomäki, A.I.; Toikkanen, V.J.; Ukkonen, M.T.; Nevalainen, R.M.; Laurikka, J.O. Long-term prognosis and causes of death after pleural infections. Scand. J. Surg. 2018, 107, 145–151. [Google Scholar] [CrossRef]
- Grijalva, C.G.; Zhu, Y.; Nuorti, J.P.; Griffin, M.R. Emergence of parapneumonic empyema in the USA. Thorax 2011, 66, 663–668. [Google Scholar] [CrossRef] [Green Version]
- Okiror, L.; Coltart, C.; Bille, A.; Guile, L.; Pilling, J.; Harrison-Phipps, K.; Routledge, T.; Lang-Lazdunski, L.; Hemsley, C.; King, J. Thoracotomy and decortication: Impact of culture-positive empyema on the outcome of surgery. Eur. J. Cardio-Thorac. Surg. 2014, 46, 901–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahn, S.A. Diagnosis and management of parapneumonic effusions and empyema. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2007, 45, 1480–1486. [Google Scholar] [CrossRef] [Green Version]
- Zettinig, D.; D’Antonoli, T.A.; Wilder-Smith, A.; Bremerich, J.; Roth, J.A.; Sexauer, R. Diagnostic accuracy of imaging findings in pleural empyema: Systematic review and meta-analysis. J. Imaging 2021, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, N.; Saraya, T.; Light, R.W.; Tsukahara, Y.; Koide, T.; Kurai, D.; Ishii, H.; Kimura, H.; Goto, H.; Takizawa, H. A simple method for differentiating complicated parapneumonic effusion/empyema from parapneumonic effusion using the split pleura sign and the amount of pleural effusion on thoracic CT. PLoS ONE 2015, 10, e0130141. [Google Scholar] [CrossRef] [Green Version]
- Colice, G.L.; Curtis, A.; Deslauriers, J.; Heffner, J.; Light, R.; Littenberg, B.; Sahn, S.; Weinstein, R.A.; Yusen, R.D. Medical and surgical treatment of parapneumonic effusions. Chest 2000, 118, 1158–1171. [Google Scholar] [CrossRef] [Green Version]
- Suárez, P.R.; Gilart, J.F.; Pérez, J.M.H.; Serhal, M.H.; Artalejo, A.L. Treatment of complicated parapneumonic pleural effusion and pleural parapneumonic empyema. Med Sci. Monit. Int. Med. J. Exp. Clin. Res. 2012, 18, CR443–CR449. [Google Scholar] [CrossRef] [Green Version]
- Light, R.W. Parapneumonic effusions and empyema. Proc. Am. Thorac. Soc. 2006, 3, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sexauer, R. Considerations on Baseline Generation for Imaging AI Studies Illustrated on the CT-Based Prediction of Empyema and Outcome Assessment. 2022. Available online: https://zenodo.org/record/5793366#.YhNPfejMLIU (accessed on 21 December 2021).
- Bossuyt, P.M.; Reitsma, J.B.; E Bruns, D.; A. Gatsonis, C.; Glasziou, P.; Irwig, L.; Lijmer, J.G.; Moher, D.; Rennie, D.; De Vet, H.C.W.; et al. STARD 2015: An updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015, 351, h5527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aquino, S.L.; Webb, W.R.; Gushiken, B.J. Pleural exudates and transudates: Diagnosis with contrast-enhanced CT. Radiology 1994, 192, 803–808. [Google Scholar] [CrossRef]
- Porcel, J.M.; Pardina, M.; Alemán, C.; Pallisa, E.; Light, R.W.; Bielsa, S. Computed tomography scoring system for discriminating between parapneumonic effusions eventually drained and those cured only with antibiotics. Respirology 2017, 22, 1199–1204. [Google Scholar] [CrossRef]
- Leung, A.N.; Müller, N.L.; Miller, R.R. CT in differential diagnosis of diffuse pleural disease. AJR Am. J. Roentgenol. 1990, 154, 487–492. [Google Scholar] [CrossRef]
- Stark, D.; Federle, M.; Goodman, P.; Podrasky, A.; Webb, W. Differentiating lung abscess and empyema: Radiography and computed tomography. AJR Am. J. Roentgenol. 1983, 141, 163–167. [Google Scholar] [CrossRef]
- Metintas, M.; Ucgun, I.; Elbek, O.; Erginel, S.; Metintas, S.; Kolsuz, M.; Harmanci, E.; Alatas, F.; Hillerdal, G.; Ozkan, R.; et al. Computed tomography features in malignant pleural mesothelioma and other commonly seen pleural diseases. Eur. J. Radiol. 2002, 41, 1–9. [Google Scholar] [CrossRef]
- Cullu, N.; Kalemci, S.; Karakaş, Ö.; Eser, I.; Yalcin, F.; Boyaci, F.N.; Karakas, E.; Yalçın, F.; Boyaci, F.N. Efficacy of CT in diagnosis of transudates and exudates in patients with pleural effusion. Diagn. Interv. Radiol. 2014, 20, 116–120. [Google Scholar] [CrossRef]
- Alonso-Charterina, S.; Lloret-Llorens, M.; Arenas-Jiménez, J.; Sánchez-Payá, J.; Fernández-Latorre, F.; Gil-Sánchez, S. Evaluation of CT findings for diagnosis of pleural effusions. Eur. Radiol. 2000, 10, 681–690. [Google Scholar] [CrossRef]
- Waite, R.J.; Carbonneau, R.J.; Balikian, J.P.; Umali, C.B.; Pezzella, A.T.; Nash, G. Parietal pleural changes in empyema: Appearances at CT. Radiology 1990, 175, 145–150. [Google Scholar] [CrossRef]
- Khatami, F.; Saatchi, M.; Zadeh, S.S.T.; Aghamir, Z.S.; Shabestari, A.N.; Reis, L.O.; Aghamir, S.M.K. A meta-analysis of accuracy and sensitivity of chest CT and RT-PCR in COVID-19 diagnosis. Sci. Rep. 2020, 10, 22402. [Google Scholar] [CrossRef] [PubMed]
- Zhan, N.; Guo, Y.; Tian, S.; Huang, B.; Tian, X.; Zou, J.; Xiong, Q.; Tang, D.; Zhang, L.; Dong, W. Clinical characteristics of COVID-19 complicated with pleural effusion. BMC Infect. Dis. 2021, 21, 176. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.P.; Morley, A.J.; Stadon, L.; De Fonseka, D.; Arnold, D.T.; Medford, A.R.; Maskell, N.A. Nonmalignant pleural effusions. Chest 2017, 151, 1099–1105. [Google Scholar] [CrossRef] [Green Version]
- DeBiasi, E.M.; Pisani, M.A.; Murphy, T.E.; Araujo, K.; Kookoolis, A.; Argento, A.C.; Puchalski, J. Mortality among patients with pleural effusion undergoing thoracentesis. Eur. Respir. J. 2015, 46, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Zamboni, M.M.; da Silva, C.T., Jr.; Baretta, R.; Cunha, E.T.; Cardoso, G.P. Important prognostic factors for survival in patients with malignant pleural effusion. BMC Pulm. Med. 2015, 15, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuvelmans, M.A.; van Ooijen, P.M.; Ather, S.; Silva, C.F.; Han, D.; Heussel, C.P.; Hickes, W.; Kauczor, H.-U.; Novotny, P.; Peschl, H.; et al. Lung cancer prediction by Deep Learning to identify benign lung nodules. Lung Cancer 2021, 154, 1–4. [Google Scholar] [CrossRef]
- Shen, K.R.; Bribriesco, A.; Crabtree, T.; Denlinger, C.; Eby, J.; Eiken, P.; Jones, D.R.; Keshavjee, S.; Maldonado, F.; Paul, S.; et al. The American Association for Thoracic Surgery consensus guidelines for the management of empyema. J. Thorac. Cardiovasc. Surg. 2017, 153, e129–e146. [Google Scholar] [CrossRef] [Green Version]
- Fjaellegaard, K.; Petersen, J.K.; Reuter, S.; Fischer, B.M.; Gerke, O.; Porcel, J.M.; Clementsen, P.F.; Laursen, C.B.; Bhatnagar, R.; Bodtger, U. Positron emission tomography-computed tomography (PET-CT) in suspected malignant pleural effusion. An updated systematic review and meta-analysis. Lung Cancer 2021, 162, 106–118. [Google Scholar] [CrossRef] [PubMed]




| Kappa * | |
|---|---|
| pleural thickening | |
| Overall | 0.68 |
| circumferential | 0.66 |
| Lung | 0.41 |
| Rib | 0.73 |
| Mediastinal | 0.71 |
| smooth | 0.65 |
| nodular | 0.61 |
| pleural mass | 0.63 |
| Enhancement * | |
| split pleura sign * | 0.79 |
| overall (incl. hemi split pleura sign) * | 0.77 |
| gas | 0.75 |
| microbubbles | 0.82 |
| pneumothorax | 0.97 |
| extrapleural fat stranding | 0.48 |
| loculation | 0.62 |
| amount | 0.80 |
| other findings | |
| rib destruction | 0.87 |
| blood | 0.38 |
| interlobar fluid | 0.47 |
| mediastinal lymphadenopathy | 0.52 |
| Chi2 | FP | TN | TP | FN | Sensitivity (95% CI) | Specificity (95% CI) | DOR (95% CI) | |
|---|---|---|---|---|---|---|---|---|
| pleural thickening | ||||||||
| overall | 92.81 * | 38 | 216 | 57 | 24 | 70.37 (59.04–79.74) | 85.04 (79.92–89.07) | 60.00 (39.68–90.73) |
| circumferential | 84.69 * | 4 | 250 | 30 | 51 | 37.03 (26.78–48.54) | 98.42 (95.75–99.49) | 52.08 (39.41–68.81) |
| lung | 96.13 * | 10 | 244 | 39 | 42 | 48.15 (37.02–59.46) | 96.06 (92.66–98.00) | 54.2 (39.62–74.14) |
| rib | 103.69 * | 10 | 244 | 41 | 40 | 50.62 (39.36–61.81) | 96.06 (92.66–97.99) | 57.08 (41.55–78.42) |
| mediastinal | 77.03 * | 7 | 247 | 31 | 50 | 38.27 (27.89–49.78) | 97.24 (94.16–98.79) | 48.46 (36.1–65.05) |
| smooth | 120.54 * | 21 | 233 | 54 | 27 | 66.67 (55.22–76.51) | 91.73 (87.46–94.69) | 69.33 (47.23–101.79) |
| nodular | 3.93 * | 18 | 236 | 1 | 80 | 1.23 (0.65–7.64) | 92.91 (88.84–95.63) | 0.21 (0.03–14.14) |
| pleural mass | 2 | 12 | 242 | 1 | 80 | 1.23 (0.06–7.64) | 95.28 (91.68–97.42) | 0.31 (0.05–20.55) |
| enhancement ** | ||||||||
| split pleura sign ** | 68.61 * | 10 | 134 | 38 | 26 | 59.38 (46.38–71.24) | 93.06 (87.26–96.43) | 48.72 (33.3–71.28) |
| hemi split pleura sign ** | 112.65 * | 13 | 131 | 50 | 14 | 78.13 (65.71–87.11) | 90.97 (84.75–94.91) | 82.2 (49.19–137.38) |
| gas | 39.14 * | 52 | 202 | 46 | 35 | 56.79 (45.33–67.60) | 79.53 (73.93–84.21) | 31.78 (21.93–46.08) |
| microbubbles | 87.93 * | 22 | 232 | 46 | 35 | 56.79 (45.33–67.60) | 91.34 (87.01–94.37) | 51.61 (36.37–73.22) |
| pneumothorax | 16.71 * | 47 | 207 | 33 | 48 | 40.74 (30.13–52.24) | 81.49 (76.05–85.96) | 21.91 (15.21–31.57) |
| extrapleural fat stranding | 59.1 * | 23 | 231 | 38 | 43 | 46.91 (35.85–58.27) | 90.94 (86.55–94.05) | 39.7 (28.34–55.59) |
| loculation | 39.14 * | 54 | 200 | 65 | 16 | 80.24 (69.61–87.95) | 78.74 (73.09–83.50) | 73.74 (44.76–121.47) |
| amount | 0.106 | 206 | 48 | 67 | 14 | 82.72 (72.36–89.90) | 19 (14.39–24.37) | 10.87 (0.66–18.02) |
| other findings | ||||||||
| rib destruction | 0.86 | 8 | 246 | 1 | 80 | 1.23 (0.06–7.64) | 96.85 (93.66–98.53) | 0.45 (0.07–29.02) |
| interlobar fluid | 5.59 * | 128 | 126 | 53 | 28 | 65.43 (53.96–75.43) | 49.61 (43.32–55.91) | 16.11 (10.75–24.13) |
| mediastinal lymphadenopathy | 5.485 * | 77 | 177 | 36 | 45 | 44.44 (33.55–55.88) | 69.69 (63.57–75.19) | 15.72 (10.8–22.87) |
| diagnosis | ||||||||
| empyema | 135.163 * | 23 | 231 | 59 | 22 | 72.84 (61.63–81.85) | 90.94 (86.55–94.05) | 82.74 (54.28–126.13) |
| pleura carcinomatosis | 141 * | 12 | 289 | 24 | 10 | 70.59 (52.33–84.29) | 96.01 (92.96–97.83) | 19.93 (10.39–38.25) |
| CT Features | Median Hospital Stay Time in All Patients | Survival Time (Kaplan-Meier-Analysis) | ||||||
|---|---|---|---|---|---|---|---|---|
| pleural thickening | with (d) | without (d) | U | p | mean with in days | mean without (d) | χ2 | p |
| overall | 20 | 14 | 10514 | 0.319 | 1094 (846–1069) | 957 (846–1069) | 1.774 | 0.183 |
| circumferential | 23 | 15 | 4220 | 0.105 | 1191 (921–1463) | 968 (867–1069) | 2.485 | 0.115 |
| lung | 22 | 14 | 6221 | 0.236 | 1238 (1012–1464) | 945 (842–1048) | 6.141 | 0.013 |
| rib | 22 | 15 | 6094 | 0.083 | 1220 (982–1459) | 955 (852–1059) | 3.369 | 0.066 |
| mediastinal | 21 | 15 | 5006 | 0.283 | 1110 (844–1376) | 976 (874–1077) | 1.371 | 0.242 |
| smooth | 20 | 15 | 9117 | 0.447 | 1242 (1045–1441) | 925 (818–1033) | 7.27 | 0.007 |
| nodular | 20 | 15 | 2721 | 0.52 | 445 (147–734) | 1026 (928–1124) | 4.131 | 0.042 |
| pleural mass | 23 | 15 | 1828 | 0.459 | 432 (85–779) | 1018 (921–1115) | 3.6 | 0.057 |
| enhancement * | ||||||||
| split pleura sign * | 20 | 15 | 3097 | 0.048 | 1214 (991–1438) | 1044 (906–1182) | 1.88 | 0.17 |
| overall (incl. hemi split pleura sign) * | 20 | 14 | 3558 | 0.014 | 1193 (995–1391) | 1032 (887–1176) | 2.362 | 0.124 |
| gas | 19 | 14 | 10630 | 0.269 | 1009 (838–1181) | 990 (875–1104) | 0.055 | 0.815 |
| microbubbles | 20 | 14 | 7976 | 0.144 | 1106 (903–1308) | 962 (855–1069) | 1.017 | 0.313 |
| pneumothorax | 18 | 15 | 9931 | 0.801 | 909 (721–1098) | 1025 (914–1135) | 1.753 | 0.186 |
| extrapleural fat stranding | 21 | 14 | 7431 | 0.203 | 1008 (902–1093) | 992 (886–1097) | 0.002 | 0.969 |
| loculation | 19 | 13 | 12054 | 0.419 | 1141 (982–1300) | 911 (793–1029) | 4.951 | 0.026 |
| amount | 16 | 14 | 7072 | 0.052 | 978 (872–1084) | 1076 (857.74–1295) | 0.555 | 0.456 |
| other findings | ||||||||
| rib destruction | 10 | 15 | 1227 | 0.416 | 264 (6–522) | 1013 (916–1110) | 2.645 | 0.104 |
| blood | 15 | 15 | 142 | 0.34 | 642 (225–1059) | 1005 (908–1103) | 0.722 | 0.396 |
| interlobar fluid | 18 | 14 | 12162 | 0.064 | 988 (902–1093) | 1009 (869–1150) | 0.018 | 0.894 |
| mediastinal lymphadenopathy | 17 | 15 | 11960 | 0.572 | 894 (732–1056) | 1040 (923–1156) | 1.512 | 0.219 |
| diagnosis | ||||||||
| empyema | 20 | 14 | 8695 | 0.035 | 1257 (1074–1440) | 911 (801–1021) | 7.617 | 0.006 |
| pleura carcinomatosis | 17 | 15 | 4631 | 0.19 | 414 (212–616) | 1062 (961–1162) | 11.535 | 0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sexauer, R.; Stieltjes, B.; Bremerich, J.; D’Antonoli, T.A.; Schmidt, N. Considerations on Baseline Generation for Imaging AI Studies Illustrated on the CT-Based Prediction of Empyema and Outcome Assessment. J. Imaging 2022, 8, 50. https://doi.org/10.3390/jimaging8030050
Sexauer R, Stieltjes B, Bremerich J, D’Antonoli TA, Schmidt N. Considerations on Baseline Generation for Imaging AI Studies Illustrated on the CT-Based Prediction of Empyema and Outcome Assessment. Journal of Imaging. 2022; 8(3):50. https://doi.org/10.3390/jimaging8030050
Chicago/Turabian StyleSexauer, Raphael, Bram Stieltjes, Jens Bremerich, Tugba Akinci D’Antonoli, and Noemi Schmidt. 2022. "Considerations on Baseline Generation for Imaging AI Studies Illustrated on the CT-Based Prediction of Empyema and Outcome Assessment" Journal of Imaging 8, no. 3: 50. https://doi.org/10.3390/jimaging8030050
APA StyleSexauer, R., Stieltjes, B., Bremerich, J., D’Antonoli, T. A., & Schmidt, N. (2022). Considerations on Baseline Generation for Imaging AI Studies Illustrated on the CT-Based Prediction of Empyema and Outcome Assessment. Journal of Imaging, 8(3), 50. https://doi.org/10.3390/jimaging8030050






