Improved Repeatability of Mouse Tibia Volume Segmentation in Murine Myelofibrosis Model Using Deep Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design and Dataset
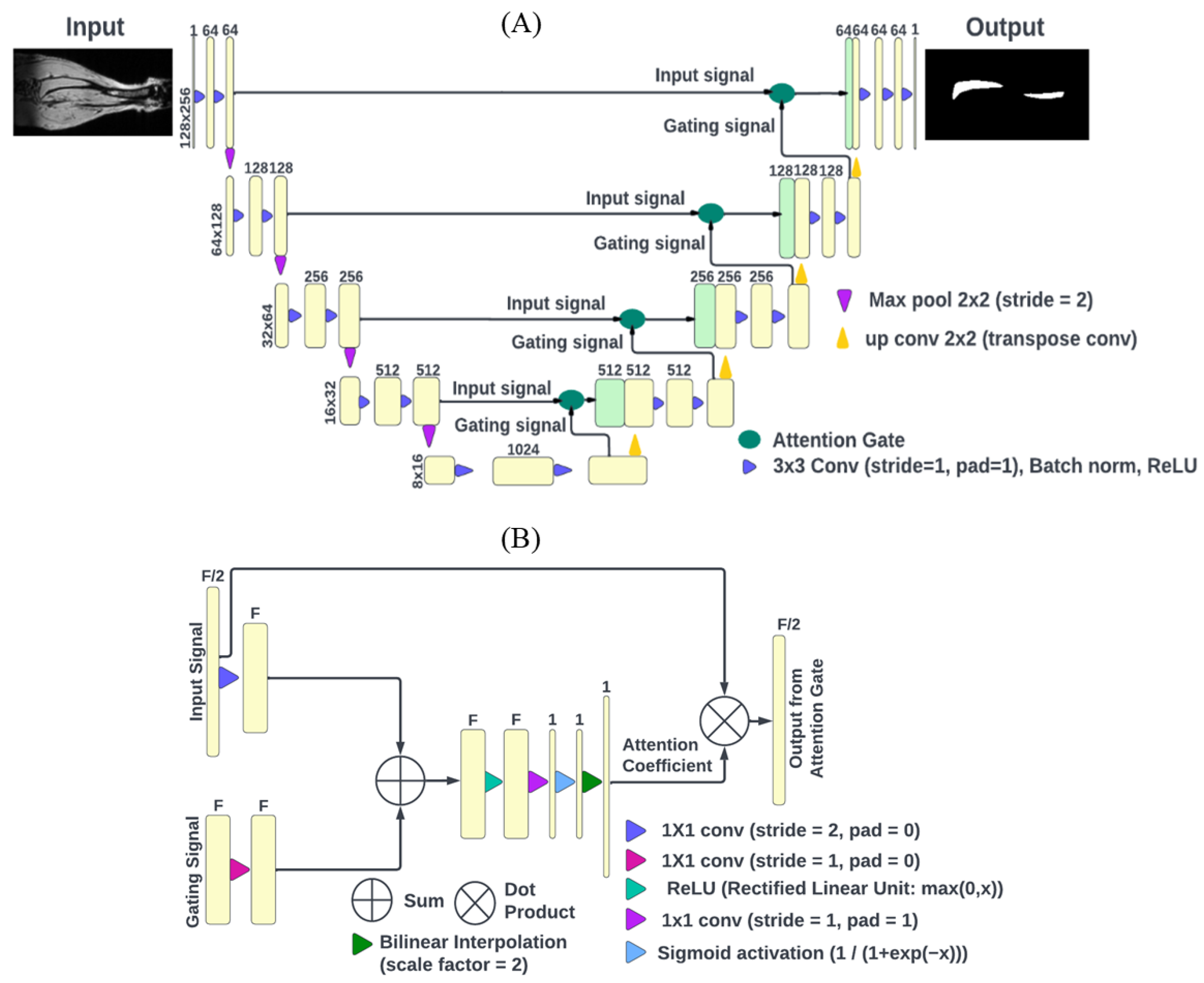
2.2. DL Model Architecture
2.3. Accuracy and Repeatability Evaluation
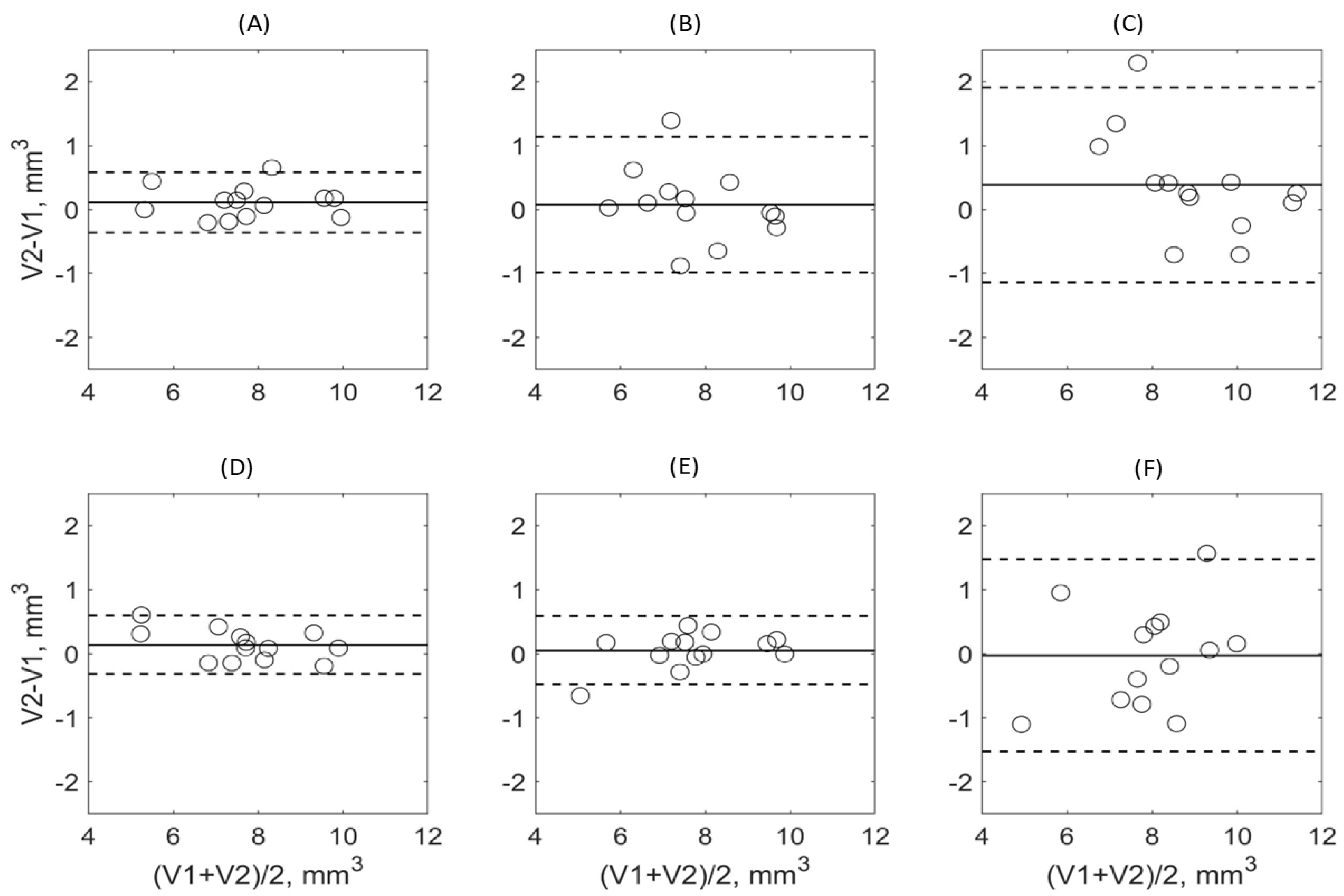
3. Results
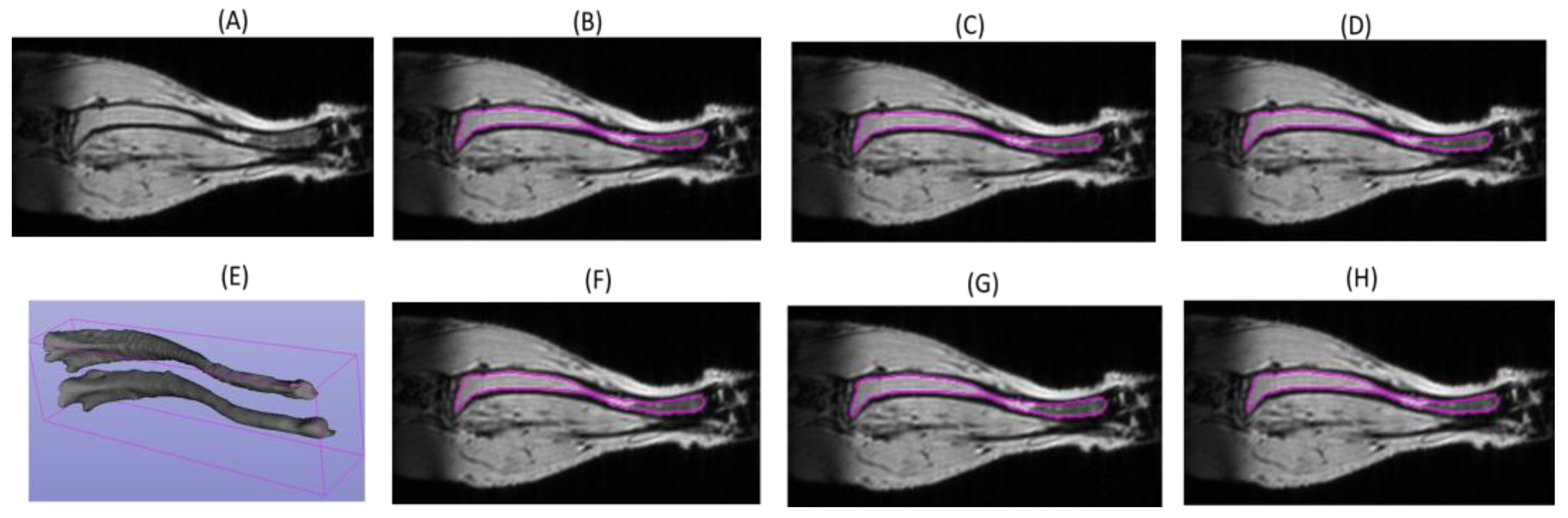
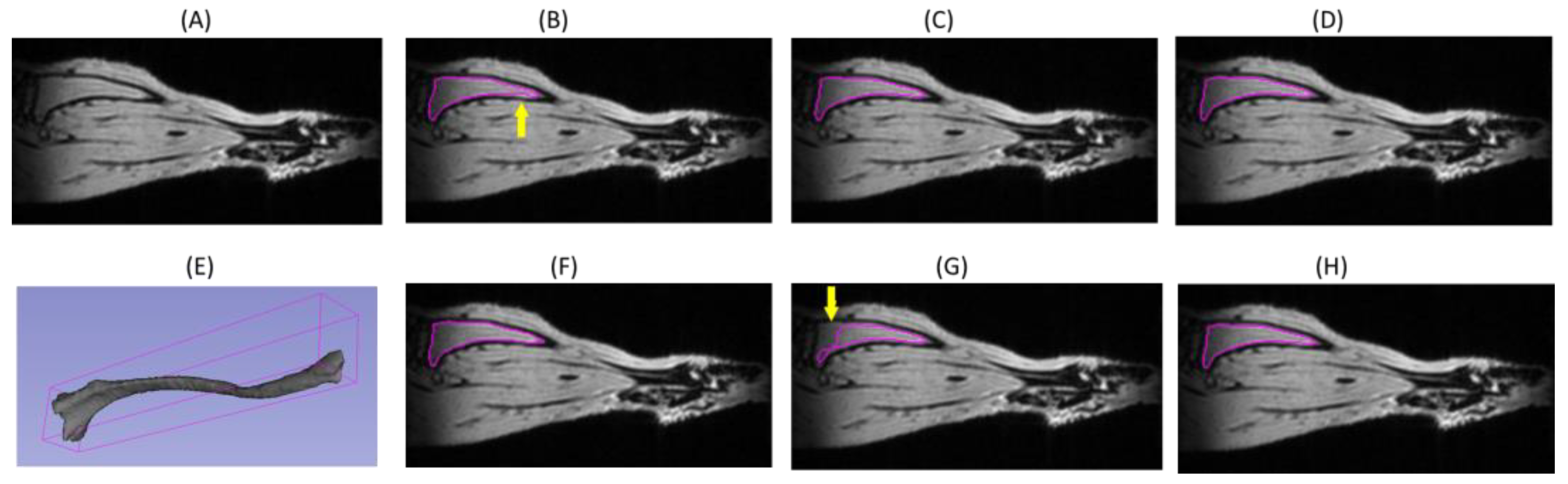
3.1. DL Model Segmenation Accuracy with Respect to Reference
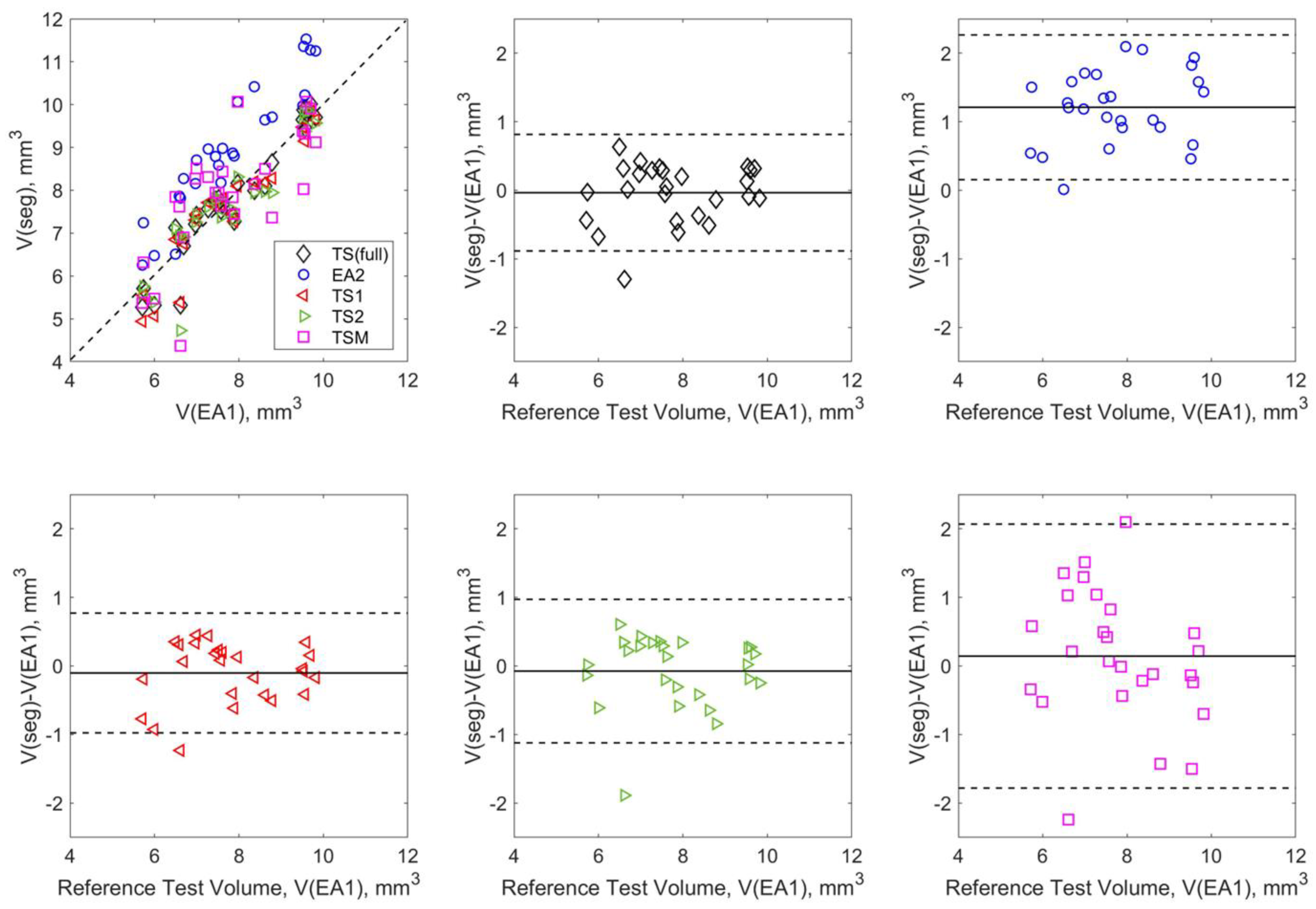
3.2. DL Model Segmenation Precision
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. MRI Acquisition Parameters
Appendix B. Performance metrics definitions
References
- Gangat, N.; Caramazza, D.; Vaidya, R.; George, G.; Begna, K.; Schwager, S.; Van Dyke, D.; Hanson, C.; Wu, W.; Pardanani, A.; et al. DIPSS Plus: A Refined Dynamic International Prognostic Scoring System for Primary Myelofibrosis That Incorporates Prognostic Information from Karyotype, Platelet Count, and Transfusion Status. J. Clin. Oncol. 2011, 29, 392–397. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Qiu, T.; Wang, H.; Yang, Q. State-of-the-Art Review on Myelofibrosis Therapies. Clin. Lymphoma Myeloma Leuk. 2022, 22, e350–e362. [Google Scholar] [CrossRef] [PubMed]
- Gianelli, U.; Vener, C.; Bossi, A.; Cortinovis, I.; Iurlo, A.; Fracchiolla, N.S.; Savi, F.; Moro, A.; Grifoni, F.; De Philippis, C.; et al. The European Consensus on grading of bone marrow fibrosis allows a better prognostication of patients with primary myelofibrosis. Mod. Pathol. 2012, 25, 1193–1202. [Google Scholar] [CrossRef] [Green Version]
- Leimkühler, N.B.; Gleitz, H.F.; Ronghui, L.; Snoeren, I.A.; Fuchs, S.N.; Nagai, J.S.; Banjanin, B.; Lam, K.H.; Vogl, T.; Kuppe, C.; et al. Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell 2021, 28, 637–652.e8. [Google Scholar] [CrossRef]
- Luker, G.D.; Nguyen, H.; Hoff, B.A.; Galbán, C.J.; Hernando, D.; Chenevert, T.L.; Talpaz, M.; Ross, B.D. A Pilot Study of Quantitative MRI Parametric Response Mapping of Bone Marrow Fat for Treatment Assessment in Myelofibrosis. Tomography 2016, 2, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Robison, T.H.; Solipuram, M.; Heist, K.; Amouzandeh, G.; Lee, W.Y.; Humphries, B.A.; Buschhaus, J.M.; Bevoor, A.; Zhang, A.; Luker, K.E.; et al. Multiparametric MRI to quantify disease and treatment response in mice with myeloproliferative neoplasms. JCI Insight 2022, 7, 161457. [Google Scholar] [CrossRef]
- Ross, B.D.; Jang, Y.; Welton, A.; Bonham, C.A.; Palagama, D.S.W.; Heist, K.; Boppisetti, J.; Imaduwage, K.P.; Robison, T.; King, L.R.; et al. A lymphatic-absorbed multi-targeted kinase inhibitor for myelofibrosis therapy. Nat. Commun. 2022, 13, 4730. [Google Scholar] [CrossRef] [PubMed]
- Obuchowski, N.A. Interpreting Change in Quantitative Imaging Biomarkers. Acad. Radiol. 2018, 25, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Raunig, D.L.; McShane, L.M.; Pennello, G.; Gatsonis, C.; Carson, P.L.; Voyvodic, J.T.; Wahl, R.L.; Kurland, B.F.; Schwarz, A.J.; Gönen, M.; et al. Quantitative imaging biomarkers: A review of statistical methods for technical performance assessment. Stat. Methods Med. Res. 2015, 24, 27–67. [Google Scholar] [CrossRef] [Green Version]
- Jha, A.K.; Mithun, S.; Jaiswar, V.; Sherkhane, U.B.; Purandare, N.C.; Prabhash, K.; Rangarajan, V.; Dekker, A.; Wee, L.; Traverso, A. Repeatability and reproducibility study of radiomic features on a phantom and human cohort. Sci. Rep. 2021, 11, 2055. [Google Scholar] [CrossRef]
- Malaih, A.A.; Dunn, J.T.; Nygård, L.; Kovacs, D.G.; Andersen, F.L.; Barrington, S.F.; Fischer, B.M. Test–retest repeatability and interobserver variation of healthy tissue metabolism using 18F-FDG PET/CT of the thorax among lung cancer patients. Nucl. Med. Commun. 2022, 43, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Shukla-Dave, A.; Obuchowski, N.A.; Chenevert, T.L.; Jambawalikar, S.; Schwartz, L.H.; Malyarenko, D.; Huang, W.; Noworolski, S.M.; Young, R.J.; Shiroishi, M.S.; et al. Quantitative imaging biomarkers alliance (QIBA) recommendations for improved precision of DWI and DCE-MRI derived biomarkers in multicenter oncology trials. J. Magn. Reson. Imaging 2019, 49, e101–e121. [Google Scholar] [CrossRef] [PubMed]
- Newitt, D.C.; Amouzandeh, G.; Partridge, S.C.; Marques, H.S.; Herman, B.A.; Ross, B.D.; Hylton, N.M.; Chenevert, T.L.; Malyarenko, D.I. Repeatability and Reproducibility of ADC Histogram Metrics from the ACRIN 6698 Breast Cancer Therapy Response Trial. Tomography 2020, 6, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Amouzandeh, G.; Heist, K.A.; Malyarenko, D.I.; Jang, J.; Robison, T.; Bonham, C.; Amirfazli, C.; Swanson, S.D.; Luker, G.D.; Ross, B.D.; et al. MR Imaging of Murine Tibia for Co-Clinical Studies of Myelofibrosis. In Proceedings of the International Society of Magnetic Resonance in Medicine Annual Meeting & Exhibition, Co-Clinical Imaging Research Symposium, Virtual, 15–20 May 2021; p. 2355. [Google Scholar]
- Becker, A.S.; Chaitanya, K.; Schawkat, K.; Muehlematter, U.J.; Hötker, A.M.; Konukoglu, E.; Donati, O.F. Variability of manual segmentation of the prostate in axial T2-weighted MRI: A multi-reader study. Eur. J. Radiol. 2019, 121, 108716. [Google Scholar] [CrossRef]
- Newitt, D.C.; Zhang, Z.; Ba, J.E.G.; Partridge, S.C.; Chenevert, T.L.; Rosen, M.A.; Bolan, P.; Marques, H.S.; Aliu, S.; Li, W.; et al. Test–retest repeatability and reproducibility of ADC measures by breast DWI: Results from the ACRIN 6698 trial. J. Magn. Reson. Imaging 2019, 49, 1617–1628. [Google Scholar] [CrossRef] [PubMed]
- Qiu, B.; van der Wel, H.; Kraeima, J.; Glas, H.H.; Guo, J.; Borra, R.J.H.; Witjes, M.J.H.; van Ooijen, P.M.A. Automatic Segmentation of Mandible from Conventional Methods to Deep Learning—A Review. J. Pers. Med. 2021, 11, 629. [Google Scholar] [CrossRef] [PubMed]
- Renard, F.; Guedria, S.; De Palma, N.; Vuillerme, N. Variability and reproducibility in deep learning for medical image segmentation. Sci. Rep. 2020, 10, 13724. [Google Scholar] [CrossRef]
- Ma, X.; Hadjiiski, L.M.; Wei, J.; Chan, H.; Cha, K.H.; Cohan, R.H.; Caoili, E.M.; Samala, R.; Zhou, C.; Lu, Y. U-Net based deep learning bladder segmentation in CT urography. Med. Phys. 2019, 46, 1752–1765. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, C.; Chan, H.-P.; Hadjiiski, L.M.; Chughtai, A. Fusion of multiple deep convolutional neural networks (DCNNs) for improved segmentation of lung nodules in CT images. In Proceedings of the SPIE Medical Imaging 2022, San Diego, CA, USA, 20–24 February 2022; Volume 12033. [Google Scholar] [CrossRef]
- Chan, H.-P.; Samala, R.K.; Hadjiiski, L.M.; Zhou, C. Deep Learning in Medical Image Analysis. Adv. Exp. Med. Biol. 2020, 1213, 3–21. [Google Scholar] [CrossRef]
- Heckelman, L.N.; Soher, B.J.; Spritzer, C.E.; Lewis, B.D.; DeFrate, L.E. Design and validation of a semi-automatic bone segmentation algorithm from MRI to improve research efficiency. Sci. Rep. 2022, 12, 7825. [Google Scholar] [CrossRef]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, A.; Mourad, R.F.; Heist, K.; Malyarenko, D.; Chan, H.P.; Chenevert, T.L.; Hadjiiski, L.M. Segmentation of Mouse Tibia on MRI using Deep Learning U-Net Models. In Proceedings of the SPIE Medical Imaging, San Deigo, CA, USA, 19–23 February 2023. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention Part III, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef] [Green Version]
- Obuchowski, N.A.; Bullen, J. Quantitative imaging biomarkers: Effect of sample size and bias on confidence interval coverage. Stat. Methods Med. Res. 2018, 27, 3139–3150. [Google Scholar] [CrossRef] [PubMed]
- Mali, S.A.; Ibrahim, A.; Woodruff, H.C.; Andrearczyk, V.; Müller, H.; Primakov, S.; Salahuddin, Z.; Chatterjee, A.; Lambin, P. Making Radiomics More Reproducible across Scanner and Imaging Protocol Variations: A Review of Harmonization Methods. J. Pers. Med. 2021, 11, 842. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, C.; Wang, R.; Liu, Z.; Wang, M.; Tan, H.; Wu, Y.; Liu, X.; Sun, H.; Yang, R.; et al. Annotation-efficient deep learning for automatic medical image segmentation. Nat. Commun. 2021, 12, 5915. [Google Scholar] [CrossRef]
- Candemir, S.; Nguyen, X.V.; Folio, L.R.; Prevedello, L.M. Training Strategies for Radiology Deep Learning Models in Data-limited Scenarios. Radiol. Artif. Intell. 2021, 3, e210014. [Google Scholar] [CrossRef]
- Madireddy, I.; Wu, T. Rule and Neural Network-Based Image Segmentation of Mice Vertebrae Images. Cureus 2022, 14, e27247. [Google Scholar] [CrossRef]
- Hadjiiski, L.; Cha, K.; Chan, H.; Drukker, K.; Morra, L.; Näppi, J.J.; Sahiner, B.; Yoshida, H.; Chen, Q.; Deserno, T.M.; et al. AAPM task group report 273: Recommendations on best practices for AI and machine learning for computer-aided diagnosis in medical imaging. Med. Phys. 2023, 50, e1–e24. [Google Scholar] [CrossRef]
- Holbrook, M.D.; Blocker, S.J.; Mowery, Y.M.; Badea, A.; Qi, Y.; Xu, E.S.; Kirsch, D.G.; Johnson, G.A.; Badea, C. MRI-Based Deep Learning Segmentation and Radiomics of Sarcoma in Mice. Tomography 2020, 6, 23–33. [Google Scholar] [CrossRef]
- Seo, S.Y.; Kim, S.-J.; Oh, J.S.; Chung, J.; Kim, S.-Y.; Oh, S.J.; Joo, S.; Kim, J.S. Unified Deep Learning-Based Mouse Brain MR Segmentation: Template-Based Individual Brain Positron Emission Tomography Volumes-of-Interest Generation Without Spatial Normalization in Mouse Alzheimer Model. Front. Aging Neurosci. 2022, 14, 807903. [Google Scholar] [CrossRef]
- Kenney, H.M.; Peng, Y.; Chen, K.L.; Ajalik, R.; Schnur, L.; Wood, R.W.; Schwarz, E.M.; Awad, H.A. A high-throughput semi-automated bone segmentation workflow for murine hindpaw micro-CT datasets. Bone Rep. 2022, 16, 101167. [Google Scholar] [CrossRef]
- Schoppe, O.; Pan, C.; Coronel, J.; Mai, H.; Rong, Z.; Todorov, M.I.; Müskes, A.; Navarro, F.; Li, H.; Ertürk, A.; et al. Deep learning-enabled multi-organ segmentation in whole-body mouse scans. Nat. Commun. 2020, 11, 5626. [Google Scholar] [CrossRef] [PubMed]

| Dataset | AJI% | AVI % | AVE% | AHD (mm) | |
|---|---|---|---|---|---|
| Attention U-Net trained on full training set | Training | 88.63 ± 1.21 | 94.64 ± 1.64 | −1.44 ± 3.07 | 0.20 ± 0.05 |
| Validation | 83.08 ± 2.68 | 91.06 ± 3.79 | −0.67 ± 6.28 | 0.52 ± 0.30 | |
| Test | 83.45 ± 5.11 | 90.21 ± 6.28 | 1.71 ± 7.94 | 0.47 ± 0.43 | |
| Attention U-Net trained on split1 | Training | 86.17 ± 2.22 | 93.51 ± 2.74 | −2.06 ± 4.92 | 0.39 ± 0.68 |
| Validation | 81.47 ± 4.58 | 89.27 ± 6.18 | 1.22 ± 8.65 | 0.57 ± 0.28 | |
| Test | 82.47 ± 6.79 | 89.15 ± 8.17 | 2.83 ± 9.89 | 0.79 ± 1.39 | |
| Attention U-Net trained on split2 | Training | 88.34 ± 1.15 | 94.79 ± 1.57 | −2.11 ± 3.19 | 0.23 ± 0.06 |
| Validation | 82.78 ± 2.65 | 91.15 ± 3.85 | −1.29 ± 6.99 | 0.78 ± 0.77 | |
| Test | 83.27 ± 5.01 | 89.94 ± 6.39 | 2.11 ± 8.35 | 0.49 ± 0.42 | |
| Attention U-Net trained on single mouse | Training | 86.38 ± 0.69 | 97.32 ± 1.09 | −9.99 ± 2.97 | 0.29 ± 0.22 |
| Validation | 76.42 ± 6.49 | 87.70 ± 8.45 | −2.41 ± 11.63 | 1.45 ± 1.41 | |
| Test | 77.99 ± 7.43 | 87.77 ± 10.05 | −0.15 ± 14.31 | 1.10 ± 1.07 | |
| EA1 vs. EA2 (EA2 reference) | Training | NA | |||
| Validation | 77.98 ± 2.63 | 97.39 ± 1.39 | −22.44 ± 6.46 | 0.36 ± 0.15 | |
| Test | 80.70 ± 2.91 | 96.70 ± 2.33 | −16.70 ± 7.66 | 0.29 ± 0.11 | |
| EA2 vs. EA1 (EA1 reference) | Training | NA | |||
| Validation | 77.98 ± 2.64 | 79.70 ± 3.36 | 18.11 ± 4.39 | 0.36 ± 0.15 | |
| Test | 80.70 ± 2.91 | 83.10 ± 3.92 | 13.94 ± 5.76 | 0.29 ± 0.11 | |
| Model | Training | Validation | Test | |
|---|---|---|---|---|
| wCV [CI] % | ||||
| A-U-net | Full | 7.7 [6.2, 10.3] 7.6 [5.5, 12.0] 8.0 [6.0, 12.3] 3.3 [2, 12] | 2.0 [1.1, 7.4] 5.3 [3.0, 20.0] 1.83 [1.0, 6.8] 11.1 [6.3, 41.6] | 2.6 [1.9, 4.3] 3.1 [2.2, 5.1] 3.2 [2.3, 5.3] 7.0 [5.0, 11.6] |
| TS1 | ||||
| TS2 | ||||
| TSM | ||||
| EA1 | Full | 7.7 [6.2, 10.3] 7.3 [5.4, 11.5] 8.0 [6.0, 12.3] 5.8 [4.6, 7.7] | 4.4 [2.5, 16.3] | 5.3 [3.8, 8.7] |
| TS1 | ||||
| TS2 | ||||
| TSM | ||||
| EA2 | Full | NA | 2.5 [1.4, 9.1] | 8.0 [5.7, 13.2] |
| TS1 | ||||
| TS2 | ||||
| TSM | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kushwaha, A.; Mourad, R.F.; Heist, K.; Tariq, H.; Chan, H.-P.; Ross, B.D.; Chenevert, T.L.; Malyarenko, D.; Hadjiiski, L.M. Improved Repeatability of Mouse Tibia Volume Segmentation in Murine Myelofibrosis Model Using Deep Learning. Tomography 2023, 9, 589-602. https://doi.org/10.3390/tomography9020048
Kushwaha A, Mourad RF, Heist K, Tariq H, Chan H-P, Ross BD, Chenevert TL, Malyarenko D, Hadjiiski LM. Improved Repeatability of Mouse Tibia Volume Segmentation in Murine Myelofibrosis Model Using Deep Learning. Tomography. 2023; 9(2):589-602. https://doi.org/10.3390/tomography9020048
Chicago/Turabian StyleKushwaha, Aman, Rami F. Mourad, Kevin Heist, Humera Tariq, Heang-Ping Chan, Brian D. Ross, Thomas L. Chenevert, Dariya Malyarenko, and Lubomir M. Hadjiiski. 2023. "Improved Repeatability of Mouse Tibia Volume Segmentation in Murine Myelofibrosis Model Using Deep Learning" Tomography 9, no. 2: 589-602. https://doi.org/10.3390/tomography9020048
APA StyleKushwaha, A., Mourad, R. F., Heist, K., Tariq, H., Chan, H. -P., Ross, B. D., Chenevert, T. L., Malyarenko, D., & Hadjiiski, L. M. (2023). Improved Repeatability of Mouse Tibia Volume Segmentation in Murine Myelofibrosis Model Using Deep Learning. Tomography, 9(2), 589-602. https://doi.org/10.3390/tomography9020048






