Synthetic Megavoltage Cone Beam Computed Tomography Image Generation for Improved Contouring Accuracy of Cardiac Pacemakers
Abstract
:1. Introduction
2. Materials and Methods
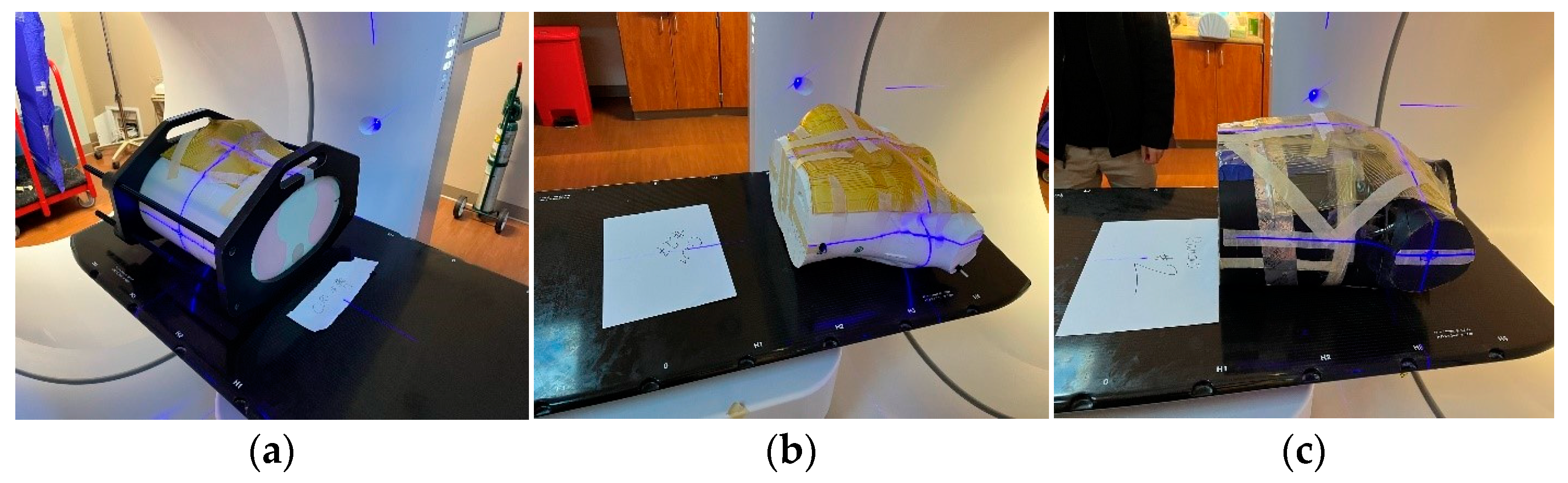
2.1. Data Acquisition
2.2. Data Preprocessing
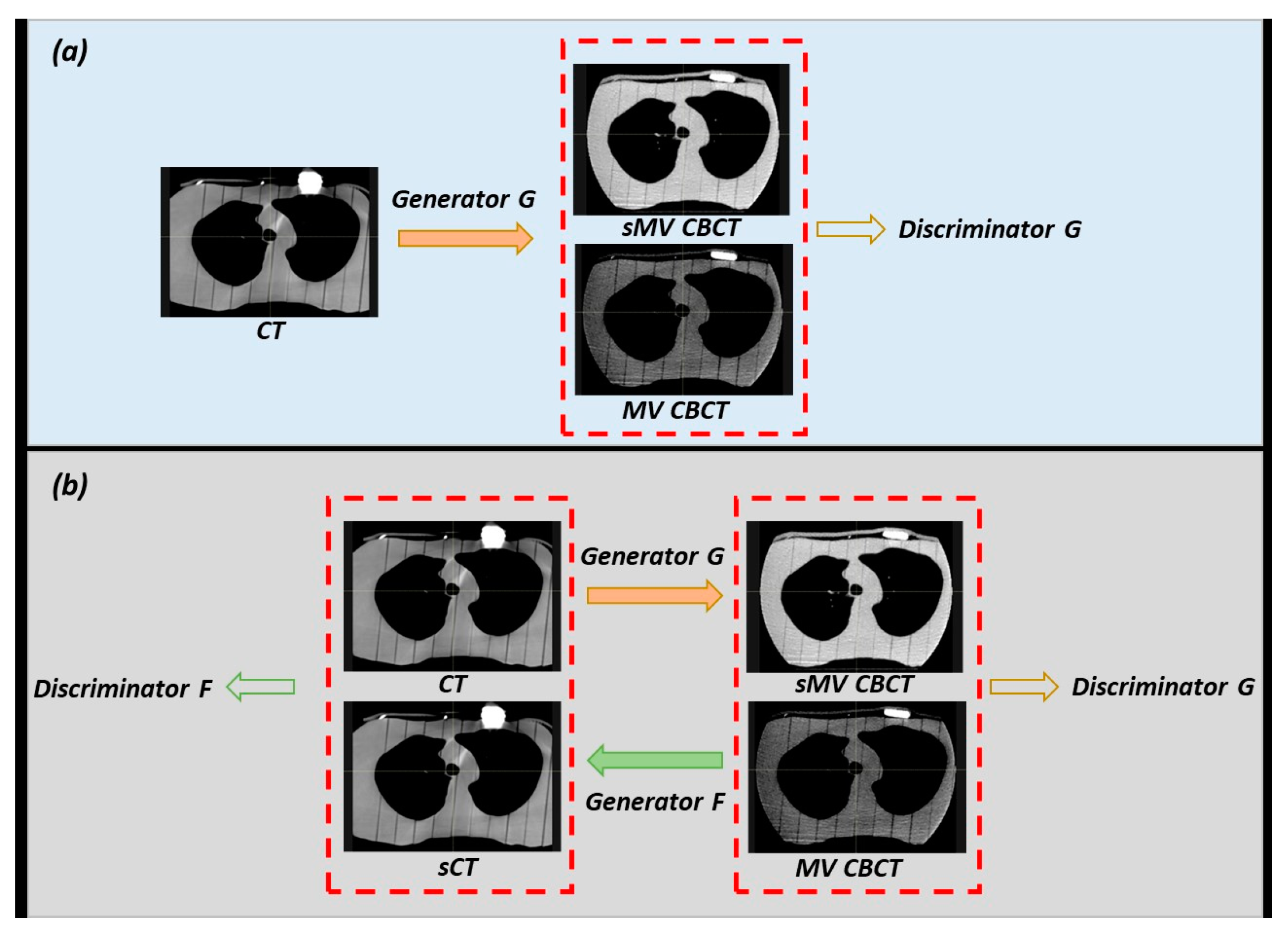
2.3. Model Training
2.4. Model Evaluation
3. Results
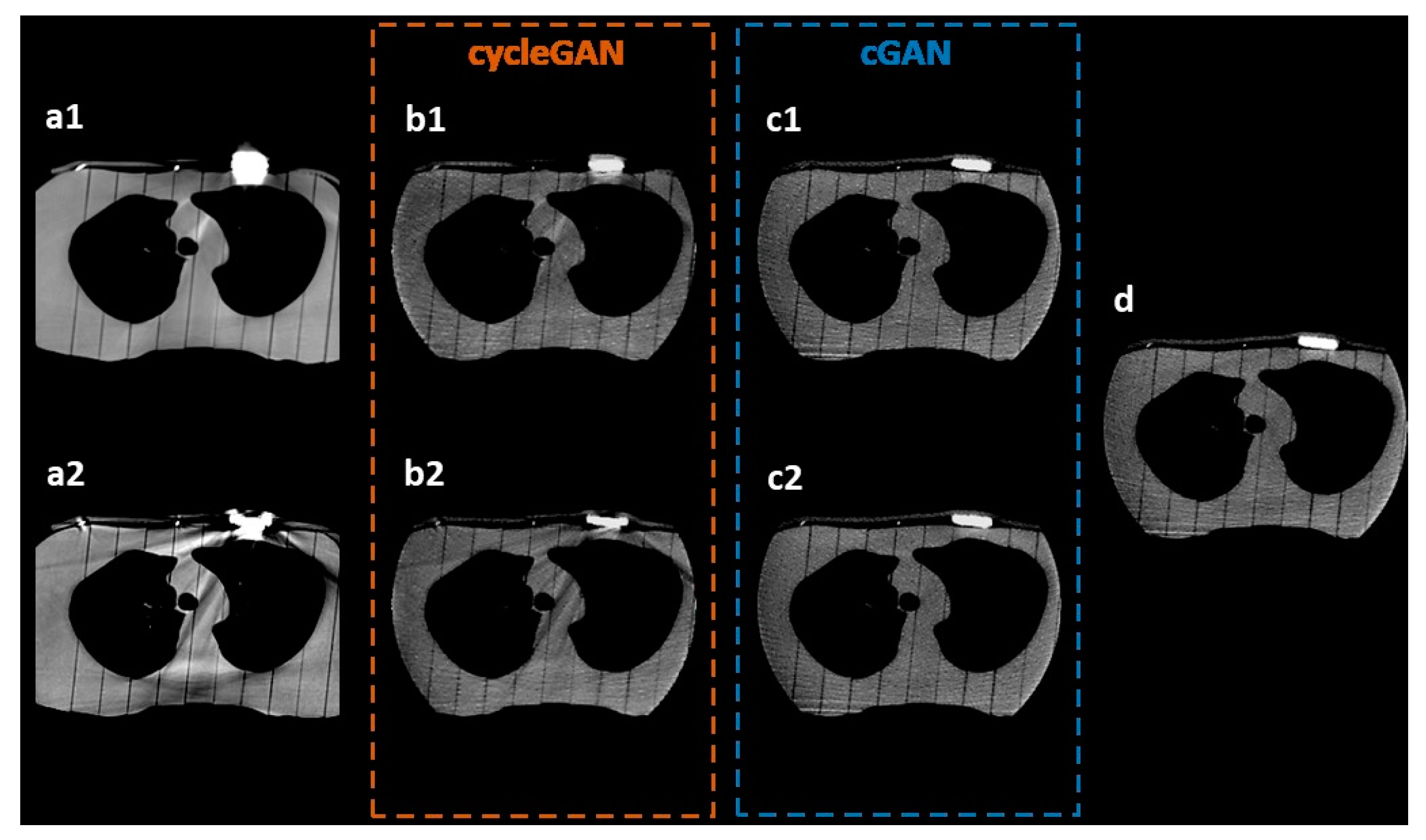
3.1. Visual Inspection
3.2. Model Comparison
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Center for Health Statistics; Heron, M. Deaths: Leading Causes for 2018; National Center for Health Statistics: Hyattsville, MD, USA, 2021. [CrossRef]
- Oldham, M.; Létourneau, D.; Watt, L.; Hugo, G.; Yan, D.; Lockman, D.; Kim, L.H.; Chen, P.Y.; Martinez, A.; Wong, J.W. Cone-beam-CT guided radiation therapy: A model for on-line application. Radiother. Oncol. 2005, 75, 271.E1–271.E8. [Google Scholar] [CrossRef] [PubMed]
- Broderick, M.; Menezes, G.; Leech, M.; Coffey, M.; Appleyard, R. A comparison of kilovoltage and megavoltage cone beam CT in radiotherapy. J. Radiother. Pract. 2007, 6, 173–178. [Google Scholar] [CrossRef]
- Chu, J.C.H.; Ni, B.; Kriz, R.; Amod Saxena, V. Applications of simulator computed tomography number for photon dose calculations during radiotherapy treatment planning. Radiother. Oncol. 2000, 55, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Kilby, W.; Sage, J.; Rabett, V. Tolerance levels for quality assurance of electron density values generated from CT in radiotherapy treatment planning. Phys. Med. Biol. 2002, 47, 1485. [Google Scholar] [CrossRef]
- Wellenberg, R.H.H.; Hakvoort, E.T.; Slump, C.H.; Boomsma, M.F.; Maas, M.; Streekstra, G.J. Metal artifact reduction techniques in musculoskeletal CT-imaging. Eur. J. Radiol. 2018, 107, 60–69. [Google Scholar] [CrossRef]
- Kim, Y.; Tomé, W.A.; Bal, M.; McNutt, T.R.; Spies, L. The impact of dental metal artifacts on head and neck IMRT dose distributions. Radiother. Oncol. 2006, 79, 198–202. [Google Scholar] [CrossRef]
- Lu, Y.; Fu, J.; Li, X.; Zhou, W.; Liu, S.; Zhang, X.; Wu, W.; Jia, C.; Liu, Y.; Chen, Z. RTN: Reinforced Transformer Network for Coronary CT Angiography Vessel-level Image Quality Assessment. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2022; Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S., Eds.; Springer Nature: Cham, Switzerland, 2022; pp. 644–653. [Google Scholar] [CrossRef]
- Wang, C.; Liang, Y.; Wu, Y.; Zhao, S.; Du, Y.P. Correction of out-of-FOV motion artifacts using convolutional neural network. Magn. Reason. Imaging 2020, 71, 93–102. [Google Scholar] [CrossRef]
- Liang, K.; Zhang, L.; Yang, H.; Yang, Y.; Chen, Z.; Xing, Y. Metal artifact reduction for practical dental computed tomography by improving interpolation-based reconstruction with deep learning. Med. Phys. 2019, 46, e823–e834. [Google Scholar] [CrossRef]
- Huang, Z.; Zhang, G.; Lin, J.; Pang, Y.; Wang, H.; Bai, T.; Zhong, L. Multi-modal feature-fusion for CT metal artifact reduction using edge-enhanced generative adversarial networks. Comput. Methods Programs Biomed. 2022, 217, 106700. [Google Scholar] [CrossRef]
- Han, C.; Kitamura, Y.; Kudo, A.; Ichinose, A.; Rundo, L.; Furukawa, Y.; Umemoto, K.; Li, Y.; Nakayama, H. Synthesizing Diverse Lung Nodules Wherever Massively: 3D Multi-Conditional GAN-Based CT Image Augmentation for Object Detection. In Proceedings of the 2019 International Conference on 3D Vision (3DV), Québec City, QC, Canada, 16–19 September 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 729–737. [Google Scholar] [CrossRef]
- Habijan, M.; Galić, I. Generation of Artificial CT Images using Patch-based Conditional Generative Adversarial Networks. In Proceedings of the 2022 7th International Conference on Smart and Sustainable Technologies (SpliTech), Split, Croatia, 5–8 July 2022; pp. 1–5. [Google Scholar] [CrossRef]
- Gossman, M.S.; Graves-Calhoun, A.R.; Wilkinson, J.D. Establishing radiation therapy treatment planning effects involvisng implantable pacemakers and implantable cardioverter-defibrillators. J. Appl. Clin. Med. Phys. 2009, 11, 33–45. [Google Scholar] [CrossRef]
- DiFilippo, F.P.; Brunken, R.C. Do Implanted Pacemaker Leads and ICD Leads Cause Metal-Related Artifact in Cardiac PET/CT? J. Nucl. Med. 2005, 46, 436–443. [Google Scholar] [PubMed]
- Do, T.; Sutter, R.; Skornitzke, S.; Weber, M.-A. CT and MRI Techniques for Imaging Around Orthopedic Hardware. RöFo-Fortschritte Röntgenstrahlen Bildgeb. Verfahr 2018, 190, 31–41. [Google Scholar] [CrossRef] [PubMed]
- McCollough, C.H.; Boedeker, K.; Cody, D.; Duan, X.; Flohr, T.; Halliburton, S.S.; Hsieh, J.; Layman, R.R.; Pelc, N.J. Principles and applications of multienergy CT: Report of AAPM Task Group 291. Med. Phys. 2020, 47, e881–e912. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yan, P.; Zhang, Y.; Yu, H.; Shi, Y.; Mou, X.; Kalra, M.K.; Zhang, Y.; Sun, L.; Wang, G. Low-Dose CT Image Denoising Using a Generative Adversarial Network With Wasserstein Distance and Perceptual Loss. IEEE Trans. Med. Imaging 2018, 37, 1348–1357. [Google Scholar] [CrossRef]
- Chou, R.; Chi, H.-Y.; Lin, Y.-H.; Ying, L.-K.; Chao, Y.-J.; Lin, C.-H. Comparison of quantitative measurements of four manufacturer’s metal artifact reduction techniques for CT imaging with a self-made acrylic phantom. Technol. Health Care 2020, 28, 273–287. [Google Scholar] [CrossRef]
- Liao, H.; Lin, W.-A.; Huo, Z.; Vogelsang, L.; Sehnert, W.J.; Zhou, S.K.; Luo, J. Generative Mask Pyramid Network for CT/CBCT Metal Artifact Reduction with Joint Projection-Sinogram Correction. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019; Shen, D., Liu, T., Peters, T.M., Staib, L.H., Essert, C., Zhou, S., Yap, P.-T., Khan, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 77–85. [Google Scholar] [CrossRef]
- Computerized Imaging Reference Systems, Inc. ATOM® Dosimetry Phantoms: Models 701–706, Reference PB ATOM 082310. Norfolk, Virginia: 2010. Available online: https://www.cirsinc.com/wp-content/uploads/2019/05/701-706-ATOM-PB-120418.pdf (accessed on 10 August 2023).
- Taddei, P.J.; Jalbout, W.; Howell, R.M.; Khater, N.; Geara, F.; Homann, K.; Newhauser, W.D. Analytical model for out-of-field dose in photon craniospinal irradiation. Phys. Med. Biol. 2013, 58, 7463–7479. [Google Scholar] [CrossRef]
- Craft, D.F.; Howell, R.M. Preparation and fabrication of a full-scale, sagittal-sliced, 3D-printed, patient-specific radiotherapy phantom. J. Appl. Clin. Med. Phys. 2017, 18, 285–292. [Google Scholar] [CrossRef]
- Mirza, M.; Osindero, S. Conditional Generative Adversarial Nets. arXiv 2014, arXiv:1411.1784. [Google Scholar]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2242–2251. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, H.; Yu, C.; Court, L.E.; Wang, X.; Wang, Q.; Pan, T.; Ding, Y.; Phan, J.; Yang, J. Compensation cycle consistent generative adversarial networks (Comp-GAN) for synthetic CT generation from MR scans with truncated anatomy. Med. Phys. 2023, 50, 4399–4414. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Xu, B.; Wang, N.; Chen, T.; Li, M. Empirical Evaluation of Rectified Activations in Convolutional Network. arXiv 2015, arXiv:1505.00853. [Google Scholar]
- Gao, L.; Xie, K.; Sun, J.; Lin, T.; Sui, J.; Yang, G.; Ni, X. Streaking artifact reduction for CBCT-based synthetic CT generation in adaptive radiotherapy. Med. Phys. 2023, 50, 879–893. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Gao, X.; Chang, Y.; Liu, G.; Pei, Y. A novel approach for eliminating metal artifacts based on MVCBCT and CycleGAN. Front. Oncol. 2022, 12, 1024160. [Google Scholar] [CrossRef] [PubMed]
- Pennig, L.; Zopfs, D.; Gertz, R.; Bremm, J.; Zaeske, C.; Große Hokamp, N.; Celik, E.; Goertz, L.; Langenbach, M.; Persigehl, T.; et al. Reduction of CT artifacts from cardiac implantable electronic devices using a combination of virtual monoenergetic images and post-processing algorithms. Eur. Radiol. 2021, 31, 7151–7161. [Google Scholar] [CrossRef] [PubMed]


| Model | DSC | Surface DSC | HD95/mm | MSD/mm | |
|---|---|---|---|---|---|
| cycleGAN | CT-to-MV | 0.89 ± 0.03 | 0.93 ± 0.04 | 1.99 ± 1.08 | 0.48 ± 0.10 |
| kV-to-MV | 0.91 ± 0.02 | 0.94 ± 0.06 | 1.75 ± 0.70 | 0.45 ± 0.17 | |
| cGAN | CT-to-MV | 0.91 ± 0.02 | 0.95 ± 0.03 | 1.38 ± 0.31 | 0.42 ± 0.07 |
| kV-to-MV | 0.92 ± 0.01 | 0.97 ± 0.01 | 1.18 ± 0.20 | 0.36 ± 0.06 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baroudi, H.; Chen, X.; Cao, W.; El Basha, M.D.; Gay, S.; Gronberg, M.P.; Hernandez, S.; Huang, K.; Kaffey, Z.; Melancon, A.D.; et al. Synthetic Megavoltage Cone Beam Computed Tomography Image Generation for Improved Contouring Accuracy of Cardiac Pacemakers. J. Imaging 2023, 9, 245. https://doi.org/10.3390/jimaging9110245
Baroudi H, Chen X, Cao W, El Basha MD, Gay S, Gronberg MP, Hernandez S, Huang K, Kaffey Z, Melancon AD, et al. Synthetic Megavoltage Cone Beam Computed Tomography Image Generation for Improved Contouring Accuracy of Cardiac Pacemakers. Journal of Imaging. 2023; 9(11):245. https://doi.org/10.3390/jimaging9110245
Chicago/Turabian StyleBaroudi, Hana, Xinru Chen, Wenhua Cao, Mohammad D. El Basha, Skylar Gay, Mary Peters Gronberg, Soleil Hernandez, Kai Huang, Zaphanlene Kaffey, Adam D. Melancon, and et al. 2023. "Synthetic Megavoltage Cone Beam Computed Tomography Image Generation for Improved Contouring Accuracy of Cardiac Pacemakers" Journal of Imaging 9, no. 11: 245. https://doi.org/10.3390/jimaging9110245
APA StyleBaroudi, H., Chen, X., Cao, W., El Basha, M. D., Gay, S., Gronberg, M. P., Hernandez, S., Huang, K., Kaffey, Z., Melancon, A. D., Mumme, R. P., Sjogreen, C., Tsai, J. Y., Yu, C., Court, L. E., Pino, R., & Zhao, Y. (2023). Synthetic Megavoltage Cone Beam Computed Tomography Image Generation for Improved Contouring Accuracy of Cardiac Pacemakers. Journal of Imaging, 9(11), 245. https://doi.org/10.3390/jimaging9110245








