Attenuation Correction Using Template PET Registration for Brain PET: A Proof-of-Concept Study
Abstract
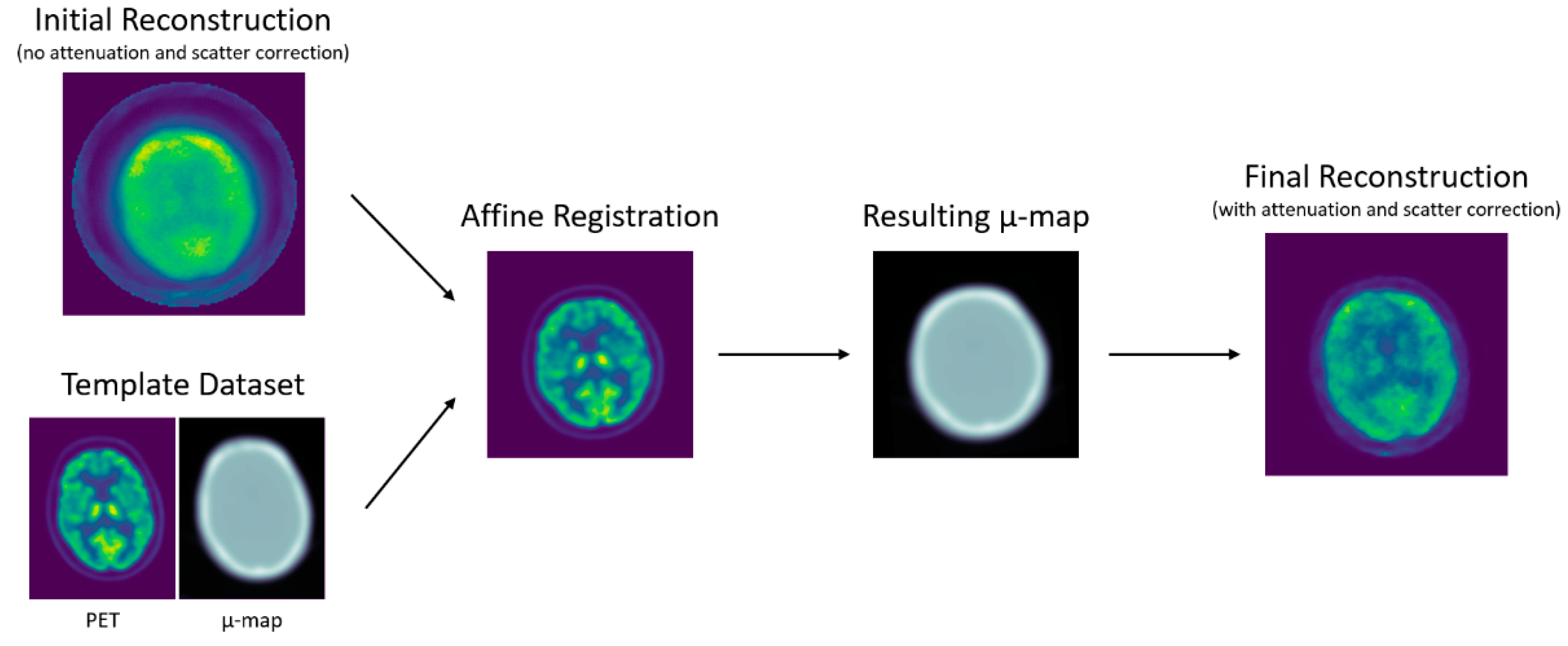
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. PET Scanner
2.1.2. Collected Data
2.2. Methods
2.2.1. Image Reconstruction
2.2.2. Attenuation Map Computation
2.2.3. Image Quality Metrics
3. Results
3.1. Comparison of Patient-Specific Attenuation Map vs. Generic Template Attenuation Map
3.2. Requirements for Template Attenuation Map
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wagenknecht, G.; Kaiser, H.-J.; Mottaghy, F.M.; Herzog, H. MRI for Attenuation Correction in PET: Methods and Challenges. Magn. Reson. Mater. Phys. Biol. Med. 2013, 26, 99–113. [Google Scholar] [CrossRef]
- Lee, J.S. A Review of Deep-Learning-Based Approaches for Attenuation Correction in Positron Emission Tomography. IEEE Trans. Radiat. Plasma Med. Sci. 2021, 5, 160–184. [Google Scholar] [CrossRef]
- Bailey, D.L. Transmission Scanning in Emission Tomography. Eur. J. Nucl. Med. Mol. Imaging 1998, 25, 774–787. [Google Scholar] [CrossRef] [PubMed]
- Ladefoged, C.N.; Law, I.; Anazodo, U.; St. Lawrence, K.; Izquierdo-Garcia, D.; Catana, C.; Burgos, N.; Cardoso, M.J.; Ourselin, S.; Hutton, B.; et al. A Multi-Centre Evaluation of Eleven Clinically Feasible Brain PET/MRI Attenuation Correction Techniques Using a Large Cohort of Patients. NeuroImage 2017, 147, 346–359. [Google Scholar] [CrossRef]
- Sekine, T.; Buck, A.; Delso, G.; ter Voert, E.E.G.W.; Huellner, M.; Veit-Haibach, P.; Warnock, G. Evaluation of Atlas-Based Attenuation Correction for Integrated PET/MR in Human Brain: Application of a Head Atlas and Comparison to True CT-Based Attenuation Correction. J. Nucl. Med. 2016, 57, 215–220. [Google Scholar] [CrossRef]
- Rezaei, A.; Defrise, M.; Nuyts, J. ML-Reconstruction for TOF-PET With Simultaneous Estimation of the Attenuation Factors. IEEE Trans. Med. Imaging 2014, 33, 1563–1572. [Google Scholar] [CrossRef]
- Benoit, D.; Ladefoged, C.N.; Rezaei, A.; Keller, S.H.; Andersen, F.L.; Højgaard, L.; Hansen, A.E.; Holm, S.; Nuyts, J. Optimized MLAA for Quantitative Non-TOF PET/MR of the Brain. Phys. Med. Biol. 2016, 61, 8854–8874. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ahnen, M.L.; Fischer, J.; Kuegler, N.; Mikhaylova, E.; Becker, R.; Lustermann, W.; Soerensen, J.; Buck, A.; Weber, B.; Sabri, O.; et al. Performance of the Ultra-Compact Fully Integrated Brain PET System BPET. In Proceedings of the 2020 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC), Boston, MA, USA, 31 October–7 November 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–4. [Google Scholar]
- Positrigo AG Exploratory Study Using a New Head-Only PET Scanner (ExploreBPET). Available online: https://clinicaltrials.gov/ct2/show/NCT04511546 (accessed on 10 December 2022).
- Thielemans, K.; Tsoumpas, C.; Mustafovic, S.; Beisel, T.; Aguiar, P.; Dikaios, N.; Jacobson, M.W. STIR: Software for Tomographic Image Reconstruction Release 2. Phys. Med. Biol. 2012, 57, 867–883. [Google Scholar] [CrossRef] [PubMed]
- Dao, V.; Mikhaylova, E.; Ahnen, M.L.; Fischer, J.; Thielemans, K.; Tsoumpas, C. Evaluation of STIR Library Adapted for PET Scanners with Non-Cylindrical Geometry. J. Imaging 2022, 8, 172. [Google Scholar] [CrossRef]
- Khateri, P.; Fischer, J.; Lustermann, W.; Tsoumpas, C.; Dissertori, G. Implementation of Cylindrical PET Scanners with Block Detector Geometry in STIR. EJNMMI Phys. 2019, 6, 15. [Google Scholar] [CrossRef]
- Stearns, C.W.; McDaniel, D.L.; Kohlmyer, S.G.; Arul, P.R.; Geiser, B.P.; Shanmugam, V. Random Coincidence Estimation from Single Event Rates on the Discovery ST PET/CT Scanner. In Proceedings of the 2003 IEEE Nuclear Science Symposium. Conference Record (IEEE Cat. No.03CH37515), Portland, OR, USA, 19–25 October 2003; IEEE: Piscataway, NJ, USA, 2003; pp. 3067–3069. [Google Scholar]
- Jan, S.; Santin, G.; Strul, D.; Staelens, S.; Assié, K.; Autret, D.; Avner, S.; Barbier, R.; Bardiès, M.; Bloomfield, P.M.; et al. GATE: A Simulation Toolkit for PET and SPECT. Phys. Med. Biol. 2004, 49, 4543–4561. [Google Scholar] [CrossRef] [PubMed]
- Modat, M.; Cash, D.M.; Daga, P.; Winston, G.P.; Duncan, J.S.; Ourselin, S. Global Image Registration Using a Symmetric Block-Matching Approach. J. Med. Imaging 2014, 1, 024003. [Google Scholar] [CrossRef]
- Ourselin, S.; Roche, A.; Subsol, G.; Pennec, X.; Ayache, N. Reconstructing a 3D Structure from Serial Histological Sections. Image Vis. Comput. 2001, 19, 25–31. [Google Scholar] [CrossRef]
- Wang, T.; Lei, Y.; Fu, Y.; Curran, W.J.; Liu, T.; Nye, J.A.; Yang, X. Machine Learning in Quantitative PET: A Review of Attenuation Correction and Low-Count Image Reconstruction Methods. Phys. Med. 2020, 76, 294–306. [Google Scholar] [CrossRef] [PubMed]
- Rundo, L.; Tangherloni, A.; Militello, C.; Gilardi, M.C.; Mauri, G. Multimodal Medical Image Registration Using Particle Swarm Optimization: A Review. In Proceedings of the 2016 IEEE Symposium Series on Computational Intelligence (SSCI), Athens, Greece, 6–9 December 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 1–8. [Google Scholar]
- Lowekamp, B.; Gabehart; Blezek, D.; Ibanez, L.; McCormick, M.; Chen, D.; Mueller, D.; Cole, D.; Johnson, H.; Marstal, K.; et al. Simpleelastix: Simpleelastix V0.9.0 2015. Available online: https://zenodo.org/record/19049#.Y6FdcX1BxZU (accessed on 10 December 2022).
- Kinahan, P.E.; Townsend, D.W.; Beyer, T.; Sashin, D. Attenuation Correction for a Combined 3D PET/CT Scanner. Med. Phys. 1998, 25, 2046–2053. [Google Scholar] [CrossRef] [PubMed]
- Leek, F.; Robinson, A.P.; Moss, R.M.; Wilson, F.J.; Hutton, B.F.; Thielemans, K. Air Fraction Correction Optimisation in PET Imaging of Lung Disease. In Proceedings of the 2020 IEEE Nuclear Science Symposium and Medical Imaging Conference (NSS/MIC), Boston, MA, USA, 31 October–7 November 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–4. [Google Scholar]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated Anatomical Labeling of Activations in SPM Using a Macroscopic Anatomical Parcellation of the MNI MRI Single-Subject Brain. NeuroImage 2002, 15, 273–289. [Google Scholar] [CrossRef]
- Collins, D.L.; Zijdenbos, A.P.; Kollokian, V.; Sled, J.G.; Kabani, N.J.; Holmes, C.J.; Evans, A.C. Design and Construction of a Realistic Digital Brain Phantom. IEEE Trans. Med. Imaging 1998, 17, 463–468. [Google Scholar] [CrossRef]
- Modat, M.; Ridgway, G.R.; Taylor, Z.A.; Lehmann, M.; Barnes, J.; Hawkes, D.J.; Fox, N.C.; Ourselin, S. Fast Free-Form Deformation Using Graphics Processing Units. Comput. Methods Programs Biomed. 2010, 98, 278–284. [Google Scholar] [CrossRef]
- Von Borczyskowski, D.; Schulte, U.; Brenner, W.; Clausen, M.; Buchert, R. Occurrence of an Artifact in Brain 18F-FDG PET with Calculated Attenuation Correction. J. Nucl. Med. Technol. 2006, 34, 232. [Google Scholar]
- Burgos, N.; Cardoso, M.J.; Thielemans, K.; Modat, M.; Pedemonte, S.; Dickson, J.; Barnes, A.; Ahmed, R.; Mahoney, C.J.; Schott, J.M.; et al. Attenuation Correction Synthesis for Hybrid PET-MR Scanners: Application to Brain Studies. IEEE Trans. Med. Imaging 2014, 33, 2332–2341. [Google Scholar] [CrossRef]
- Alessio, A.M.; Kinahan, P.E.; Champley, K.M. Attenuation-Emission Alignment in Cardiac PET/CT Based on Consistency Conditions: Attenuation-Emission Alignment in Cardiac PET/CT. Med. Phys. 2010, 37, 1191–1200. [Google Scholar] [CrossRef] [PubMed]













| Patient ID | Correct µ | T1 | T2 | T3 | T4 |
|---|---|---|---|---|---|
| P1 | 0.97x + 0.02 | 0.93x + 0.05 | 1.00x − 0.01 | 0.98x + 0.01 | 0.97x + 0.01 |
| P2 | 1.02x − 0.02 | 1.05x − 0.05 | 0.94x + 0.06 | 1.11x − 0.10 1 | 1.13x − 0.13 |
| P3 | 1.07x − 0.07 | 1.04x − 0.04 | 0.95x + 0.04 | 1.23x − 0.22 1 | 1.11x − 0.11 |
| P4 | 0.98x + 0.01 | 0.94x + 0.04 | 0.96x + 0.03 | 0.96x + 0.03 | 0.98x + 0.01 |
| Patient ID | Correct µ | T1 | T2 | T3 | T4 |
|---|---|---|---|---|---|
| P1 | 0.851 | 0.855 | 0.881 | 0.830 | 0.834 |
| P2 | 0.932 | 0.936 | 0.918 | 0.912 | 0.928 |
| P3 | 0.916 | 0.904 | 0.803 | 0.885 | 0.919 |
| P4 | 0.934 | 0.913 | 0.919 | 0.924 | 0.925 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jehl, M.; Mikhaylova, E.; Treyer, V.; Hofbauer, M.; Hüllner, M.; Kaufmann, P.A.; Buck, A.; Warnock, G.; Dao, V.; Tsoumpas, C.; et al. Attenuation Correction Using Template PET Registration for Brain PET: A Proof-of-Concept Study. J. Imaging 2023, 9, 2. https://doi.org/10.3390/jimaging9010002
Jehl M, Mikhaylova E, Treyer V, Hofbauer M, Hüllner M, Kaufmann PA, Buck A, Warnock G, Dao V, Tsoumpas C, et al. Attenuation Correction Using Template PET Registration for Brain PET: A Proof-of-Concept Study. Journal of Imaging. 2023; 9(1):2. https://doi.org/10.3390/jimaging9010002
Chicago/Turabian StyleJehl, Markus, Ekaterina Mikhaylova, Valerie Treyer, Marlena Hofbauer, Martin Hüllner, Philipp A. Kaufmann, Alfred Buck, Geoff Warnock, Viet Dao, Charalampos Tsoumpas, and et al. 2023. "Attenuation Correction Using Template PET Registration for Brain PET: A Proof-of-Concept Study" Journal of Imaging 9, no. 1: 2. https://doi.org/10.3390/jimaging9010002
APA StyleJehl, M., Mikhaylova, E., Treyer, V., Hofbauer, M., Hüllner, M., Kaufmann, P. A., Buck, A., Warnock, G., Dao, V., Tsoumpas, C., Deidda, D., Thielemans, K., Ahnen, M. L., & Fischer, J. (2023). Attenuation Correction Using Template PET Registration for Brain PET: A Proof-of-Concept Study. Journal of Imaging, 9(1), 2. https://doi.org/10.3390/jimaging9010002








