PyPore3D: An Open Source Software Tool for Imaging Data Processing and Analysis of Porous and Multiphase Media
Abstract
:1. Introduction
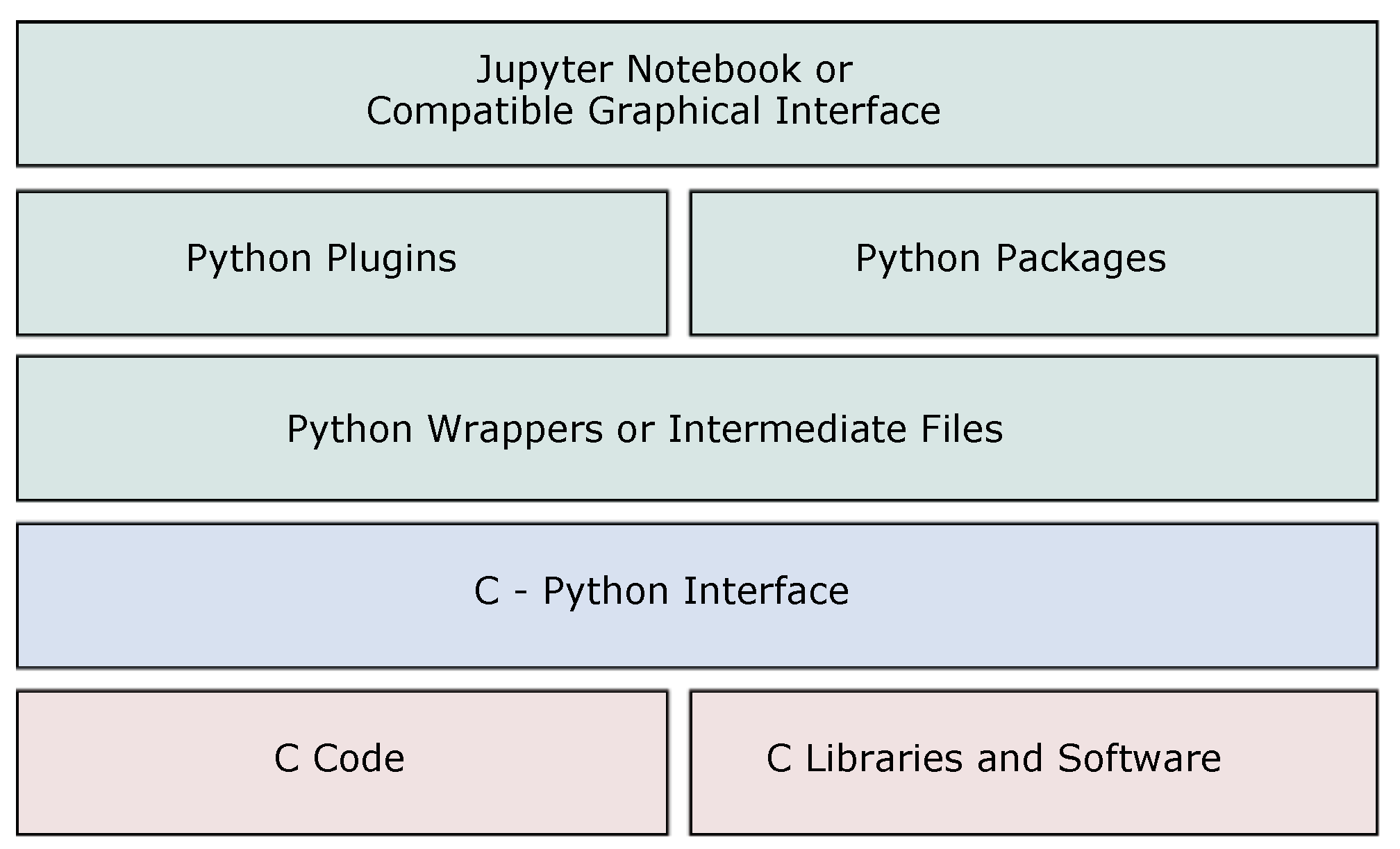
2. Software Architecture
- Combining the benefits of open source tools and the simplicity of Python language with the powerful performance of C (especially with respect to integration with parallel computing and memory management);
- Providing easy means for pipeline customisation through a modular architecture.
2.1. Pore3D C Core and C Libraries
2.2. C-Python Interface
2.3. Python Wrappers
2.4. Python Plugins and Packages
2.5. Jupyter Notebook or Compatible Graphical Interface
3. Application Examples
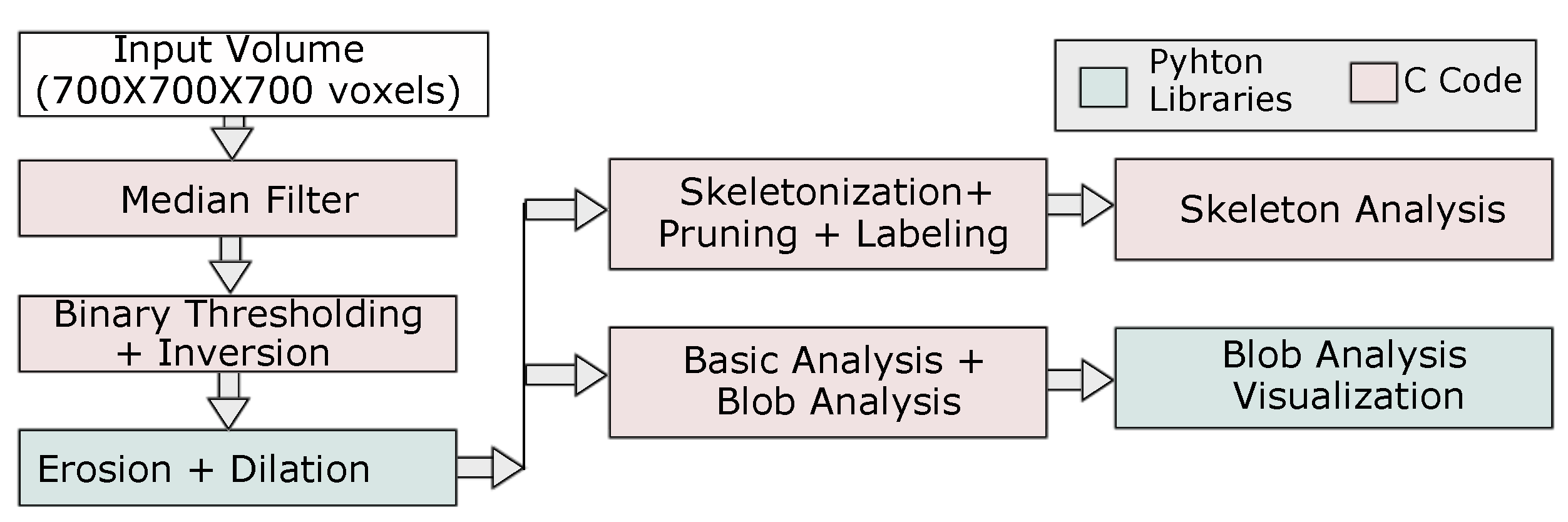
3.1. Statistical and Topological Pore Analysis
3.1.1. Basic Analysis
- Volume density (VV): quantifies the sample porosity. This is computed by the percentage of object voxels with respect to the considered volume.
- Specific surface area (SV): is a measure of the surface of the object with respect to the total volume.
- Mean curvature (MV): is an index of the dominance of convex or concave shapes.
- Euler characteristics (): is an index for the connectivity of the object network [49].
3.1.2. Connected Components (Blob) Analysis
3.1.3. Skeleton Analysis
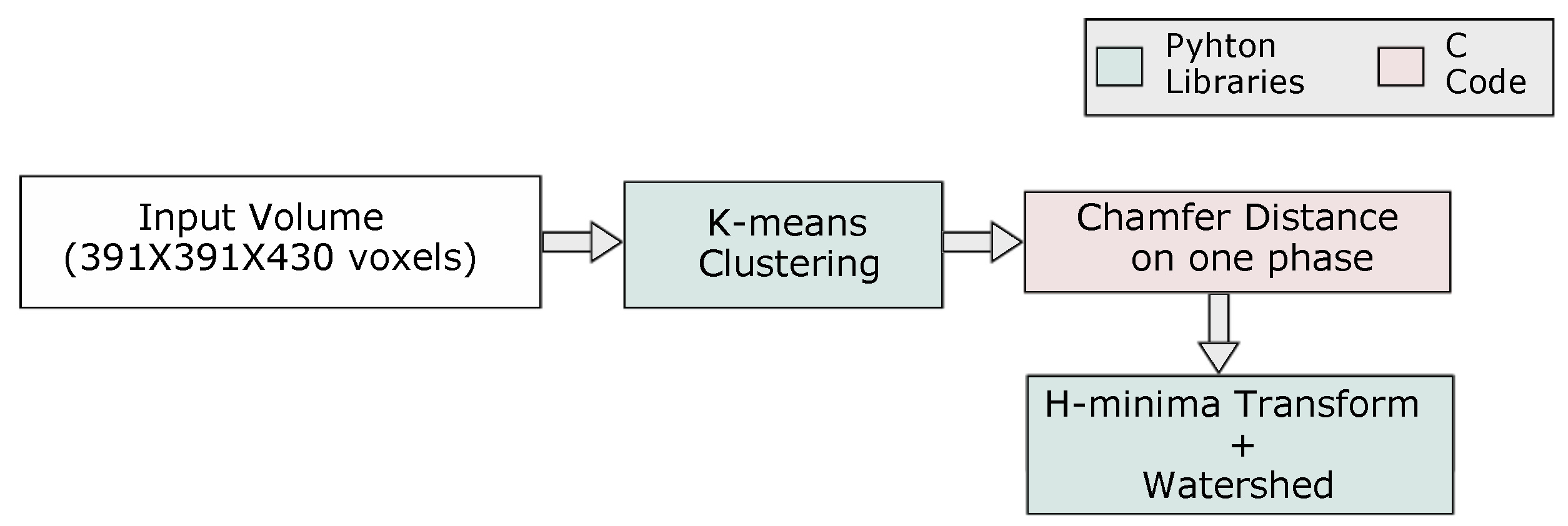
3.2. Analysis of Multiphase Materials
3.3. Morphometric Analysis
- BvTv: The ratio between bone volume and total volume;
- BsBv: The ratio between bone surface and bone volume;
- TbN: The trabecular thickness;
- TbTh: The trabecular separation of the solid phase objects;
- TbSp: Trabecular number measuring the number of traversals across a solid structure.
4. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maire, E.; Withers, P.J. Quantitative X-ray tomography. Int. Mater. Rev. 2014, 59, 1–43. [Google Scholar] [CrossRef] [Green Version]
- Oliphant, T.E. Python for Scientific Computing. Comput. Sci. Eng. 2007, 9, 10–20. [Google Scholar] [CrossRef] [Green Version]
- Beazley, D. Automated scientific software scripting with SWIG. Future Gener. Comput. Syst. 2003, 19, 599–609. [Google Scholar] [CrossRef]
- Behnel, S.; Bradshaw, R.; Citro, C.; Dalcin, L.; Seljebotn, D.S.; Smith, K. Cython: The Best of Both Worlds. Comput. Sci. Eng. 2011, 13, 31–39. [Google Scholar] [CrossRef]
- Volumegraphics. VG Studio Max. Available online: https://www.volumegraphics.com/en/products/vgsm.html (accessed on 13 June 2022).
- Thermo Fisher. Imaging Data Visualization, Analysis, and Management Software Solutions. Available online: https://www.thermofisher.com/it/en/home/electron-microscopy/products/software-em-3d-vis/3d-visualization-analysis-software.html (accessed on 13 June 2022).
- Mavi Home Page. Available online: http://www.mavi-3d.de (accessed on 13 June 2022).
- DIPlib Home Page. Available online: https://diplib.org/ (accessed on 13 June 2022).
- ITK Home Page. Available online: http://www.itk.org/ (accessed on 13 June 2022).
- PoreSpy Home Page. Available online: https://porespy.org/index.html (accessed on 13 June 2022).
- Lindquist, W.; Venkatarangan, A. Investigating 3D geometry of porous media from high resolution images. Phys. Chem. Earth Part A Solid Earth Geod. 1999, 24, 593–599. [Google Scholar] [CrossRef]
- Ketcham, R.A. Computational methods for quantitative analysis of three-dimensional features in geological specimens. Geosphere 2005, 1, 32–41. [Google Scholar] [CrossRef]
- Ketcham, R.A.; Ryan, T.M. Quantification and visualization of anisotropy in trabecular bone. J. Microsc. 2004, 213, 158–171. [Google Scholar] [CrossRef] [Green Version]
- iMorph Home Page. Available online: http://imorph.sourceforge.net (accessed on 13 June 2022).
- Tromba, G.; Abrami, A.; Casarin, K.; Chenda, V.; Dreossi, D.; Mancini, L.; Menk, R.H.; Quai, E.; Sodini, N.; Vascotto, A.; et al. The SYRMEP Beamline of Elettra: Clinical Mammography and Bio-medical Applications. AIP Conf. Proc. 2010, 1266, 18–23. [Google Scholar] [CrossRef]
- Batenburg, J.; De Carlo, F.; Mancini, L.; Sijbers, J. Advanced x-ray tomography: Experiment, modeling, and algorithms. Meas. Sci. Technol. 2018, 29, 080101. [Google Scholar] [CrossRef] [Green Version]
- Salvo, L.; Cloetens, P.; Maire, E.; Zabler, S.; Blandin, J.; Buffière, J.; Ludwig, W.; Boller, E.; Bellet, D.; Josserond, C. X-ray micro-tomography an attractive characterisation technique in materials science. Nucl. Instrum. Methods Phys. Res. Sect. B Beam Interact. Mater. Atoms 2003, 200, 273–286. [Google Scholar] [CrossRef]
- Simpleitk Home Page. Available online: https://simpleitk.org (accessed on 13 June 2022).
- LaRue, A.; Baker, D.R.; Polacci, M.; Allard, P.; Sodini, N. Can vesicle size distributions assess eruption intensity during volcanic activity? Solid Earth 2013, 4, 373–380. [Google Scholar] [CrossRef] [Green Version]
- Mancini, L.; Arzilli, F.; Polacci, M.; Voltolini, M. Editorial: Recent Advancements in X-Ray and Neutron Imaging of Dynamic Processes in Earth Sciences. Front. Earth Sci. 2020, 8, 588463. [Google Scholar] [CrossRef]
- Giuliani, A. Advanced High-Resolution Tomography in Regenerative Medicine: Three-Dimensional Exploration into the Interactions between Tissues, Cells, and Biomaterials; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Brun, F.; Turco, G.; Accardo, A.; Paoletti, S. Automated quantitative characterization of alginate/hydroxyapatite bone tissue engineering scaffolds by means of micro-CT image analysis. J. Mater. Sci. Mater. Med. 2011, 22, 2617–2629. [Google Scholar] [CrossRef] [PubMed]
- Tavella, S.; Ruggiu, A.; Giuliani, A.; Brun, F.; Canciani, B.; Manescu, A.; Marozzi, K.; Cilli, M.; Costa, D.; Liu, Y.; et al. Bone turnover in wild type and pleiotrophin-transgenic mice housed for three months in the International Space Station (ISS). PLoS ONE 2012, 7, e33179. [Google Scholar] [CrossRef]
- ORS. Dragonfly. Available online: https://www.theobjects.com/dragonfly/index.html (accessed on 13 June 2022).
- ImageJ Home Page. Available online: https://imagej.nih.gov/ij/ (accessed on 13 June 2022).
- Brun, F.; Mancini, L.; Kasae, P.; Favretto, S.; Dreossi, D.; Tromba, G. Pore3D: A software library for quantitative analysis of porous media. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrometers Detect. Assoc. Equip. 2010, 615, 326–332. [Google Scholar] [CrossRef]
- Zandomeneghi, D.; Voltolini, M.; Mancini, L.; Brun, F.; Dreossi, D.; Polacci, M. Quantitative analysis of X-ray microtomography images of geomaterials: Application to volcanic rocks. Geosphere 2010, 6, 793–804. [Google Scholar] [CrossRef] [Green Version]
- SWIG Home Page. Available online: http://www.swig.org/ (accessed on 13 June 2022).
- Plotly Home Page. Available online: https://plotly.com (accessed on 13 June 2022).
- Elettra Home Page. Available online: https://www.elettra.trieste.it/elettra-beamlines/syrmep.html (accessed on 13 June 2022).
- Stroeven, P.; Hu, J.; Guo, Z. Shape assessment of particles in concrete technology: 2D image analysis and 3D stereological extrapolation. Cem. Concr. Compos. 2009, 31, 84–91. [Google Scholar] [CrossRef]
- Baker, D.; Mancini, L.; Polacci, M.; Higgins, M.; Gualda, G.; Hill, R.; Rivers, M. An introduction to the application of X-ray microtomography to the three-dimensional study of igneous rocks. Lithos 2012, 148, 262–276. [Google Scholar] [CrossRef]
- Lanzafame, G.; Iezzi, G.; Mancini, L.; Lezzi, F.; Mollo, S.; Ferlito, C. Solidification and Turbulence (Non-laminar) during Magma Ascent: Insights from 2D and 3D Analyses of Bubbles and Minerals in an Etnean Dyke. J. Petrol. 2017, 58, 1511–1533. [Google Scholar] [CrossRef]
- Liedl, A.; Buono, G.; Lanzafame, G.; Dabagov, S.B.; Della Ventura, G.; Hampai, D.; Mancini, L.; Marcelli, A.; Pappalardo, L. A 3D imaging textural characterization of pyroclastic products from the 1538 AD Monte Nuovo eruption (Campi Flegrei, Italy). Lithos 2019, 340, 316–331. [Google Scholar] [CrossRef]
- Voltolini, M.; Barnard, H.; Creux, P.; Ajo-Franklin, J. A new mini-triaxial cell for combined high-pressure and high-temperature in situ synchrotron X-ray microtomography experiments up to 400 °C and 24 MPa. J. Synchrotron Radiat. 2019, 26, 238–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kudrna Prašek, M.; Pistone, M.; Baker, D.R.; Sodini, N.; Marinoni, N.; Lanzafame, G.; Mancini, L. A compact and flexible induction furnace for in situ X-ray microradiograhy and computed microtomography at Elettra: Design, characterization and first tests. J. Synchrotron Radiat. 2018, 25, 1172–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arzilli, F.; Spina, G.L.; Burton, M.R.; Polacci, M.; Gall, N.L.; Hartley, M.E.; Genova, D.D.; Cai, B.; Vo, N.T.; Bamber, E.C.; et al. Magma fragmentation in highly explosive basaltic eruptions induced by rapid crystallization. Nat. Geosci. 2019, 12, 1023–1028. [Google Scholar] [CrossRef]
- Dobson, K.J.; Allabar, A.; Bretagne, E.; Coumans, J.; Cassidy, M.; Cimarelli, C.; Coats, R.; Connolley, T.; Courtois, L.; Dingwell, D.B.; et al. Quantifying Microstructural Evolution in Moving Magma. Front. Earth Sci. 2020, 8, 287. [Google Scholar] [CrossRef]
- Le Gall, N.; Arzilli, F.; La Spina, G.; Polacci, M.; Cai, B.; Hartley, M.E.; Vo, N.T.; Atwood, R.C.; Di Genova, D.; Nonni, S.; et al. In situ quantification of crystallisation kinetics of plagioclase and clinopyroxene in basaltic magma: Implications for lava flow. Earth Planet. Sci. Lett. 2021, 568, 117016. [Google Scholar] [CrossRef]
- Polacci, M.; Mancini, L.; Baker, D.R. The contribution of synchrotron X-ray computed microtomography to understanding volcanic processes. J. Synchrotron Radiat. 2010, 17, 215–221. [Google Scholar] [CrossRef] [Green Version]
- Lanzafame, G.; Casetta, F.; Giacomoni, P.P.; Donato, S.; Mancini, L.; Coltorti, M.; Ntaflos, T.; Ferlito, C. The Skaros effusive sequence at Santorini (Greece): Petrological and geochemical constraints on an interplinian cycle. Lithos 2020, 362–363, 105504. [Google Scholar] [CrossRef]
- Lanzafame, G.; Ferlito, C.; Donato, S. Combining chemical and X-Ray microtomography investigations on crustal xenoliths at Mount Etna: Evidence of volcanic gas fluxing. Ann. Geophys. 2018, 61, VO672. [Google Scholar] [CrossRef]
- Lanzafame, G.; Casetta, F.; Giacomoni, P.P.; Coltorti, M.; Ferlito, C. The Rare Trachyandesitic Lavas at Mount Etna: A Case Study to Investigate Eruptive Process and Propose a New Interpretation for Magma Genesis. Minerals 2021, 11, 333. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Nikopoulos, N.; Pitas, I. An efficient algorithm for 3d binary morphological transformations with 3d structuring elements for arbitrary size and shape. IEEE Workshop Nonlinear Signal Image Process. 2000, 9, 283–286. [Google Scholar]
- Whitehouse, W. The quantitative morphology of anisotropic trabecular bone. J. Microsc. 1974, 101, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Harrigan, T.P.; Mann, R.W. Characterization of microstructural anisotropy in orthotropic materials using a second rank tensor. J. Mater. Sci. 1984, 19, 761–767. [Google Scholar] [CrossRef]
- Ohser, J.; Schladitz, K. Front Matter. In 3D Images of Materials Structures; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar] [CrossRef]
- Arns, C.H.; Knackstedt, M.A.; Pinczewski, W.V.; Mecke, K.R. Euler-Poincaré characteristics of classes of disordered media. Phys. Rev. E 2001, 63, 031112. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.; Kashyap, R.; Chu, C. Building Skeleton Models via 3-D Medial Surface Axis Thinning Algorithms. CVGIP Graph. Model. Image Process. 1994, 56, 462–478. [Google Scholar] [CrossRef]
- Pudney, C. Distance-Ordered Homotopic Thinning: A Skeletonization Algorithm for 3D Digital Images. Comput. Vis. Image Underst. 1998, 72, 404–413. [Google Scholar] [CrossRef]
- Palágyi, K.; Kuba, A. A Parallel 3D 12-Subiteration Thinning Algorithm. Graph. Models Image Process. 1999, 61, 199–221. [Google Scholar] [CrossRef] [Green Version]
- Svensson, S.; Sanniti di Baja, G. Simplifying curve skeletons in volume images. Comput. Vis. Image Underst. 2003, 90, 242–257. [Google Scholar] [CrossRef]
- Vincent, L.; Soille, P. Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Trans. Pattern Anal. Mach. Intell. 1991, 13, 583–598. [Google Scholar] [CrossRef] [Green Version]
- Soille, P. Morphological Image Analysis: Principles and Applications; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Beare, R.; Lehmann, G.G. The watershed transform in ITK-discussion and new developments. Afr. Insight 2006, 1–24. [Google Scholar] [CrossRef]
- Paganin, D.; Mayo, S.; Gureyev, T.; Miller, P.; Wilkins, S. Simultaneous Phase and Amplitude Extraction from a Single Defocused Image of a Homogeneous Object. J. Microsc. 2002, 206, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Parfitt, A. Bone histomorphometry: Standardization of nomenclature, symbols and units (summary of proposed system). Bone 1988, 9, 67–69. [Google Scholar] [CrossRef]
- Simmons, C.A.; Hipp, J.A. Method-based differences in the automated analysis of the three-dimensional morphology of trabecular bone. Off. J. Am. Soc. Bone Miner. Res. 1997, 12, 942–947. [Google Scholar] [CrossRef] [PubMed]
- Mancini, L.; Aboulhassan, A.; Brun, F.; Contillo, A.; Kourousias, G.; Lanzafame, G.N.; Mancini, L.; Voltolini, M. PyPore3D [Data Set]. 2022. Elettra Sincrotrone Trieste. Available online: https://vuo.elettra.eu/pls/vuo/open_access_data_portal.show_view_investigation?FRM_ID=10099 (accessed on 13 June 2022).

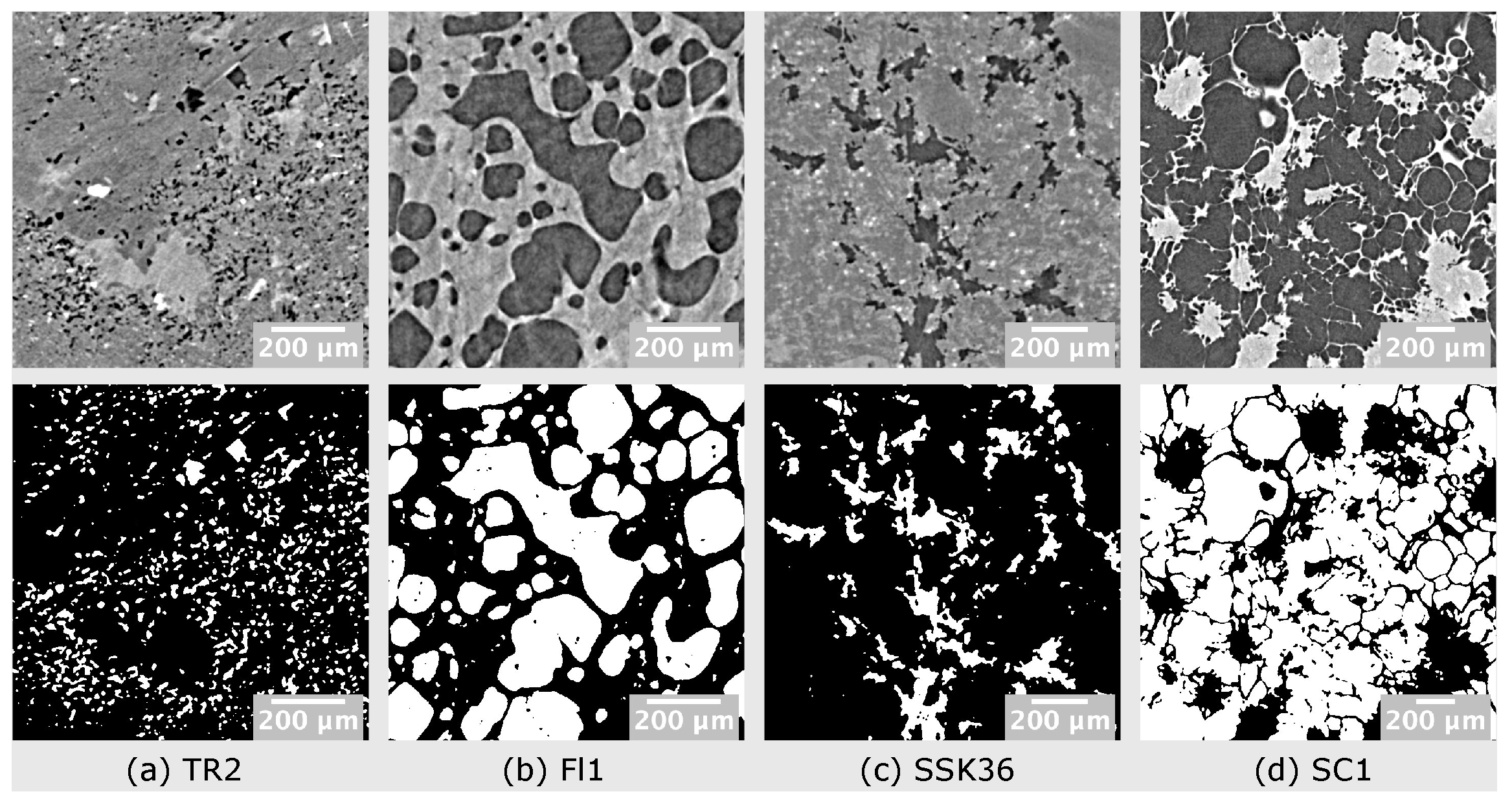
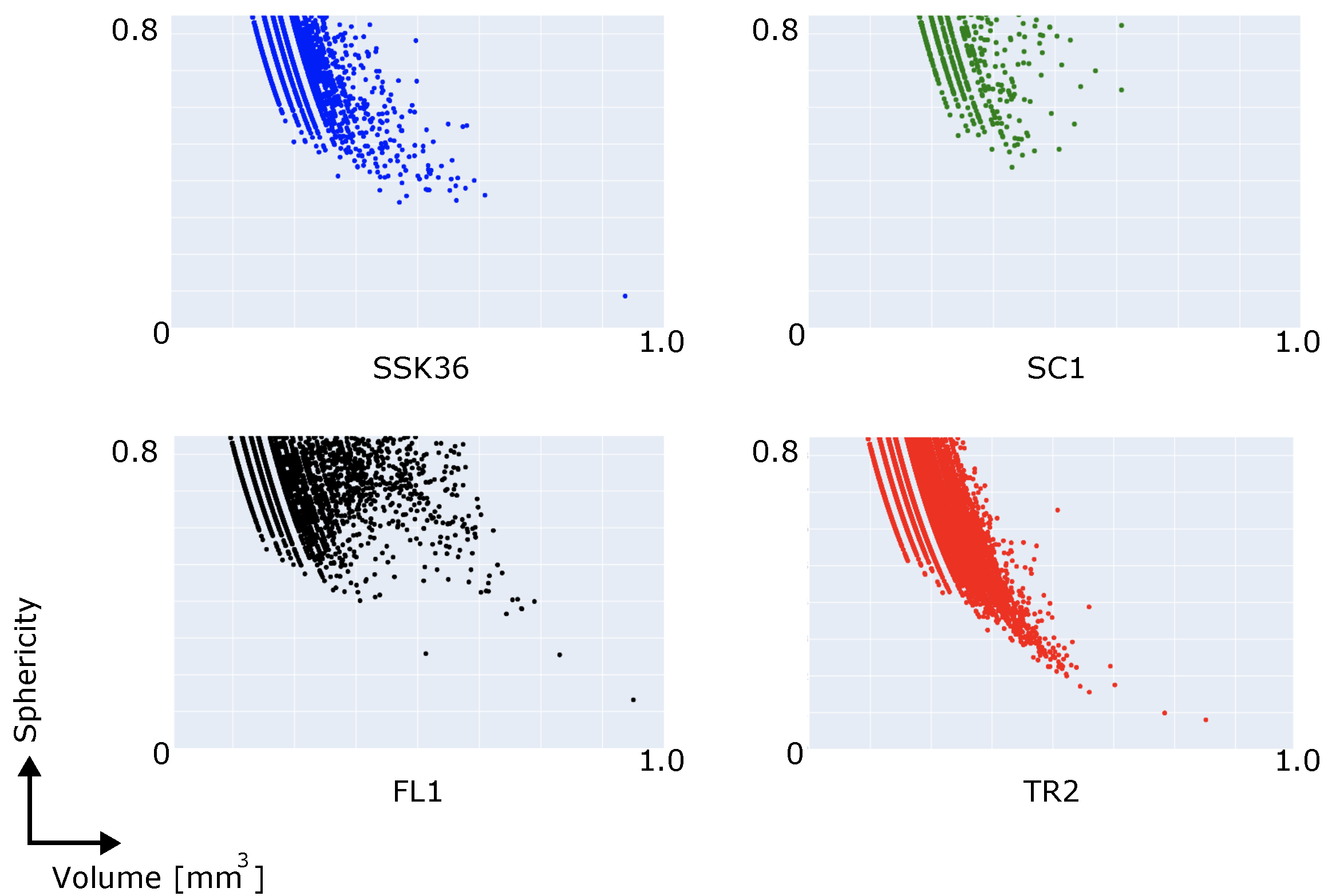



| TR2 | FL1 | SSK36 | SC1 | |
|---|---|---|---|---|
| Sample to detector dist. (mm) | 150 | 150 | 200 | 100 |
| Pixel Size (μm) | 1.37 | 1.37 | 1.8 | 2.6 |
| N° of Projections | 1800 | 1800 | 1800 | 1800 |
| Exposure time per projection (s) | 2 | 2 | 2 | 1 |
| TR2 | FL1 | SSK36 | SC1 | |
|---|---|---|---|---|
| Median width | 3 | 3 | 3 | 3 |
| Automatic 3D single Otsu threshold | 104 | 125 | 105 | 122 |
| Erosion/Dilation width | 3 | 3 | 3 | 3 |
| LKC Pruning | 5 | 5 | 5 | 5 |
| Isotropic voxel size (m) | 1.37 | 1.37 | 1.8 | 2.6 |
| TR2 | FL1 | SSK36 | SC1 | |
|---|---|---|---|---|
| (mm3) | 37,436 | 9842 | 1873 | −2557 |
| MV (mm2) | 5585 | 611 | 593 | 60 |
| SV (mm1) | 43 | 29 | 18 | 27 |
| VV | 0.1 | 0.5 | 0.1 | 0.7 |
| EA | 0.08 | 0.08 | 0.11 | 0.03 |
| I | 0.89 | 0.89 | 0.85 | 0.85 |
| Connectivity Density | 5214 | 644 | 3273 | 3193 |
| Method | Time (s) |
|---|---|
| K-means (4 classes) | 78 |
| Inverted Distance Field | 2 |
| H-minima Filter | 2 |
| Watershed Filtering | 16 |
| Morphometric Indicator | Value |
|---|---|
| BvTv | 0.1 |
| BsBv (mm−1) | 32 |
| TbN (mm) | 1.61 |
| TbTh (mm) | 0.06 |
| TbSp (mm−1) | 0.56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aboulhassan, A.; Brun, F.; Kourousias, G.; Lanzafame, G.; Voltolini, M.; Contillo, A.; Mancini, L. PyPore3D: An Open Source Software Tool for Imaging Data Processing and Analysis of Porous and Multiphase Media. J. Imaging 2022, 8, 187. https://doi.org/10.3390/jimaging8070187
Aboulhassan A, Brun F, Kourousias G, Lanzafame G, Voltolini M, Contillo A, Mancini L. PyPore3D: An Open Source Software Tool for Imaging Data Processing and Analysis of Porous and Multiphase Media. Journal of Imaging. 2022; 8(7):187. https://doi.org/10.3390/jimaging8070187
Chicago/Turabian StyleAboulhassan, Amal, Francesco Brun, George Kourousias, Gabriele Lanzafame, Marco Voltolini, Adriano Contillo, and Lucia Mancini. 2022. "PyPore3D: An Open Source Software Tool for Imaging Data Processing and Analysis of Porous and Multiphase Media" Journal of Imaging 8, no. 7: 187. https://doi.org/10.3390/jimaging8070187
APA StyleAboulhassan, A., Brun, F., Kourousias, G., Lanzafame, G., Voltolini, M., Contillo, A., & Mancini, L. (2022). PyPore3D: An Open Source Software Tool for Imaging Data Processing and Analysis of Porous and Multiphase Media. Journal of Imaging, 8(7), 187. https://doi.org/10.3390/jimaging8070187









