AI in Breast Cancer Imaging: A Survey of Different Applications
Abstract
:1. Introduction
1.1. Breast Cancer: Statistics and Risk Factors
1.2. Screening and Commonly Found Lesions
1.3. The Role of Artificial Intelligence in Medical Imaging
1.4. Related Work
2. Traditional AI in Lesion Detection and Tissue Interpretation
2.1. Texture Features
- N1—Lowest Risk, parenchyma is mainly composed by fatty tissue without visible ducts.
- P1—Low Risk, ducts may occupy as far as a quadrant of the breast.
- P2—High Risk, there is a “severe involvement” of ducts that occupy more than 25% of the breast.
- DY—Highest Risk, the severe involvement seen in P2 is accompanied by dysplasia.
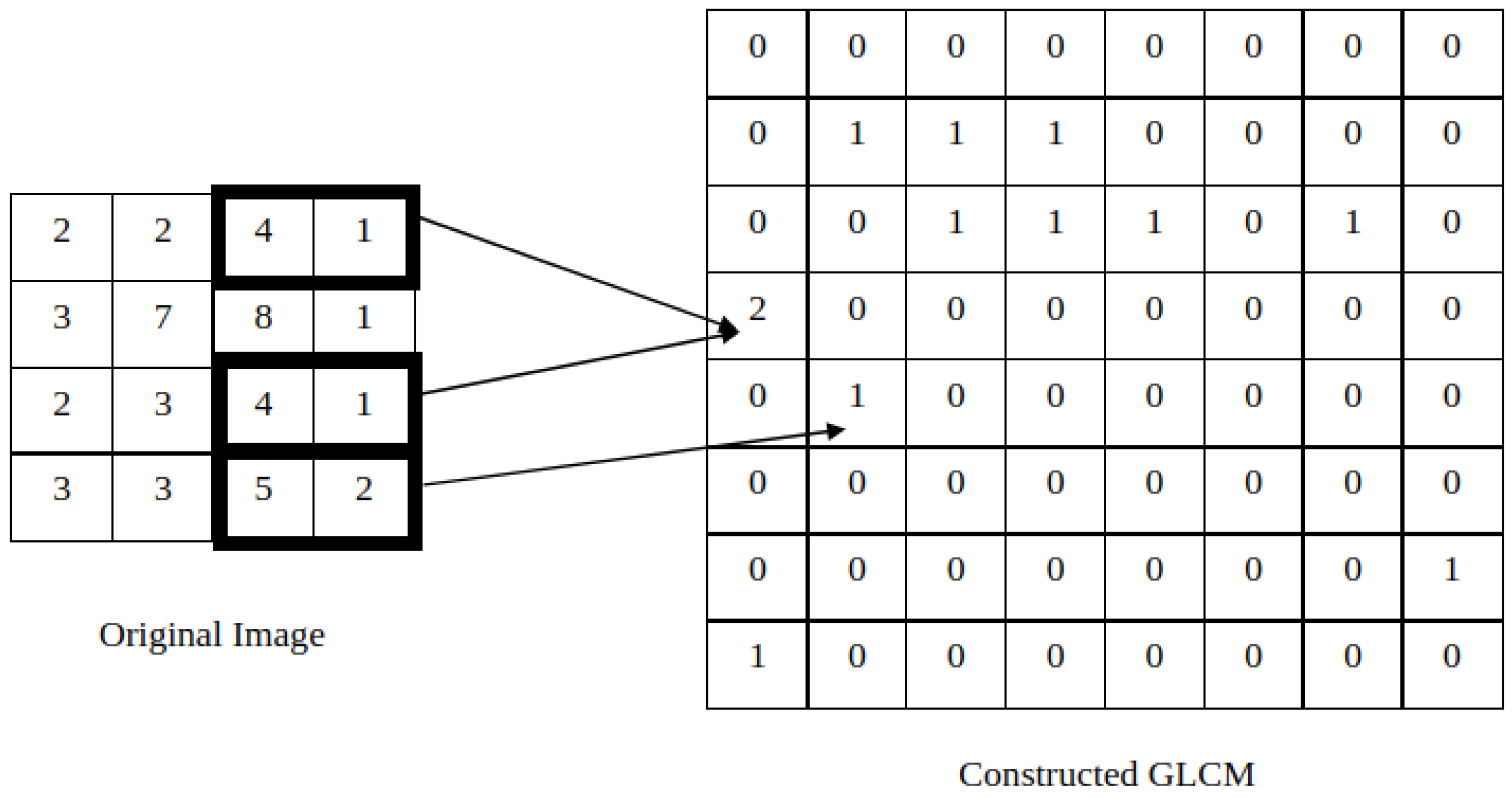
2.1.1. Co-Occurrence Features
2.1.2. Run-Length Features
2.2. Additional Features
2.3. AI in Breast Imaging Analysis
2.3.1. Machine Learning in Mammography Lesion Interpretation
2.3.2. Lesion Interpretation with Alternative Imaging Modalities
2.3.3. AI in Breast Cancer Risk Prediction
3. AI and Data Augmentation
4. Self-Supervised Learning
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA-Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA-Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA-Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Singletary, S. Rating the Risk Factors for Breast Cancer. Ann. Surg. 2003, 237, 474–482. [Google Scholar] [CrossRef]
- Pike, M.C.; Krailo, M.; Henderson, B.; Casagrande, J.; Hoel, D. ‘Hormonal’ risk factors, ‘breast tissue age’and the age-incidence of breast cancer. Nature 1983, 303, 767–770. [Google Scholar] [CrossRef]
- Barnard, M.E.; Boeke, C.E.; Tamimi, R.M. Established breast cancer risk factors and risk of intrinsic tumor subtypes. Biochim. Biophys. Acta Rev. Cancer 2015, 1856, 73–85. [Google Scholar] [CrossRef]
- Boyd, N.F.; Martin, L.J.; Yaffe, M.J.; Minkin, S. Mammographic density and breast cancer risk: Current understanding and future prospects. Breast Cancer Res. 2011, 13, 1–12. [Google Scholar] [CrossRef]
- Huo, Z.; Giger, M.L.; Wolverton, D.E.; Zhong, W.; Cumming, S.; Olopade, O.I. Computerized analysis of mammographic parenchymal patterns for breast cancer risk assessment: Feature selection. Med. Phys. 2000, 27, 4–12. [Google Scholar] [CrossRef]
- Jakes, R.; Duffy, S.; Ng, F.; Gao, F.; Ng, E. Mammographic parenchymal patterns and risk of breast cancer at and after a prevalence screen in Singaporean women. Int. J. Epidemiol. 2000, 29, 11–19. [Google Scholar] [CrossRef]
- Niell, B.L.; Freer, P.E.; Weinfurtner, R.J.; Arleo, E.K.; Drukteinis, J.S. Screening for breast cancer. Radiol. Clin. 2017, 55, 1145–1162. [Google Scholar] [CrossRef]
- Coleman, C. Early detection and screening for breast cancer. Semin. Oncol. Nurs. 2017, 33, 141–155. [Google Scholar] [CrossRef] [PubMed]
- Løberg, M.; Lousdal, M.L.; Bretthauer, M.; Kalager, M. Benefits and harms of mammography screening. Breast Cancer Res. 2015, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Geisel, J.; Raghu, M.; Hooley, R. The role of ultrasound in breast cancer screening: The case for and against ultrasound. Semin. Ultrasound CT MR 2018, 39, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Morrow, M.; Waters, J.; Morris, E. MRI for breast cancer screening, diagnosis, and treatment. Lancet 2011, 378, 1804–1811. [Google Scholar] [CrossRef]
- Rocha García, A.; Mera Fernández, D. Breast tomosynthesis: State of the art. Radiología 2019, 61, 274–285. [Google Scholar] [CrossRef]
- Ikeda, D.; Miyake, K. Breast Imaging: The Requisites; Requisites in Radiology Series; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Fischer, U.; Baum, F. Interventional Breast Imaging: Ultrasound, Mammography, and MR Guidance Techniques; Thieme: New York, NY, USA, 2011. [Google Scholar]
- Stoitsis, J.; Valavanis, I.; Mougiakakou, S.G.; Golemati, S.; Nikita, A.; Nikita, K.S. Computer aided diagnosis based on medical image processing and artificial intelligence methods. Nucl. Instrum. Methods Phys. Res. A Accel. Spectrom. Detect. Assoc. Equip. 2006, 569, 591–595. [Google Scholar] [CrossRef]
- Ongsulee, P. Artificial intelligence, machine learning and deep learning. In Proceedings of the 2017 15th International Conference on ICT and Knowledge Engineering (ICT&KE), Bangkok, Thailand, 22–24 November 2017; pp. 1–6. [Google Scholar]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine learning and deep learning. Electron. Mark. 2021, 31, 685–695. [Google Scholar] [CrossRef]
- Choy, G.; Khalilzadeh, O.; Michalski, M.; Do, S.; Samir, A.E.; Pianykh, O.S.; Geis, J.R.; Pandharipande, P.V.; Brink, J.A.; Dreyer, K.J. Current applications and future impact of machine learning in radiology. Radiology 2018, 288, 318–328. [Google Scholar] [CrossRef]
- Tuceryan, M.; Jain, A.K. Texture analysis. In Handbook of Pattern Recognition and Computer Vision; World Scientific Publishing Co., Inc.: Hackensack, NJ, USA, 1993; pp. 235–276. [Google Scholar]
- Wolfe, J.N. Breast patterns as an index of risk for developing breast cancer. AJR Am. J. Roentgenol. 1976, 126, 1130–1137. [Google Scholar] [CrossRef]
- Manduca, A.; Carston, M.J.; Heine, J.J.; Scott, C.G.; Pankratz, V.S.; Brandt, K.R.; Sellers, T.A.; Vachon, C.M.; Cerhan, J.R. Texture features from mammographic images and risk of breast cancer. Cancer Epidemiol. Biomark. Prev. 2009, 18, 837–845. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Mendel, K.R.; Lan, L.; Sheth, D.; Giger, M.L. Digital mammography in breast cancer: Additive value of radiomics of breast parenchyma. Radiology 2019, 291, 15. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. Man Cybern. Syst. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Vadakkenveettil, B. Grey Level Co-Occurrence Matrices: Generalisation and Some New Features. Int. J. Comput. Sci. Eng. Technol. 2012, 2, 151–157. [Google Scholar]
- Galloway, M.M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Li, H.; Giger, M.L.; Huo, Z.; Olopade, O.I.; Lan, L.; Weber, B.L.; Bonta, I. Computerized analysis of mammographic parenchymal patterns for assessing breast cancer risk: Effect of ROI size and location. Med. Phys. 2004, 31, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Shi, Y.; Chin, L.K.; Hutchinson, P.E.; Zhang, Y.; Chierchia, G.; Talbot, H.; Jiang, X.; Bourouina, T.; Liu, A.Q. Machine-Learning-Assisted Intelligent Imaging Flow Cytometry: A Review. Adv. Intell. Syst. 2021, 3, 2100073. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chiou, H.J.; Chou, S.Y.; Chiou, S.Y.; Wang, H.K.; Chou, Y.H.; Chiang, H.K. Computer-aided diagnosis of soft-tissue tumors using sonographic morphologic and texture features. Acad. Radiol. 2009, 16, 1531–1538. [Google Scholar] [CrossRef]
- Kayode, A.A.; Akande, N.O.; Adegun, A.A.; Adebiyi, M.O. An automated mammogram classification system using modified support vector machine. Med. Devices 2019, 12, 275. [Google Scholar] [CrossRef]
- Mohanty, A.K.; Senapati, M.R.; Beberta, S.; Lenka, S.K. Texture-based features for classification of mammograms using decision tree. Neural. Comput. Appl. 2013, 23, 1011–1017. [Google Scholar] [CrossRef]
- Wei, M.; Du, Y.; Wu, X.; Zhu, J. Automatic classification of benign and malignant breast tumors in ultrasound image with texture and morphological features. In Proceedings of the 2019 IEEE 13th International Conference on Anti-counterfeiting, Security, and Identification (ASID), Xiamen, China, 25–27 October 2019; pp. 126–130. [Google Scholar]
- Nie, K.; Chen, J.H.; Hon, J.Y.; Chu, Y.; Nalcioglu, O.; Su, M.Y. Quantitative analysis of lesion morphology and texture features for diagnostic prediction in breast MRI. Acad. Radiol. 2008, 15, 1513–1525. [Google Scholar] [CrossRef]
- Mendes, J.; Matela, N. Breast cancer risk assessment: A review on mammography-based approaches. J. Imaging 2021, 7, 98. [Google Scholar] [CrossRef]
- Tan, M.; Zheng, B.; Ramalingam, P.; Gur, D. Prediction of near-term breast cancer risk based on bilateral mammographic feature asymmetry. Acad. Radiol. 2013, 20, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Zheng, Y.; Keller, B.M.; Ray, S.; Wang, Y.; Conant, E.F.; Gee, J.C.; Kontos, D. Parenchymal texture analysis in digital mammography: A fully automated pipeline for breast cancer risk assessment. Med. Phys. 2015, 42, 4149–4160. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Mariapun, S.; Yip, C.H.; Ng, K.H.; Teo, S.H. A novel method of determining breast cancer risk using parenchymal textural analysis of mammography images on an Asian cohort. Phys. Med. Biol. 2019, 64, 035016. [Google Scholar] [CrossRef]
- Qiu, Y.; Wang, Y.; Yan, S.; Tan, M.; Cheng, S.; Liu, H.; Zheng, B. An initial investigation on developing a new method to predict short-term breast cancer risk based on deep learning technology. In Medical Imaging 2016: Computer-Aided Diagnosis; SPIE: Bellingham, WA, USA, 2016; Volume 9785, pp. 517–522. [Google Scholar]
- Yala, A.; Lehman, C.; Schuster, T.; Portnoi, T.; Barzilay, R. A deep learning mammography-based model for improved breast cancer risk prediction. Radiology 2019, 292, 60–66. [Google Scholar] [CrossRef]
- Wang, K.; Gou, C.; Duan, Y.; Lin, Y.; Zheng, X.; Wang, F.Y. Generative adversarial networks: Introduction and outlook. IEEE/CAA J. Autom. Sin. 2017, 4, 588–598. [Google Scholar] [CrossRef]
- Goodfellow, I.J. NIPS 2016 Tutorial: Generative Adversarial Networks. arXiv 2017, arXiv:1701.00160. [Google Scholar]
- Osuala, R.; Kushibar, K.; Garrucho, L.; Linardos, A.; Szafranowska, Z.; Klein, S.; Glocker, B.; Diaz, O.; Lekadir, K. A review of generative adversarial networks in cancer imaging: New applications, new solutions. arXiv 2021, arXiv:2107.09543. [Google Scholar]
- Korkinof, D.; Rijken, T.; O’Neill, M.; Yearsley, J.; Harvey, H.; Glocker, B. High-resolution mammogram synthesis using progressive generative adversarial networks. arXiv 2018, arXiv:1807.03401. [Google Scholar]
- Korkinof, D.; Heindl, A.; Rijken, T.; Harvey, H.; Glocker, B. MammoGAN: High-resolution synthesis of realistic mammograms. In Proceedings of the International Conference on Medical Imaging with Deep Learning–Extended Abstract Track, London, UK, 8–10 July 2019. [Google Scholar]
- Heusel, M.; Ramsauer, H.; Unterthiner, T.; Nessler, B.; Hochreiter, S. GANs trained by a two time-scale update rule converge to a local nash equilibrium. In Proceedings of the Advances in Neural Information Processing Systems 30 (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Man, R.; Yang, P.; Xu, B. Classification of breast cancer histopathological images using discriminative patches screened by generative adversarial networks. IEEE Access 2020, 8, 155362–155377. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Ma, X.; Wang, J.; Zheng, X.; Liu, Z.; Long, W.; Zhang, Y.; Wei, J.; Lu, Y. Automated fibroglandular tissue segmentation in breast MRI using generative adversarial networks. Phys. Med. Biol. 2020, 65, 105006. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted, Munich, Germany, 5–9 October 2015; Springer: Amsterdam, The Netherlands, 2015; pp. 234–241. [Google Scholar]
- Wu, E.; Wu, K.; Cox, D.; Lotter, W. Conditional infilling GANs for data augmentation in mammogram classification. In Image Analysis for Moving Organ, Breast, and Thoracic Images; Springer: Amsterdam, The Netherlands, 2018; pp. 98–106. [Google Scholar]
- Senaras, C.; Niazi, M.K.K.; Sahiner, B.; Pennell, M.P.; Tozbikian, G.; Lozanski, G.; Gurcan, M.N. Optimized generation of high-resolution phantom images using cGAN: Application to quantification of Ki67 breast cancer images. PLoS ONE 2018, 13, e0196846. [Google Scholar] [CrossRef] [PubMed]
- Fujioka, T.; Mori, M.; Kubota, K.; Kikuchi, Y.; Katsuta, L.; Adachi, M.; Oda, G.; Nakagawa, T.; Kitazume, Y.; Tateishi, U. Breast ultrasound image synthesis using deep convolutional generative adversarial networks. Diagnostics 2019, 9, 176. [Google Scholar] [CrossRef] [Green Version]
- Oyelade, O.N.; Ezugwu, A.E.; Almutairi, M.S.; Saha, A.K.; Abualigah, L.; Chiroma, H. A generative adversarial network for synthetization of regions of interest based on digital mammograms. Sci. Rep. 2022, 12, 1–30. [Google Scholar] [CrossRef]
- Negi, A.; Raj, A.N.J.; Nersisson, R.; Zhuang, Z.; Murugappan, M. RDA-UNET-WGAN: An accurate breast ultrasound lesion segmentation using wasserstein generative adversarial networks. Arab. J. Sci. Eng. 2020, 45, 6399–6410. [Google Scholar] [CrossRef]
- Li, Z.; Cui, Z.; Wang, S.; Qi, Y.; Ouyang, X.; Chen, Q.; Yang, Y.; Xue, Z.; Shen, D.; Cheng, J.Z. Domain Generalization for Mammography Detection via Multi-style and Multi-view Contrastive Learning. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Amsterdam, The Netherlands, 2021; pp. 98–108. [Google Scholar]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Gao, Y.; Wang, X.; Zhang, T.; Han, L.; Beets-Tan, R.; Mann, R. Self-supervised learning of mammograms with pathology aware. In Proceedings of the Medical Imaging with Deep Learning, Zurich, Switzerland, 6–8 July 2022. [Google Scholar]
- Miller, J.D.; Arasu, V.A.; Pu, A.X.; Margolies, L.R.; Sieh, W.; Shen, L. Self-Supervised Deep Learning to Enhance Breast Cancer Detection on Screening Mammography. arXiv 2022, arXiv:2203.08812. [Google Scholar]
- Ouyang, X.; Che, J.; Chen, Q.; Li, Z.; Zhan, Y.; Xue, Z.; Wang, Q.; Cheng, J.Z.; Shen, D. Self-adversarial learning for detection of clustered microcalcifications in mammograms. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September 27–1 October 2021; Springer: Amsterdam, The Netherlands, 2021; pp. 78–87. [Google Scholar]
- Srinidhi, C.L.; Martel, A.L. Improving Self-supervised Learning with Hardness-aware Dynamic Curriculum Learning: An Application to Digital Pathology. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 562–571. [Google Scholar]
- Truong, T.; Mohammadi, S.; Lenga, M. How Transferable Are Self-supervised Features in Medical Image Classification Tasks? In Proceedings of the Machine Learning for Health, PMLR, Virtual Event, 6–7 August 2021; pp. 54–74. [Google Scholar]
- Chan, H.P.; Samala, R.K.; Hadjiiski, L.M. CAD and AI for breast cancer—Recent development and challenges. Br. J. Radiol. 2019, 93, 20190580. [Google Scholar] [CrossRef]
- Hassan, N.M.; Hamad, S.; Mahar, K. Mammogram breast cancer CAD systems for mass detection and classification: A review. Multimed. Tools. Appl. 2022, 81, 20043–20075. [Google Scholar] [CrossRef]
- Le, E.; Wang, Y.; Huang, Y.; Hickman, S.; Gilbert, F. Artificial intelligence in breast imaging. Clin. Radiol. 2019, 74, 357–366. [Google Scholar] [CrossRef]


| Authors | Goal | Method/Algorithm | Imaging Modality | Results |
|---|---|---|---|---|
| Kayode et al. [32] | Benign/Malignant Lesion Differentiation | Texture Features with SVM | Mammography | Sensitivity = 94.47%; Specificity = 91.3% |
| Mohanty et al. [33] | Benign/Malignant Lesion Differentiation | Texture Features with Decistion Tree | Mammography | AUC = 0.995 |
| Wei et al. [34] | Benign/Malignant Lesion Differentiation | Texture Features/LBP with SVM | Ultrasound | Sensitivity = 87.04%; Specificity = 87.62%; AUC = 0.9342 |
| Nie et al. [35] | Benign/Malignant Lesion Differentiation | Texture/Morphology Features with Artificial Neural Network | MRI | AUC = 0.82 |
| Huo et al. [8] | High-Risk/Low-Risk group Differentiation | Texture Features with Linear Discriminant Analysis | Mammography | AUC = 0.91 |
| Tan et al. [37] | Risk Prediction based on a “prior” evaluation | Asymmetry Texture Features/risk-factors with SVM | Mammography | AUC = 0.725 |
| Zheng et al. [39] | Differentiate contra-lateral healthy images from diseased women from normal cases | Texture Features with Logistic Regression | Mammography | AUC = 0.85 |
| Qiu et al. [41] | Risk Prediction based on a “prior” evaluation | CNN | Mammography | Sensitivity = 70.3%; Specificity = 60%; AUC = 0.697 |
| Yala et al. [42] | Single-Image + Risk Factors Risk Prediction | CNN (ResNet18) | Mammography | AUC = 0.7 |
| Dimitrios Korkinof et al. [46,47] | Mammogram Synthesis | PGGAN | Mammography | ≈50% probability of identifying synthetic samples |
| Rui Man et al. [49] | Mammogram Patches Synthesis | AnoGAN | Histopathological | Classifiers with >99% accuracy |
| Xiangyuan Ma et al. [51] | Segmentation Masks Synthesis | GAN | Mammography Segmentation Masks | Dice-Coefficient > 87%; Jaccard Index > 77% |
| Eric Wu et al. [53] | Mammogram Variation | GAN | Mammography | Classifiers with accuracy of 89.6% |
| Caglar Senaras et al. [54] | Image-to-Image Mammogram Synthesis | GAN | Mammography | ≈50% probability of identifying synthetic samples |
| Li et al. [58] | Lesion Detection | SSL, GAN and CNN | Mammography | Improvements of ≈3 pp on accuracy |
| Gao et al. [60] | Normalization, classification and segmentation | SSL and CNN | Mammography | Improvements of ≈10 to 15 pp on AUC scores |
| Miller et al. [61] | Breast cancer detection | SSL and CNN | Mammography | Improved 4-fold data efficiency and ≈3 pp on accuracy |
| Ouyang et al. [62] | Detection of clustered microcalcifications | SLL and CNN | Mammography | Improvements of ≈5 pp on AUC scores |
| Srinidhi and Martel [63] | Classification | SSL, curriculum learning, CNN | Histology | Improvements of ≈2 pp on AUC scores |
| Truong et al. [64] | Classification and Detection | SSL and CNN | lymph node images, fundus images, and chest X-ray images | Improvements of ≈2 pp on AUC scores |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, J.; Domingues, J.; Aidos, H.; Garcia, N.; Matela, N. AI in Breast Cancer Imaging: A Survey of Different Applications. J. Imaging 2022, 8, 228. https://doi.org/10.3390/jimaging8090228
Mendes J, Domingues J, Aidos H, Garcia N, Matela N. AI in Breast Cancer Imaging: A Survey of Different Applications. Journal of Imaging. 2022; 8(9):228. https://doi.org/10.3390/jimaging8090228
Chicago/Turabian StyleMendes, João, José Domingues, Helena Aidos, Nuno Garcia, and Nuno Matela. 2022. "AI in Breast Cancer Imaging: A Survey of Different Applications" Journal of Imaging 8, no. 9: 228. https://doi.org/10.3390/jimaging8090228
APA StyleMendes, J., Domingues, J., Aidos, H., Garcia, N., & Matela, N. (2022). AI in Breast Cancer Imaging: A Survey of Different Applications. Journal of Imaging, 8(9), 228. https://doi.org/10.3390/jimaging8090228









