ADHD-AID: Aiding Tool for Detecting Children’s Attention Deficit Hyperactivity Disorder via EEG-Based Multi-Resolution Analysis and Feature Selection
Abstract
:1. Introduction
1.1. Research Gaps
1.2. Contributions and Novelty
- Extracting features from multiple domains including, time and time–frequency, and then combining them instead of relying on a single domain.
- Utilizing several multi-resolution analysis methods to analyze EEG signals and remove noise such as discrete wavelet transform (DWT), variational mode decomposition (VMD), and empirical wavelet transform (EWT).
- Employing multiple feature extraction approaches such as nonlinear features, band-power features, entropy-based features, and statistical features.
- Exploring the best electrode placement site that influences the identification performance.
- Introducing various FS approaches to select the highly significant features, thus diminishing the complexity of the classification models.
1.3. This Paper’s Structure
2. Related Works
2.1. ML-Based Frameworks
2.2. DL-Based Frameworks
2.3. Limitations of Previous Frameworks
3. Materials and Methods
3.1. Multi-Resolution Analysis
3.1.1. Variational Mode Decomposition
- where M is the overall amount of modes, wm is the frequency that corresponds to the mth mode, and z(t) is the original signal. Utilizing Lagrangian multipliers () and the quadratic penalty factor (), the restricted problem is transformed into an unrestricted one. This results in the addition of and for improved convergence properties. The enhanced Lagrangian is represented by the following equation [41]:
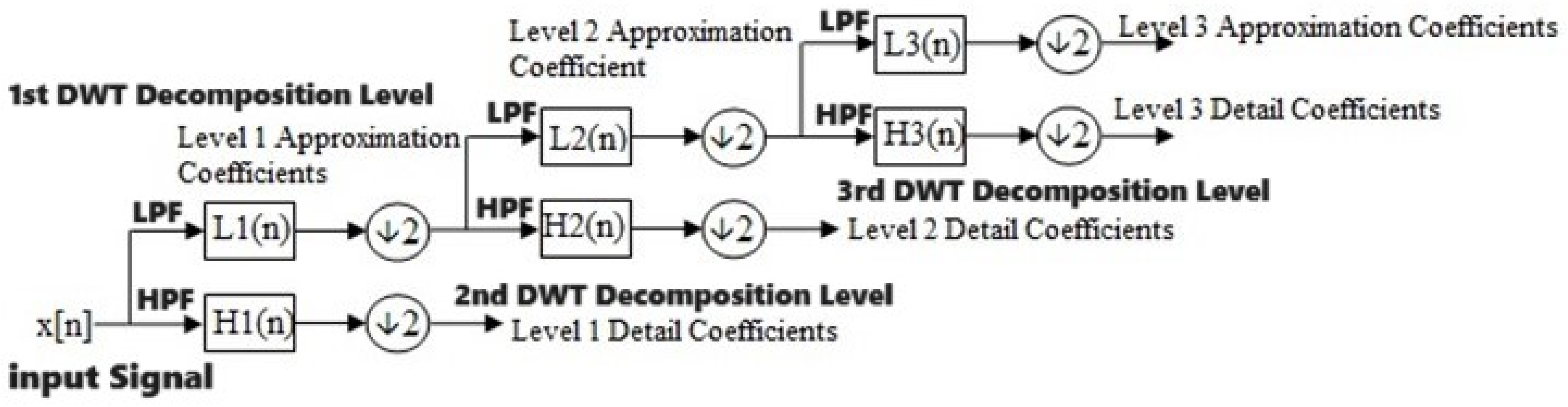
3.1.2. Discrete Wavelet Transform
- where k is the instance/sample and n is the decomposition level.
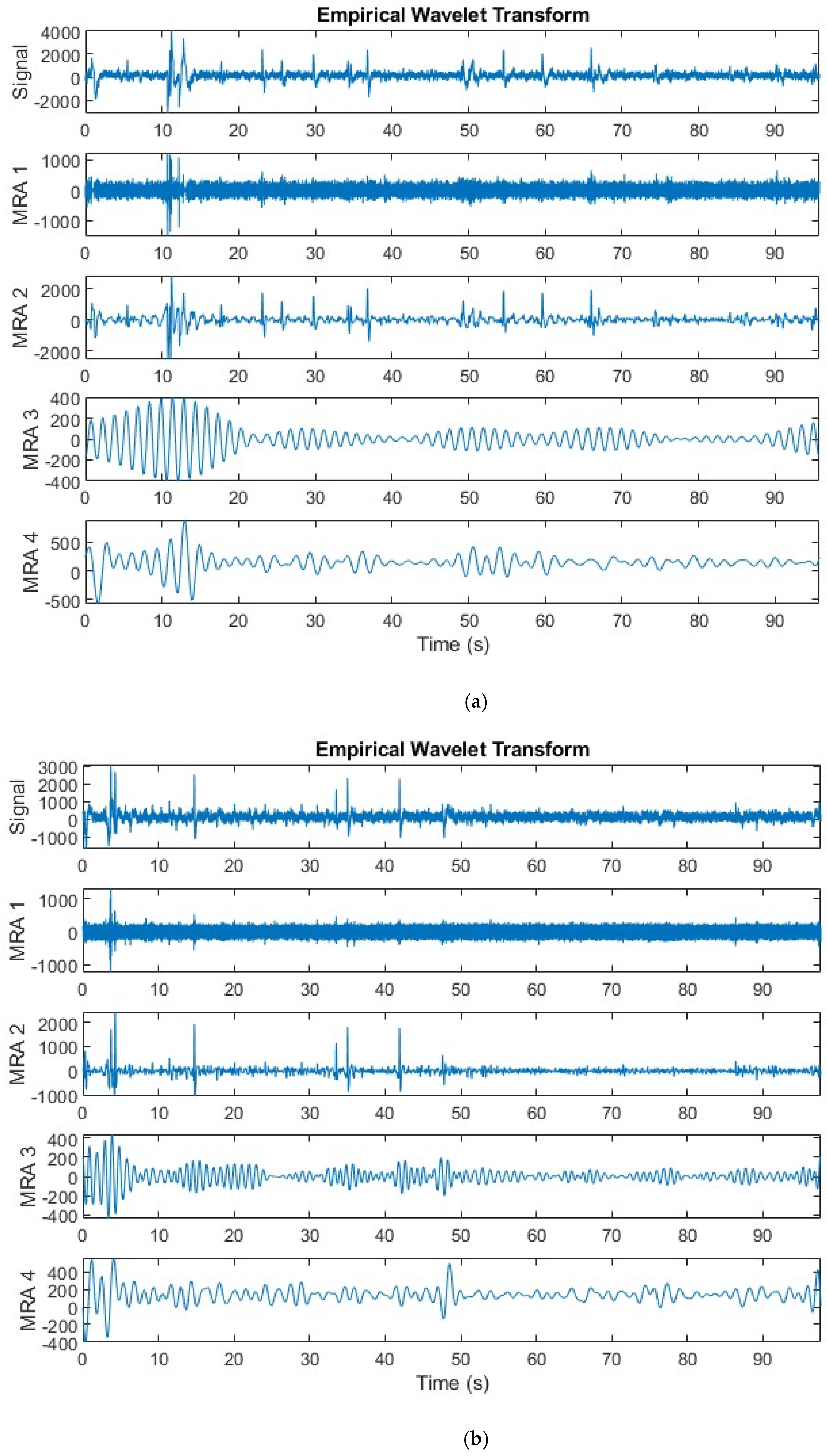
3.1.3. Empirical Wavelet Decomposition
3.2. EEG Dataset
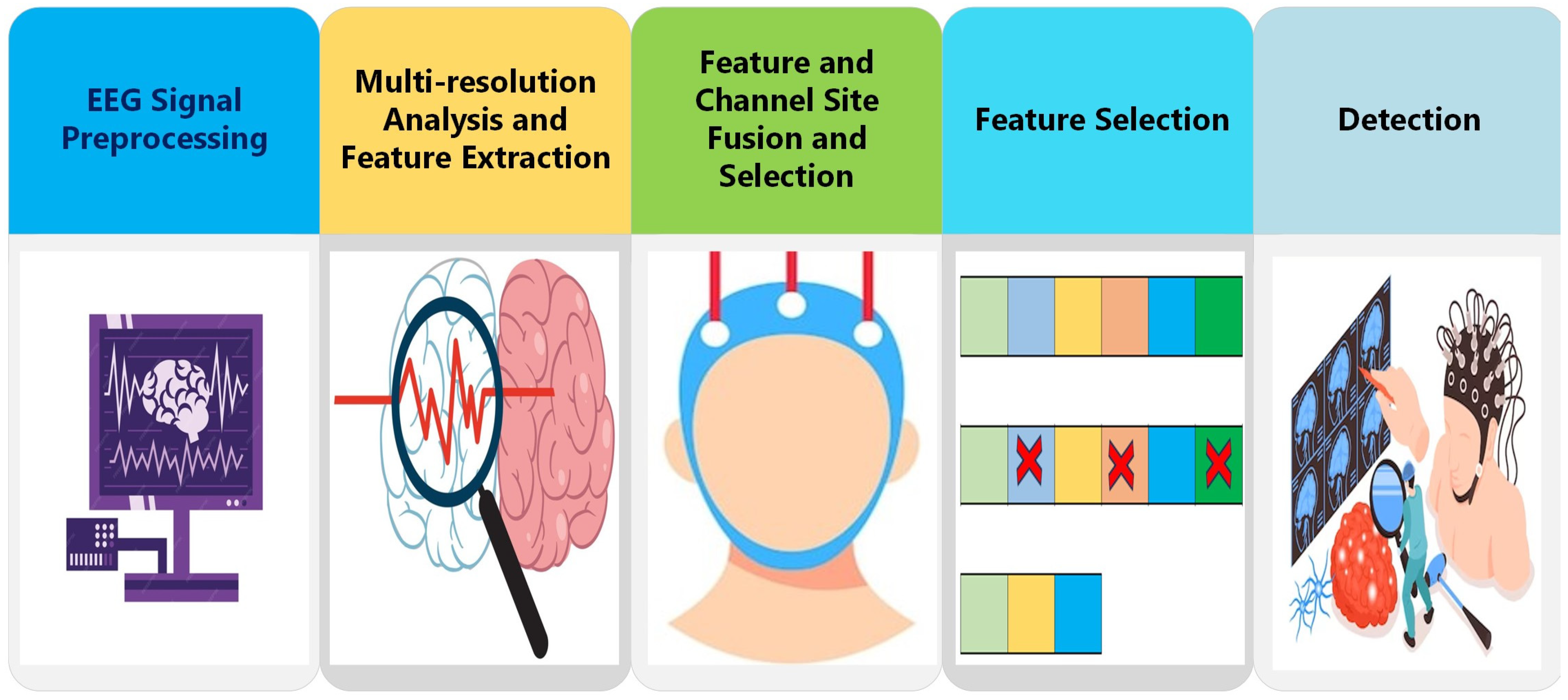
3.3. Proposed ADHD-AID Tool
3.3.1. EEG Signal Pre-Processing
3.3.2. Multi-Resolution Analysis and Feature Extraction
3.3.3. Feature and Channel Site Fusion and Selection
3.3.4. Feature Selection
3.3.5. Detection
4. Parameter Setting
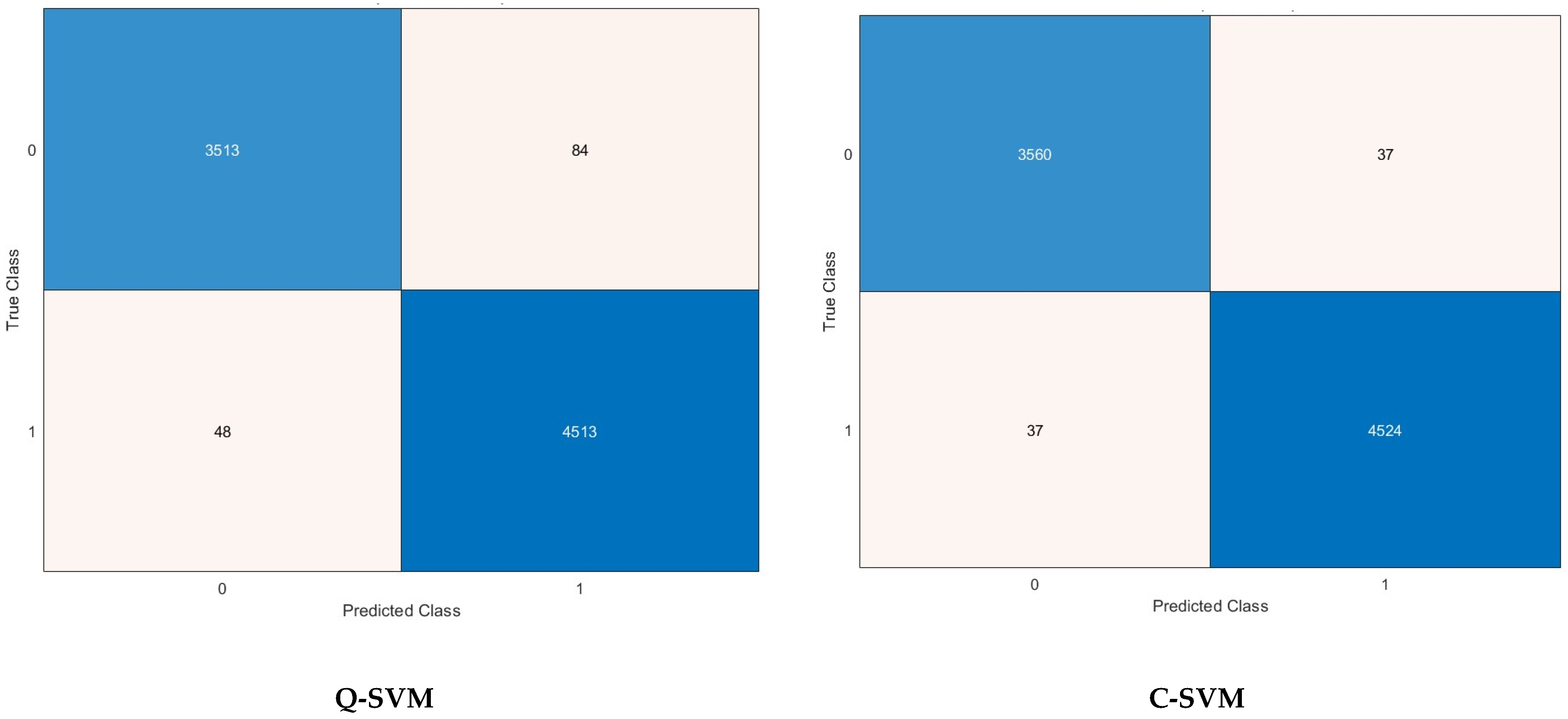
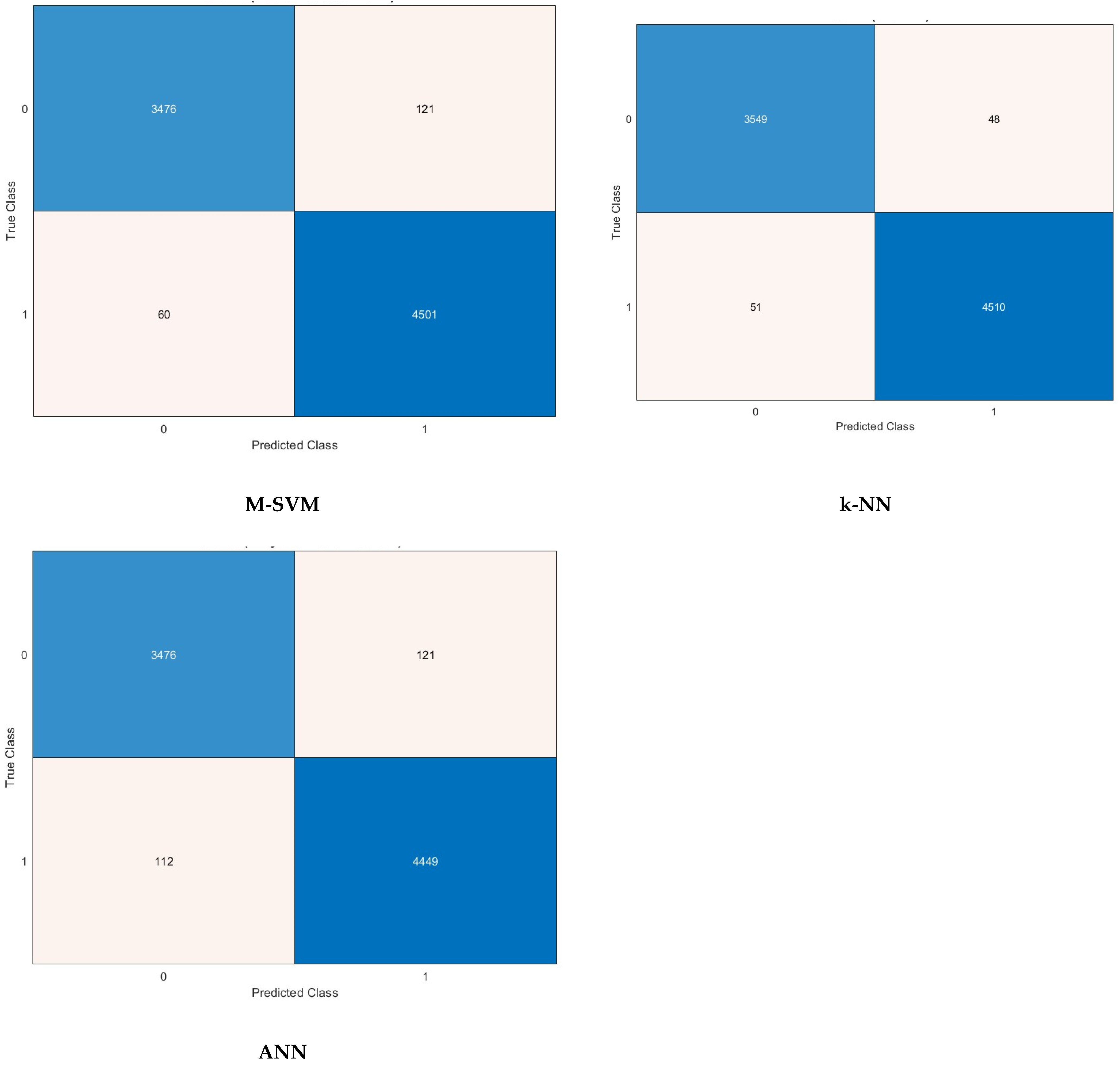
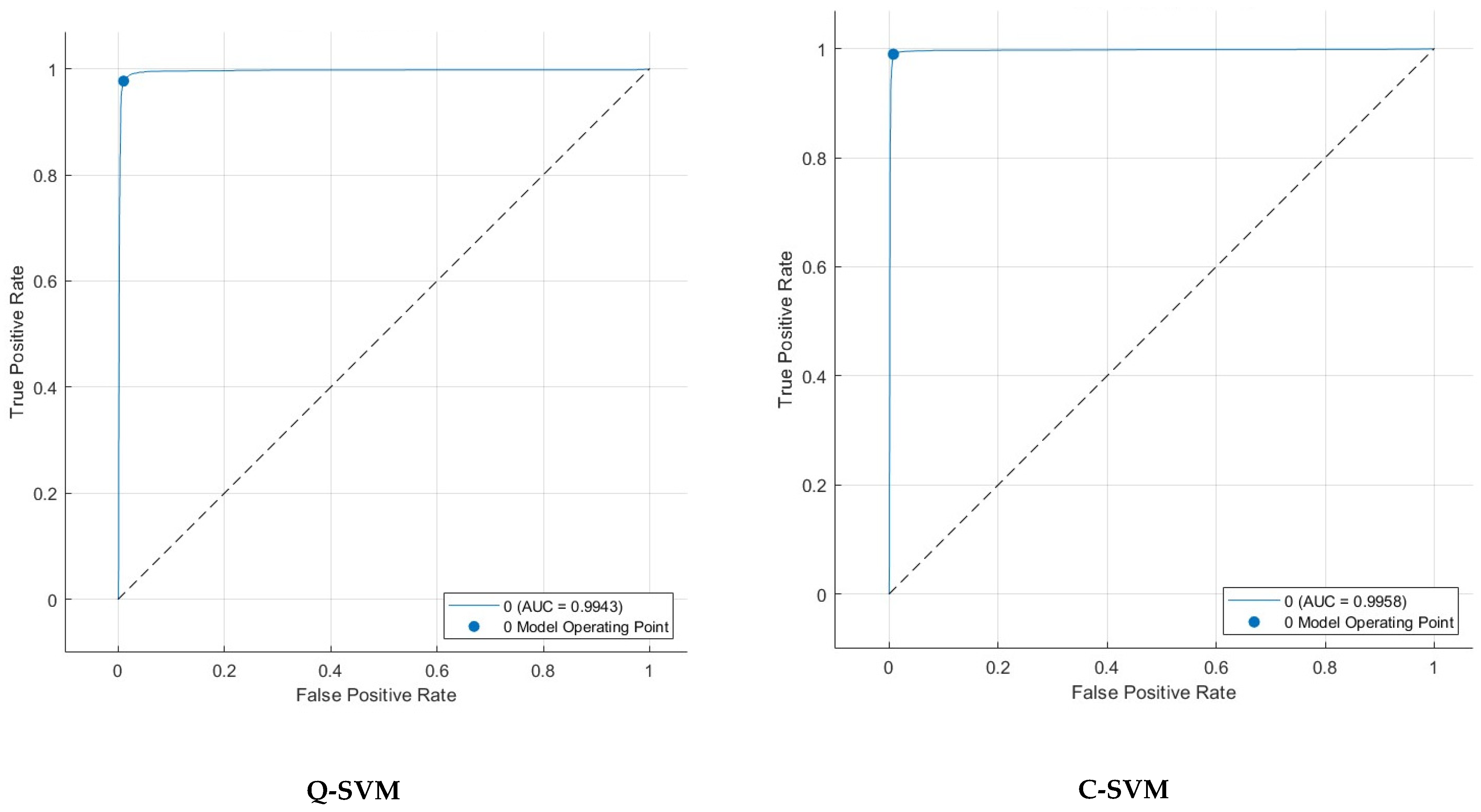
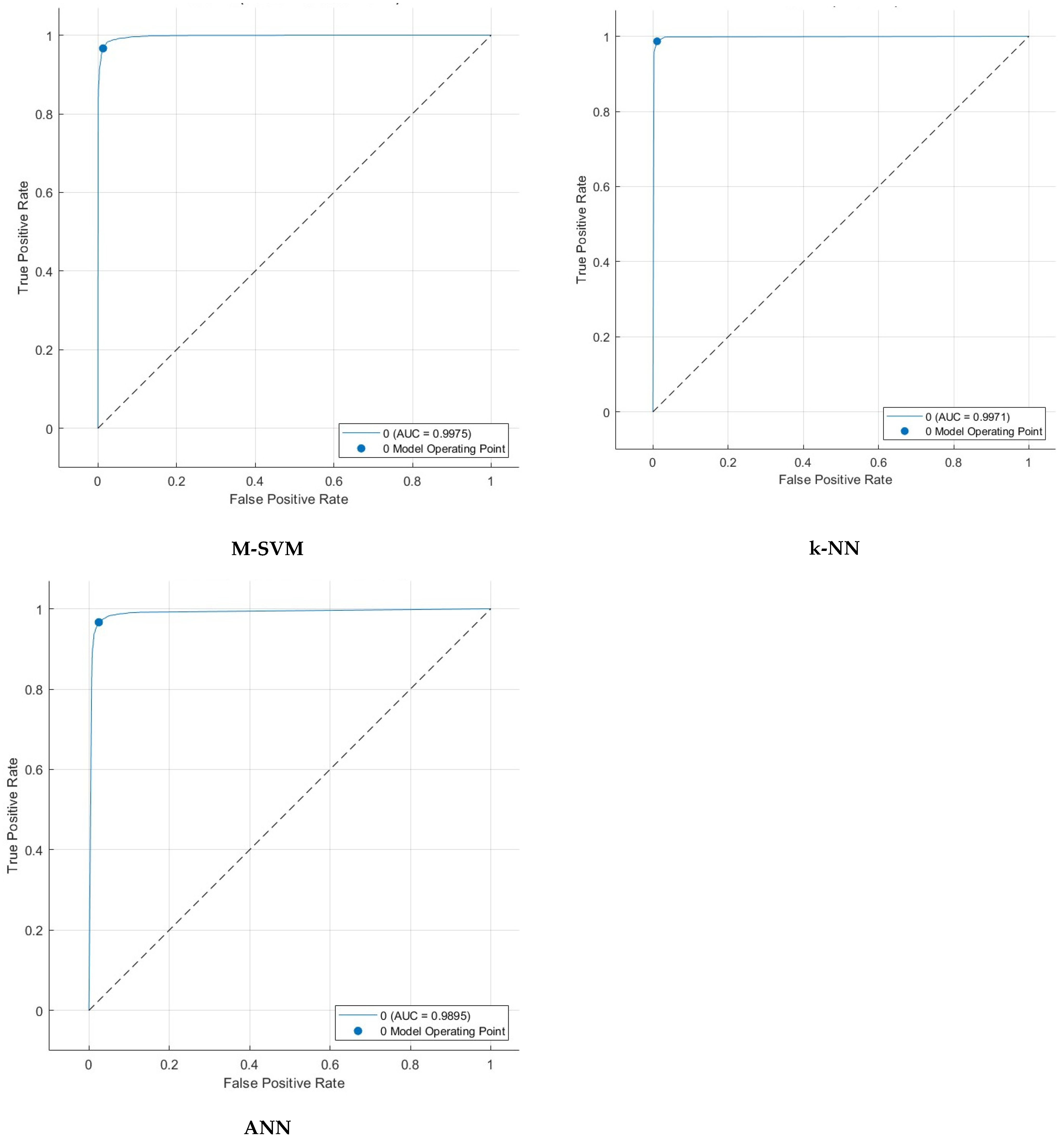
5. Detection Results
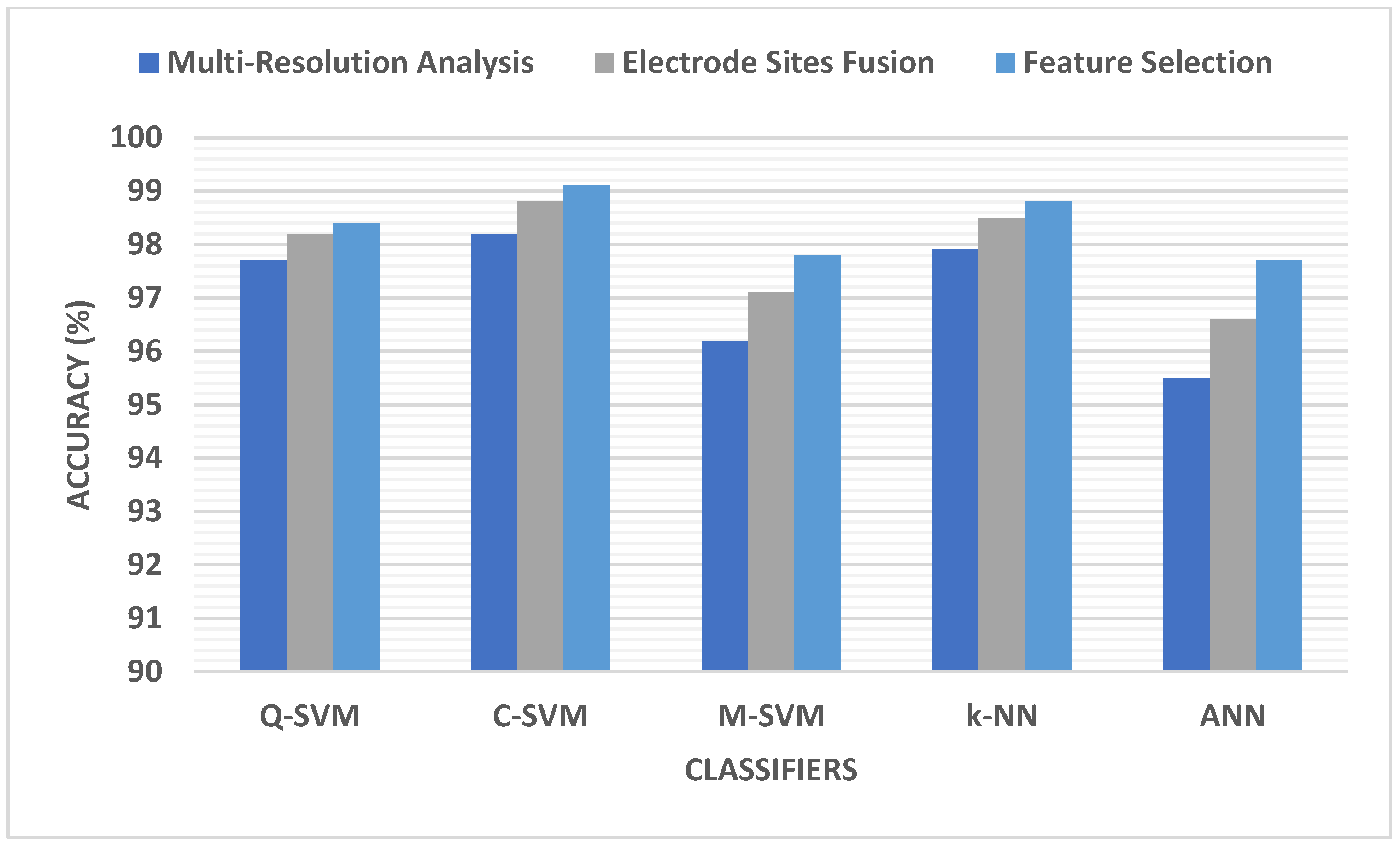
5.1. Multi-Domain Feature Extraction Results
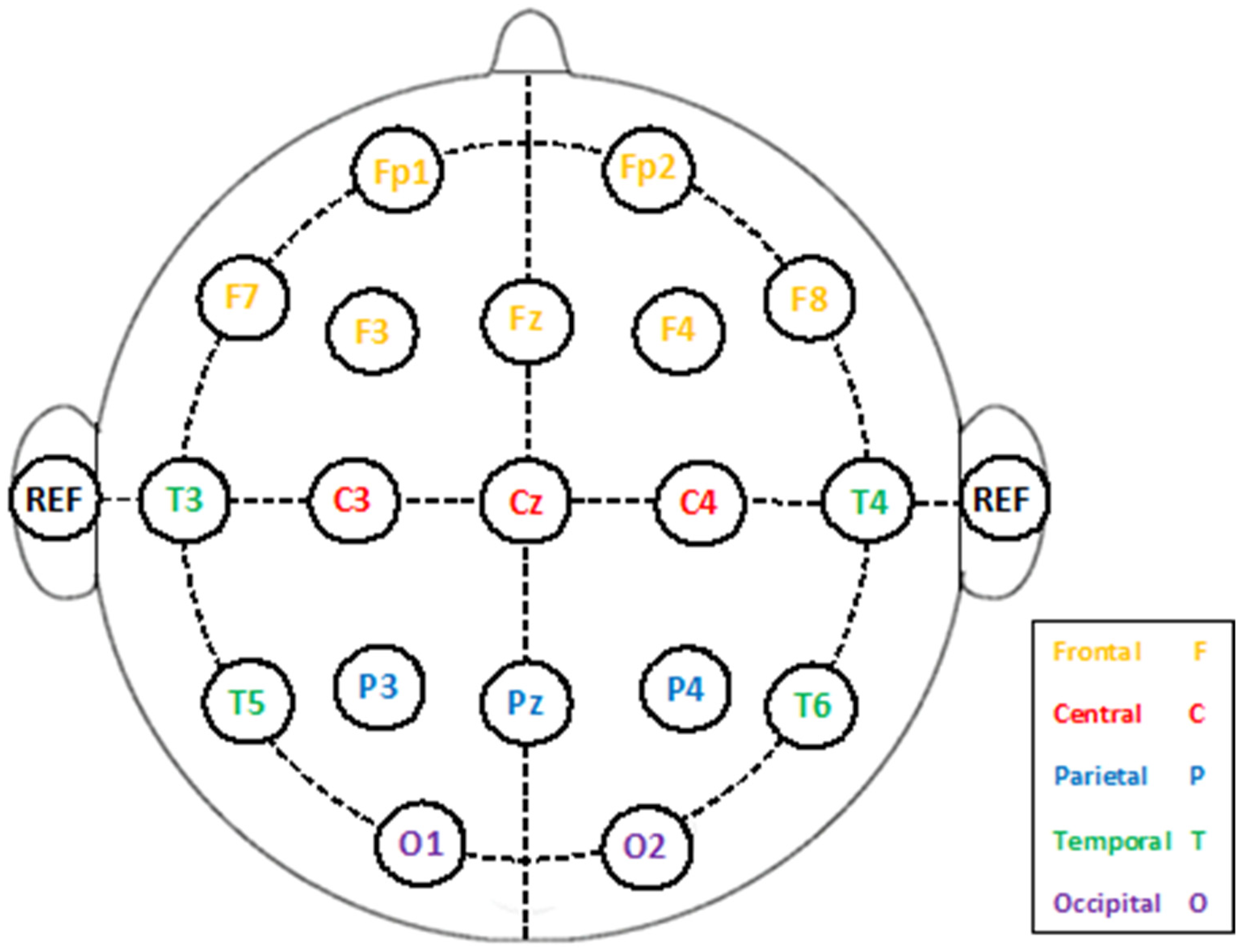
5.2. Electrode Placement Site Results
5.3. Electrode Site Selection Results
5.4. Feature Selection Results
6. Discussion
6.1. Comparative Analysis
6.2. Limitations and Upcoming Works
7. Conclusions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| ADHD | Attention deficit hyperactivity disorder |
| ANN | Artificial neural networks |
| ANOVA | Analysis of variance |
| Chi2 | Chi squared |
| Conv-LSTM | Convolutional long short-term memory |
| CNN | Convolutional neural network |
| C-SVM | Cubic support vector machine |
| dPTE | Directed phase transfer entropy |
| DWT | Discrete wavelet transform |
| EBM | Explainable boosted machine |
| ECM | Effective connectivity matrices |
| EEG | Electroencephalography |
| EMD | Empirical mode decomposition |
| EWD | Empirical wavelet transform |
| EVD | Empirical variational decomposition |
| FFT | Fast Fourier transform |
| fMRI | Functional magnetic resonance imaging |
| FN | False negative |
| FP | False positive |
| FS | Feature selection |
| GA | Genetic algorithm |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| GRU | Gated recurrent network |
| HT | Hilbert transform |
| ITD | Intrinsic time-scale decomposition |
| k-NN | k-nearest neighbors |
| KW | Kruskal–Wallis |
| LRM | Layer-wise Relevance Propagation |
| LSTM | Long short-term memory |
| MCC | Mathew correlation coefficient |
| ML | Machine learning |
| MRA | Multi-resolution analysis |
| MRI | Magnetic resonance imaging |
| mRMR | Minimum redundancy and maximal relevance |
| M-SVM | Medium Gaussian support vector machine |
| PCA | Principal component analysis |
| Q-SVM | Quadratic support vector machine |
| RLMD | Robust local mode decomposition |
| SVM | Support vector machine |
| TN | True negative |
| TP | True positive |
| VMD | Variational mode decomposition |
References
- Goldstein, S.; DeVries, M. (Eds.) Handbook of DSM-5 Disorders in Children and Adolescents; Springer International Publishing: Cham, Switzerland, 2017; ISBN 978-3-319-57194-2. [Google Scholar]
- Sridhar, C.; Bhat, S.; Acharya, U.R.; Adeli, H.; Bairy, G.M. Diagnosis of Attention Deficit Hyperactivity Disorder Using Imaging and Signal Processing Techniques. Comput. Biol. Med. 2017, 88, 93–99. [Google Scholar] [CrossRef]
- Khare, S.K.; March, S.; Barua, P.D.; Gadre, V.M.; Acharya, U.R. Application of Data Fusion for Automated Detection of Children with Developmental and Mental Disorders: A Systematic Review of the Last Decade. Inf. Fusion 2023, 99, 101898. [Google Scholar] [CrossRef]
- Khare, S.K.; Gaikwad, N.B.; Bajaj, V. VHERS: A Novel Variational Mode Decomposition and Hilbert Transform-Based EEG Rhythm Separation for Automatic ADHD Detection. IEEE Trans. Instrum. Meas. 2022, 71, 4008310. [Google Scholar] [CrossRef]
- Lola, H.M.; Belete, H.; Gebeyehu, A.; Zerihun, A.; Yimer, S.; Leta, K. Attention Deficit Hyperactivity Disorder (ADHD) among Children Aged 6 to 17 Years Old Living in Girja District, Rural Ethiopia. Behav. Neurol. 2019, 2019, 1753580. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, W.; Song, Y.; Sun, L.; Li, X. EEG Characteristics of Children with Attention-Deficit/Hyperactivity Disorder. Neuroscience 2019, 406, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Sumi, S.; Watanabe, H.; Suzuki, F.; Kuru, Y.; Shiino, T.; Kimura, T.; Wang, C.; Lin, Y.; Ichiyanagi, M. The Kiddie Schedule for Affective Disorders and Schizophrenia Present and Lifetime Version (K-SADS-PL) for DSM-5: A Validation for Neurodevelopmental Disorders in Japanese Outpatients. Compr. Psychiatry 2020, 96, 152148. [Google Scholar] [CrossRef]
- Moreno-García, I.; Delgado-Pardo, G.; Roldán-Blasco, C. Attention and Response Control in ADHD. Evaluation through Integrated Visual and Auditory Continuous Performance Test. Span. J. Psychol. 2015, 18, E1. [Google Scholar] [CrossRef]
- Yaghoobi Karimu, R.; Azadi, S. Diagnosing the ADHD Using a Mixture of Expert Fuzzy Models. Int. J. Fuzzy Syst. 2018, 20, 1282–1296. [Google Scholar] [CrossRef]
- Marshall, P.; Hoelzle, J.; Nikolas, M. Diagnosing Attention-Deficit/Hyperactivity Disorder (ADHD) in Young Adults: A Qualitative Review of the Utility of Assessment Measures and Recommendations for Improving the Diagnostic Process. Clin. Neuropsychol. 2021, 35, 165–198. [Google Scholar] [CrossRef]
- Attallah, O. Cervical Cancer Diagnosis Based on Multi-Domain Features Using Deep Learning Enhanced by Handcrafted Descriptors. Appl. Sci. 2023, 13, 1916. [Google Scholar] [CrossRef]
- Attallah, O. A Deep Learning-Based Diagnostic Tool for Identifying Various Diseases via Facial Images. Digit. Health 2022, 8, 20552076221124432. [Google Scholar] [CrossRef]
- Attallah, O. An Intelligent ECG-Based Tool for Diagnosing COVID-19 via Ensemble Deep Learning Techniques. Biosensors 2022, 12, 299. [Google Scholar] [CrossRef]
- Attallah, O. A Computer-Aided Diagnostic Framework for Coronavirus Diagnosis Using Texture-Based Radiomics Images. Digit. Health 2022, 8, 20552076221092543. [Google Scholar] [CrossRef]
- Attallah, O. MB-AI-His: Histopathological Diagnosis of Pediatric Medulloblastoma and Its Subtypes via AI. Diagnostics 2021, 11, 359. [Google Scholar] [CrossRef]
- Attallah, O.; Zaghlool, S. AI-Based Pipeline for Classifying Pediatric Medulloblastoma Using Histopathological and Textural Images. Life 2022, 12, 232. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. CoMB-Deep: Composite Deep Learning-Based Pipeline for Classifying Childhood Medulloblastoma and Its Classes. Front. Neuroinform. 2021, 15, 663592. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O.; Sharkas, M.A.; Gadelkarim, H. Deep Learning Techniques for Automatic Detection of Embryonic Neurodevelopmental Disorders. Diagnostics 2020, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Maniruzzaman, M.; Hasan, M.A.M.; Asai, N.; Shin, J. Optimal Channels and Features Selection Based ADHD Detection From EEG Signal Using Statistical and Machine Learning Techniques. IEEE Access 2023, 11, 33570–33583. [Google Scholar] [CrossRef]
- Atila, O.; Deniz, E.; Ari, A.; Sengur, A.; Chakraborty, S.; Barua, P.D.; Acharya, U.R. LSGP-USFNet: Automated Attention Deficit Hyperactivity Disorder Detection Using Locations of Sophie Germain’s Primes on Ulam’s Spiral-Based Features with Electroencephalogram Signals. Sensors 2023, 23, 7032. [Google Scholar] [CrossRef]
- Sharma, Y.; Singh, B.K. Attention Deficit Hyperactivity Disorder Detection in Children Using Multivariate Empirical EEG Decomposition Approaches: A Comprehensive Analytical Study. Expert Syst. Appl. 2023, 213, 119219. [Google Scholar] [CrossRef]
- Khare, S.K.; Acharya, U.R. An Explainable and Interpretable Model for Attention Deficit Hyperactivity Disorder in Children Using EEG Signals. Comput. Biol. Med. 2023, 155, 106676. [Google Scholar] [CrossRef]
- Parashar, A.; Kalra, N.; Singh, J.; Goyal, R.K. Machine Learning Based Framework for Classification of Children with ADHD and Healthy Controls. Intellligent Autom. Soft Comput. 2021, 28, 669–682. [Google Scholar] [CrossRef]
- Alim, A.; Imtiaz, M.H. Automatic Identification of Children with ADHD from EEG Brain Waves. Signals 2023, 4, 193–205. [Google Scholar] [CrossRef]
- Cura, O.K.; Atli, S.K.; Akan, A. Attention Deficit Hyperactivity Disorder Recognition Based on Intrinsic Time-Scale Decomposition of EEG Signals. Biomed. Signal Process. Control 2023, 81, 104512. [Google Scholar]
- Ekhlasi, A.; Nasrabadi, A.M.; Mohammadi, M. Classification of the Children with ADHD and Healthy Children Based on the Directed Phase Transfer Entropy of EEG Signals. Front. Biomed. Technol. 2021, 8, 115–122. [Google Scholar] [CrossRef]
- Hernández-Andrade, L.; Hermosillo-Abundis, A.C.; Betancourt-Navarrete, B.L.; Ruge, D.; Trenado, C.; Lemuz-López, R.; Pelayo-González, H.J.; López-Cortés, V.A.; Bonilla-Sánchez, M.D.R.; García-Flores, M.A.; et al. EEG Global Coherence in Scholar ADHD Children during Visual Object Processing. Int. J. Environ. Res. Public Health 2022, 19, 5953. [Google Scholar] [CrossRef]
- Maniruzzaman, M.; Shin, J.; Hasan, M.A.M.; Yasumura, A. Efficient Feature Selection and Machine Learning Based ADHD Detection Using EEG Signal. Comput. Mater. Contin. 2022, 72, 5179–5195. [Google Scholar] [CrossRef]
- Miao, B.; Zhang, Y. A Feature Selection Method for Classification of ADHD. In Proceedings of the 2017 4th International Conference on Information, Cybernetics and Computational Social Systems (ICCSS), Dalian, China, 24–26 July 2017; IEEE: New York, NY, USA, 2017; pp. 21–25. [Google Scholar]
- Mercado-Aguirre, I.M.; Gutierrez-Ruiz, K.P.; Contreras-Ortiz, S.H. EEG Feature Selection for ADHD Detection in Children. In Proceedings of the 16th International Symposium on Medical Information Processing and Analysis, Lima, Peru, 3–4 October 2020; SPIE: Cergy-Pontoise, France, 2020; Volume 11583, pp. 238–246. [Google Scholar]
- Aggarwal, S.; Chugh, N.; Balyan, A. Identification of ADHD Disorder in Children Using EEG Based on Visual Attention Task by Ensemble Deep Learning. In Proceedings of the International Conference on Data Science and Applications: ICDSA 2022, Kolkata, India, 26–27 March 2022; Springer: Singapore, 2023; Volume 2, pp. 243–259. [Google Scholar]
- Moghaddari, M.; Lighvan, M.Z.; Danishvar, S. Diagnose ADHD Disorder in Children Using Convolutional Neural Network Based on Continuous Mental Task EEG. Comput. Methods Programs Biomed. 2020, 197, 105738. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Song, Y.; Li, X. A Deep Learning Framework for Identifying Children with ADHD Using an EEG-Based Brain Network. Neurocomputing 2019, 356, 83–96. [Google Scholar] [CrossRef]
- Bakhtyari, M.; Mirzaei, S. ADHD Detection Using Dynamic Connectivity Patterns of EEG Data and ConvLSTM with Attention Framework. Biomed. Signal Process. Control 2022, 76, 103708. [Google Scholar] [CrossRef]
- Vahid, A.; Bluschke, A.; Roessner, V.; Stober, S.; Beste, C. Deep Learning Based on Event-Related EEG Differentiates Children with ADHD from Healthy Controls. J. Clin. Med. 2019, 8, 1055. [Google Scholar] [CrossRef]
- Esas, M.Y.; Latifoğlu, F. Detection of ADHD from EEG Signals Using New Hybrid Decomposition and Deep Learning Techniques. J. Neural Eng. 2023, 20, 036028. [Google Scholar] [CrossRef]
- Nouri, A.; Tabanfar, Z. Detection of ADHD Disorder in Children Using Layer-Wise Relevance Propagation and Convolutional Neural Network: An EEG Analysis. Front. Biomed. Technol. 2023, 11, 14–21. [Google Scholar] [CrossRef]
- Nasrabadi, A.M.; Allahverdy, A.; Samavati, M.; Mohammadi, M.R. EEG Data for ADHD/Control Children. IEEE Dataport 2020. [Google Scholar] [CrossRef]
- Rezaeezadeh, M.; Shamekhi, S.; Shamsi, M. Attention Deficit Hyperactivity Disorder Diagnosis Using Non-Linear Univariate and Multivariate EEG Measurements: A Preliminary Study. Phys. Eng. Sci. Med. 2020, 43, 577–592. [Google Scholar] [CrossRef] [PubMed]
- Allahverdy, A.; Moghadam, A.K.; Mohammadi, M.R.; Nasrabadi, A.M. Detecting Adhd Children Using the Attention Continuity as Nonlinear Feature of Eeg. Front. Biomed. Technol. 2016, 3, 28–33. [Google Scholar]
- Dragomiretskiy, K.; Zosso, D. Variational Mode Decomposition. IEEE Trans. Signal Process. 2013, 62, 531–544. [Google Scholar] [CrossRef]
- Mallat, S.G. A Theory for Multiresolution Signal Decomposition: The Wavelet Representation. IEEE Trans. Pattern Anal. Mach. Intell. 1989, 11, 674–693. [Google Scholar] [CrossRef]
- Zhang, D. Wavelet Transform. In Fundamentals of Image Data Mining; Springer: Cham, Switzerland, 2019; pp. 35–44. [Google Scholar]
- Mallat, S. A Wavelet Tour of Signal Processing, 2nd ed.; New York Academic: New York, NY, USA, 1999. [Google Scholar]
- Attallah, O. CerCan· Net: Cervical Cancer Classification Model via Multi-Layer Feature Ensembles of Lightweight CNNs and Transfer Learning. Expert Syst. Appl. 2023, 229 Pt B, 120624. [Google Scholar] [CrossRef]
- Ekhlasi, A.; Nasrabadi, A.M.; Mohammadi, M.R. Direction of Information Flow between Brain Regions in ADHD and Healthy Children Based on EEG by Using Directed Phase Transfer Entropy. Cogn. Neurodyn. 2021, 15, 975–986. [Google Scholar] [CrossRef]
- Mohammadi, M.R.; Khaleghi, A.; Nasrabadi, A.M.; Rafieivand, S.; Begol, M.; Zarafshan, H. EEG Classification of ADHD and Normal Children Using Non-Linear Features and Neural Network. Biomed. Eng. Lett. 2016, 6, 66–73. [Google Scholar] [CrossRef]
- Attallah, O. An Effective Mental Stress State Detection and Evaluation System Using Minimum Number of Frontal Brain Electrodes. Diagnostics 2020, 10, 292. [Google Scholar] [CrossRef]
- Ismail, L.E.; Karwowski, W. Applications of EEG Indices for the Quantification of Human Cognitive Performance: A Systematic Review and Bibliometric Analysis. PLoS ONE 2020, 15, e0242857. [Google Scholar] [CrossRef] [PubMed]
- Khaleghi, A.; Birgani, P.M.; Fooladi, M.F.; Mohammadi, M.R. Applicable Features of Electroencephalogram for ADHD Diagnosis. Res. Biomed. Eng. 2020, 36, 1–11. [Google Scholar] [CrossRef]
- Memar, P.; Faradji, F. A Novel Multi-Class EEG-Based Sleep Stage Classification System. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 26, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Hariharan, M.; Sindhu, R.; Vijean, V.; Yazid, H.; Nadarajaw, T.; Yaacob, S.; Polat, K. Improved Binary Dragonfly Optimization Algorithm and Wavelet Packet Based Non-Linear Features for Infant Cry Classification. Comput. Methods Programs Biomed. 2018, 155, 39–51. [Google Scholar] [CrossRef]
- Baygin, M. An Accurate Automated Schizophrenia Detection Using TQWT and Statistical Moment Based Feature Extraction. Biomed. Signal Process. Control 2021, 68, 102777. [Google Scholar] [CrossRef]
- Gupta, V.; Priya, T.; Yadav, A.K.; Pachori, R.B.; Acharya, U.R. Automated Detection of Focal EEG Signals Using Features Extracted from Flexible Analytic Wavelet Transform. Pattern Recognit. Lett. 2017, 94, 180–188. [Google Scholar] [CrossRef]
- Subasi, A. Classification of EMG Signals Using Combined Features and Soft Computing Techniques. Appl. Soft Comput. 2012, 12, 2188–2198. [Google Scholar] [CrossRef]
- Şen, B.; Peker, M.; Çavuşoğlu, A.; Çelebi, F.V. A Comparative Study on Classification of Sleep Stage Based on EEG Signals Using Feature Selection and Classification Algorithms. J. Med. Syst. 2014, 38, 18. [Google Scholar] [CrossRef]
- Jenke, R.; Peer, A.; Buss, M. Feature Extraction and Selection for Emotion Recognition from EEG. IEEE Trans. Affect. Comput. 2014, 5, 327–339. [Google Scholar] [CrossRef]
- Sudarshan, V.K.; Acharya, U.R.; Oh, S.L.; Adam, M.; Tan, J.H.; Chua, C.K.; Chua, K.P.; San Tan, R. Automated Diagnosis of Congestive Heart Failure Using Dual Tree Complex Wavelet Transform and Statistical Features Extracted from 2 s of ECG Signals. Comput. Biol. Med. 2017, 83, 48–58. [Google Scholar] [CrossRef]
- Kemp, F. The Handbook of Parametric and Nonparametric Statistical Procedures. J. R. Stat. Soc. Ser. D Stat. 2003, 52, 248–249. [Google Scholar] [CrossRef]
- Liu, H.; Setiono, R. Chi2: Feature Selection and Discretization of Numeric Attributes. In Proceedings of the 7th IEEE International Conference on Tools with Artificial Intelligence, Herndon, VA, USA, 5–8 November 1995; IEEE: New York, NY, USA, 1995; pp. 388–391. [Google Scholar]
- Khan, S.A.; Hussain, A.; Basit, A.; Akram, S. Kruskal-Wallis-Based Computationally Efficient Feature Selection for Face Recognition. Sci. World J. 2014, 2014, 672630. [Google Scholar]
- Aceves-Fernandez, M.A. Methodology Proposal of ADHD Classification of Children Based on Cross Recurrence Plots. Nonlinear Dyn. 2021, 104, 1491–1505. [Google Scholar] [CrossRef]
- Wåhlstedt, C.; Thorell, L.B.; Bohlin, G. Heterogeneity in ADHD: Neuropsychological Pathways, Comorbidity and Symptom Domains. J. Abnorm. Child Psychol. 2009, 37, 551–564. [Google Scholar] [CrossRef]
- Attallah, O. DIAROP: Automated Deep Learning-Based Diagnostic Tool for Retinopathy of Prematurity. Diagnostics 2021, 11, 2034. [Google Scholar] [CrossRef]
- Attallah, O. RADIC: A Tool for Diagnosing COVID-19 from Chest CT and X-Ray Scans Using Deep Learning and Quad-Radiomics. Chemom. Intell. Lab. Syst. 2023, 233, 104750. [Google Scholar] [CrossRef]
- Khare, S.K.; Acharya, U.R. Adazd-Net: Automated Adaptive and Explainable Alzheimer’s Disease Detection System Using EEG Signals. Knowl.-Based Syst. 2023, 278, 110858. [Google Scholar] [CrossRef]
- El-Dahshan, E.-S.A.; Bassiouni, M.M.; Khare, S.K.; Tan, R.-S.; Acharya, U.R. ExHyptNet: An Explainable Diagnosis of Hypertension Using EfficientNet with PPG Signals. Expert Syst. Appl. 2023, 239, 122388. [Google Scholar] [CrossRef]
- Khare, S.K.; Gadre, V.M.; Acharya, R. ECGPsychNet: An Optimized Hybrid Ensemble Model for Automatic Detection of Psychiatric Disorders Using ECG Signals. Physiol. Meas. 2023, 44, 115004. [Google Scholar] [CrossRef]
- Assari, S.; Caldwell, C.H. Family Income at Birth and Risk of Attention Deficit Hyperactivity Disorder at Age 15: Racial Differences. Children 2019, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhao, X.; Zhong, J.; Zhou, Y. Localization and Diagnosis of Attention-Deficit/Hyperactivity Disorder. Healthcare 2021, 9, 372. [Google Scholar] [CrossRef] [PubMed]
- Arns, M.; Heinrich, H.; Strehl, U. Evaluation of Neurofeedback in ADHD: The Long and Winding Road. Biol. Psychol. 2014, 95, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Bazanova, O.M.; Nikolenko, E.D.; Barry, R.J. Reactivity of Alpha Rhythms to Eyes Opening (the Berger Effect) during Menstrual Cycle Phases. Int. J. Psychophysiol. 2017, 122, 56–64. [Google Scholar] [CrossRef]
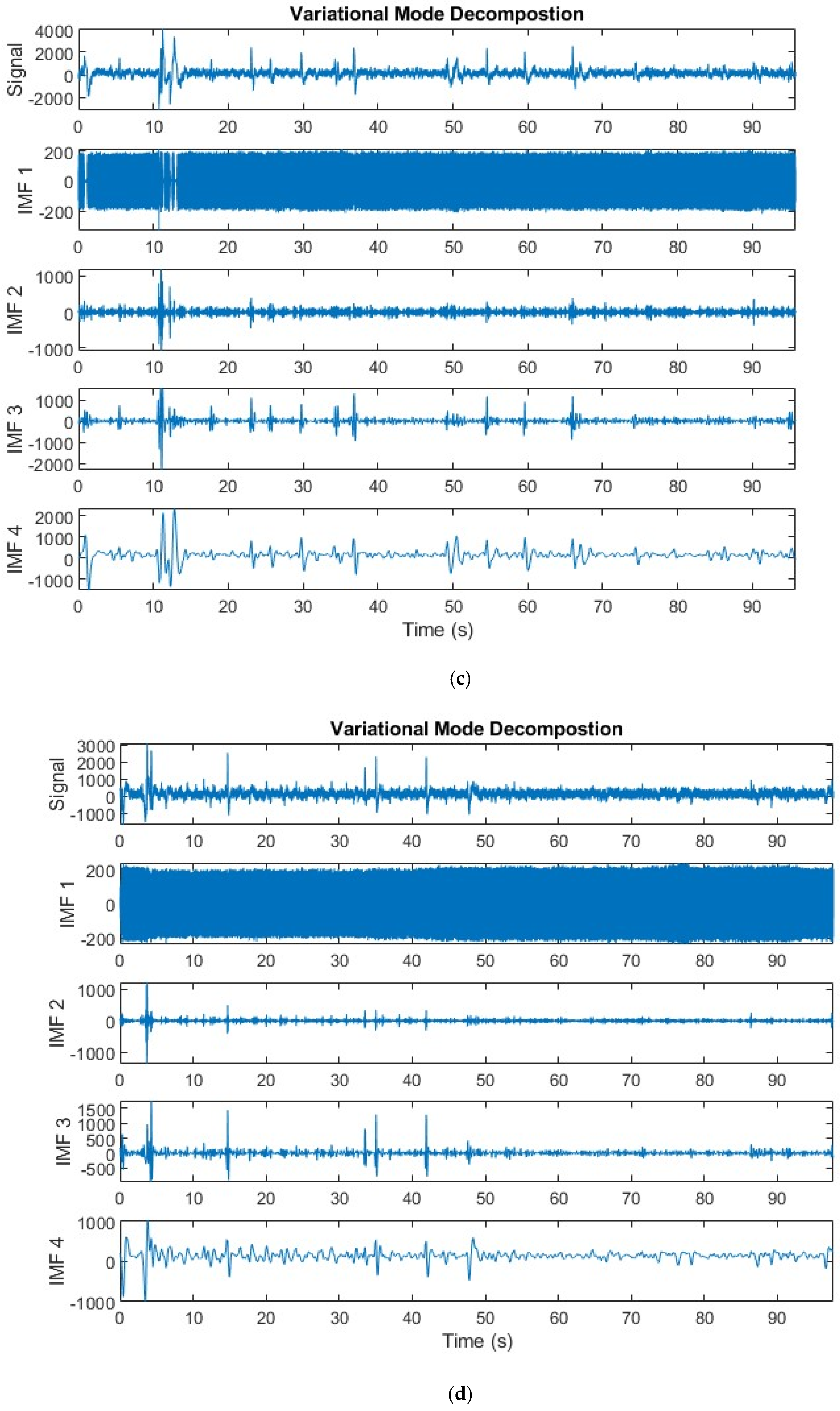
| Article | Dataset | Feature Extraction | Feature Selection | Models | Accuracy | Limitations |
|---|---|---|---|---|---|---|
| [21] | IEEE Dataport [38] 60 Healthy 61 ADHD | A total of 15 features including power, energy, entropy, and statistical-based features obtained from EMD | GA | ANN | 96.16% |
|
| [22] | IEEE Dataport [38] 60 Healthy 61 ADHD | A total of 41 statistical features from the fifth mode of VMD-HT | N/A | EBM | 99.81% |
|
| [25] | Private Dataset 15 Healthy 18 ADHD | A total of 15 connectivity-based features from different modes of ITD | N/A | Bagging Trees | 99.46% |
|
| [26] | IEEE Dataport [38] 60 Healthy 61 ADHD | ECMs using dPTE | GA | ANN | 89.1% |
|
| [23] | IEEE Dataport [38] 60 Healthy 61 ADHD | Employed EEG signals directly to training models | N/A | Adaboost | 84% |
|
| [6] | Private Dataset 50 ADHD 58 Healthy | PSD + entropy features + bi-spectral features | mRMR | SVM | 84.59% |
|
| [24] | IEEE Dataport [38] 60 Healthy 61 ADHD | A total of 10 statistical, power spectral density, and entropy-based features from the time domain | PCA | SVM | 94.2% |
|
| [39] | Private Dataset 12 ADHD 12 Healthy | Linear univariate and multi-variate features + nonlinear univariate and multi-variate features | N/A | SVM | 99.58% |
|
| [31] | IEEE Dataport [38] 60 Healthy 61 ADHD | N/A | N/A | LSTM+GRU | 95.33% |
|
| [32] | Private Dataset [40] 15 Healthy 15 ADHD | N/A | N/A | CNN | 97.47% |
|
| [33] | Private Dataset 51 Healthy 50 ADHD | A total of 13 connectivity-based features from the brain network | N/A | CNN | 94.67% |
|
| [34] | IEEE Dataport [38] 46 ADHD 45 Healthy | Dynamic connectivity tensor | N/A | Conv-LSTM | 99.34% |
|
| [35] | Private Dataset 44 Healthy 100 ADHD | N/A | N/A | EEGNet | 83% |
|
| [36] | IEEE Dataport [38] 60 Healthy 61 ADHD | VMD+RLMD | N/A | CNN | 95.24% |
|
| [37] | IEEE Dataport [38] 30 Healthy 31 ADHD | PSD frequency features using FT | N/A | CNN | 94.52% |
|
| Hjorth Activity [51] | Renyi Entropy [52] | Skewness [53] |
| Hjorth Mobility [51] | Shanon Entropy [52] | Kurtosis [53] |
| Hjorth Complexity [51] | Log Energy Entropy [54] | Auto Regressive Model [55] |
| Log Root Sum of Sequential Variation [51] | Tsallis Entropy [52] | Band-Power Alpha |
| Mean Curve Length [56] | First Difference [57] | Band-Power Beta |
| Mean Energy [56] | Second Difference [57] | Band-Power Theta |
| Mean Teager Energy [56] | Normalized First Difference [57] | Band-Power Gamma |
| Median [56] | Normalized Second Difference [57] | Band-Power Delta |
| Minimum [56] | Variance [58] | Ratio Band-Power Alpha Beta |
| Maximum [56] | Standard Deviation [58] | Arithmetic Mean [56] |
| Method | Q-SVM | C-SVM | M-SVM | k-NN | ANN |
|---|---|---|---|---|---|
| Time | 97.9 | 98.5 | 96.3 | 97.7 | 96.4 |
| DWT | 97.7 | 98.2 | 96.2 | 97.9 | 95.5 |
| EWT | 96.8 | 97.9 | 95.6 | 98.1 | 94.9 |
| VMD | 96.3 | 97.3 | 94.5 | 97.1 | 94.1 |
| Electrode Locations | Q-SVM | C-SVM | M-SVM | k-NN | ANN |
|---|---|---|---|---|---|
| Pre-Frontal | 82.9 | 86.1 | 81.6 | 86.9 | 81.0 |
| Frontal | 91.2 | 93.5 | 90.1 | 95.1 | 88.8 |
| Central | 84.3 | 87.4 | 83.6 | 88.3 | 82.0 |
| Parietal | 88.1 | 90.6 | 87.3 | 92.7 | 85.9 |
| Temporal | 89.5 | 91.6 | 87.5 | 92.4 | 86.9 |
| Occipital | 83.5 | 84.2 | 82.3 | 86.1 | 80.6 |
| Electrode Locations | Q-SVM | C-SVM | M-SVM | k-NN | ANN |
|---|---|---|---|---|---|
| Frontal | 93.5 | 90.1 | 95.1 | 88.8 | 91.2 |
| Temporal + Frontal | 95.7 | 97.0 | 94.4 | 97.2 | 93.7 |
| Temporal + Frontal + Parietal | 97.5 | 98.1 | 95.7 | 97.9 | 95.4 |
| Temporal + Frontal + Parietal + Central | 97.4 | 98.4 | 96.5 | 98.1 | 95.9 |
| Temporal + Frontal + Parietal + Central + Occipital | 97.8 | 98.6 | 96.9 | 98.2 | 96.0 |
| Temporal + Frontal + Parietal + Central + Occipital + Pre-Frontal | 98.2 | 98.8 | 97.1 | 98.5 | 96.6 |
| Features | Q-SVM | C-SVM | M-SVM | k-NN | ANN |
|---|---|---|---|---|---|
| Chi2 | |||||
| 1000 | 98.0 | 98.7 | 96.6 | 98.7 | 96.6 |
| 1500 | 98.3 | 98.9 | 96.9 | 98.8 | 96.8 |
| 2000 | 98.4 | 98.9 | 97.3 | 98.7 | 96.8 |
| ANOVA | |||||
| 1000 | 98.3 | 99.1 | 97.8 | 98.6 | 96.9 |
| 1500 | 98.3 | 98.9 | 97.8 | 98.5 | 96.7 |
| 2000 | 98.4 | 98.8 | 97.3 | 98.6 | 96.4 |
| Kruskal–Wallis (KW) | |||||
| 1000 | 98.1 | 99.0 | 97.0 | 98.7 | 96.7 |
| 1500 | 98.4 | 99.0 | 97.5 | 98.7 | 97.1 |
| 2000 | 98.4 | 98.9 | 97.3 | 98.6 | 96.4 |
| FS Method | Features Number | Sensitivity | Specificity | Precision | F1-Score | MCC | |
|---|---|---|---|---|---|---|---|
| Q-SVM | KW | 1000 | 97.2 | 98.8 | 98.5 | 97.8 | 96.2 |
| Q-SVM | KW | 1500 | 97.7 | 98.9 | 98.7 | 98.2 | 96.7 |
| Q-SVM | KW | 2000 | 97.5 | 98.8 | 98.5 | 98.0 | 96.4 |
| C-SVM | ANOVA | 1000 | 98.9 | 99.2 | 98.9 | 98.9 | 98.2 |
| C-SVM | ANOVA | 1500 | 98.6 | 99.1 | 98.9 | 98.7 | 97.7 |
| C-SVM | ANOVA | 2000 | 98.5 | 99.0 | 98.7 | 98.6 | 97.5 |
| M-SVM | ANOVA | 1000 | 96.6 | 98.7 | 98.3 | 97.5 | 95.5 |
| M-SVM | ANOVA | 1500 | 97.0 | 98.5 | 98.1 | 97.5 | 95.6 |
| M-SVM | ANOVA | 2000 | 96.5 | 98.2 | 97.6 | 97.1 | 94.8 |
| k-NN | Chi2 | 1000 | 98.3 | 98.9 | 98.6 | 98.6 | 97.2 |
| k-NN | Chi2 | 1500 | 98.7 | 98.9 | 98.6 | 98.6 | 97.5 |
| k-NN | Chi2 | 2000 | 98.6 | 98.9 | 98.6 | 98.6 | 97.4 |
| ANN | KW | 1000 | 96.4 | 96.9 | 96.2 | 96.3 | 93.3 |
| ANN | KW | 1500 | 96.6 | 97.5 | 96.9 | 96.8 | 94.2 |
| ANN | KW | 2000 | 96.4 | 97.0 | 96.3 | 96.4 | 93.5 |
| Article | Feature Extraction | Feature Selection | Models | Accuracy | Sensitivity | Specificity | F1-Score | MCC |
|---|---|---|---|---|---|---|---|---|
| [21] | A total of 15 features including power, energy, entropy, and statistical-based features obtained from EMD | GA | ANN | 96.16% | - | - | 96.32% | 92.0% |
| [26] | ECMs using dPTE | GA | ANN | 89.1% | - | - | - | - |
| [23] | Employed EEG signals directly with training models | N/A | Adaboost | 84% | 96.0% | 70.0% | - | - |
| [24] | A total of 10 statistical, power spectral density, and entropy-based features from time domain | PCA | SVM | 94.2% | - | - | - | - |
| [31] | N/A | N/A | LSTM+GRU | 95.33% | 96.20% | 95.80% | ||
| [36] | VMD + RLMD | N/A | CNN | 95.24% | - | - | - | - |
| [62] | Cross-recurrence plots + Welch power spectral distribution | N/A | N/A | 97.24% | 97.0% | 94.0% | - | - |
| Proposed ADHD-AID | A total of 30 features (nonlinear features, band-power features, entropy-based features, and statistical features) from time and time–frequency domains of VMD, DWT, and EWT | ANOVA | C-SVM | 99.10% | 98.9% | 99.2% | 98.9% | 98.2% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Attallah, O. ADHD-AID: Aiding Tool for Detecting Children’s Attention Deficit Hyperactivity Disorder via EEG-Based Multi-Resolution Analysis and Feature Selection. Biomimetics 2024, 9, 188. https://doi.org/10.3390/biomimetics9030188
Attallah O. ADHD-AID: Aiding Tool for Detecting Children’s Attention Deficit Hyperactivity Disorder via EEG-Based Multi-Resolution Analysis and Feature Selection. Biomimetics. 2024; 9(3):188. https://doi.org/10.3390/biomimetics9030188
Chicago/Turabian StyleAttallah, Omneya. 2024. "ADHD-AID: Aiding Tool for Detecting Children’s Attention Deficit Hyperactivity Disorder via EEG-Based Multi-Resolution Analysis and Feature Selection" Biomimetics 9, no. 3: 188. https://doi.org/10.3390/biomimetics9030188
APA StyleAttallah, O. (2024). ADHD-AID: Aiding Tool for Detecting Children’s Attention Deficit Hyperactivity Disorder via EEG-Based Multi-Resolution Analysis and Feature Selection. Biomimetics, 9(3), 188. https://doi.org/10.3390/biomimetics9030188






