A Novel and Quantitative Detection Assay (effluxR) for Identifying Efflux-Associated Resistance Genes Using Multiplex Digital PCR in Clinical Isolates of Pseudomonas aeruginosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Genomic DNA Extraction
2.3. Detection of the Mex Efflux Pump Genes Using Multiplex Quantitative PCR (mqPCR)
2.3.1. Investigation of Optimal mqPCR Annealing/Extension Temperature
2.3.2. Investigation of Optimal gDNA Concentration of P. aeruginosa Strains
2.4. DNA Agarose Gel Electrophoresis
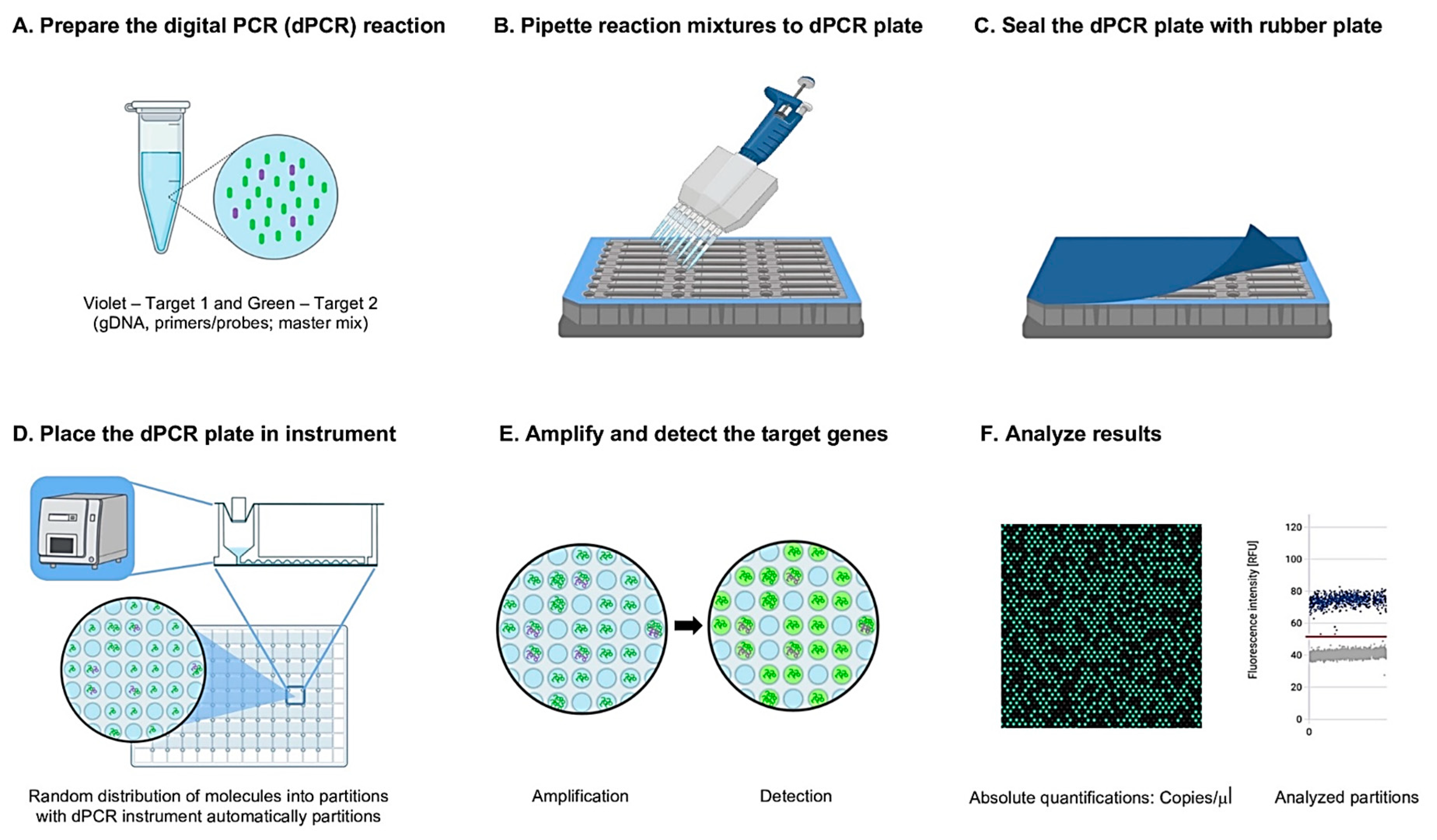
2.5. Development of the EffluxR Detection Assay to Detect the mex Efflux Pump Genes Using Multiplex Digital PCR (mdPCR)
2.5.1. Optimization of gDNA Concentration in P. aeruginosa
2.5.2. Limit of Detection (LOD) of the effluxR Detection Assay Using the mdPCR System
2.5.3. Sensitivity and Specificity Determination of the effluxR Detection Assay Using the mdPCR System
2.6. Statistical Analysis
3. Results
3.1. Optimising Annealing/Extension Temperatures for Amplifying the RND Genes Using mqPCR
3.2. Optimal gDNA Concentration of P. aeruginosa ATCC for Amplifying the Mex Genes Using mqPCR
3.3. MexB, mexD, mexY, and 16s rRNA Bands were Detected in All P. aeruginosa Strains Using Agarose Gel Electrophoresis
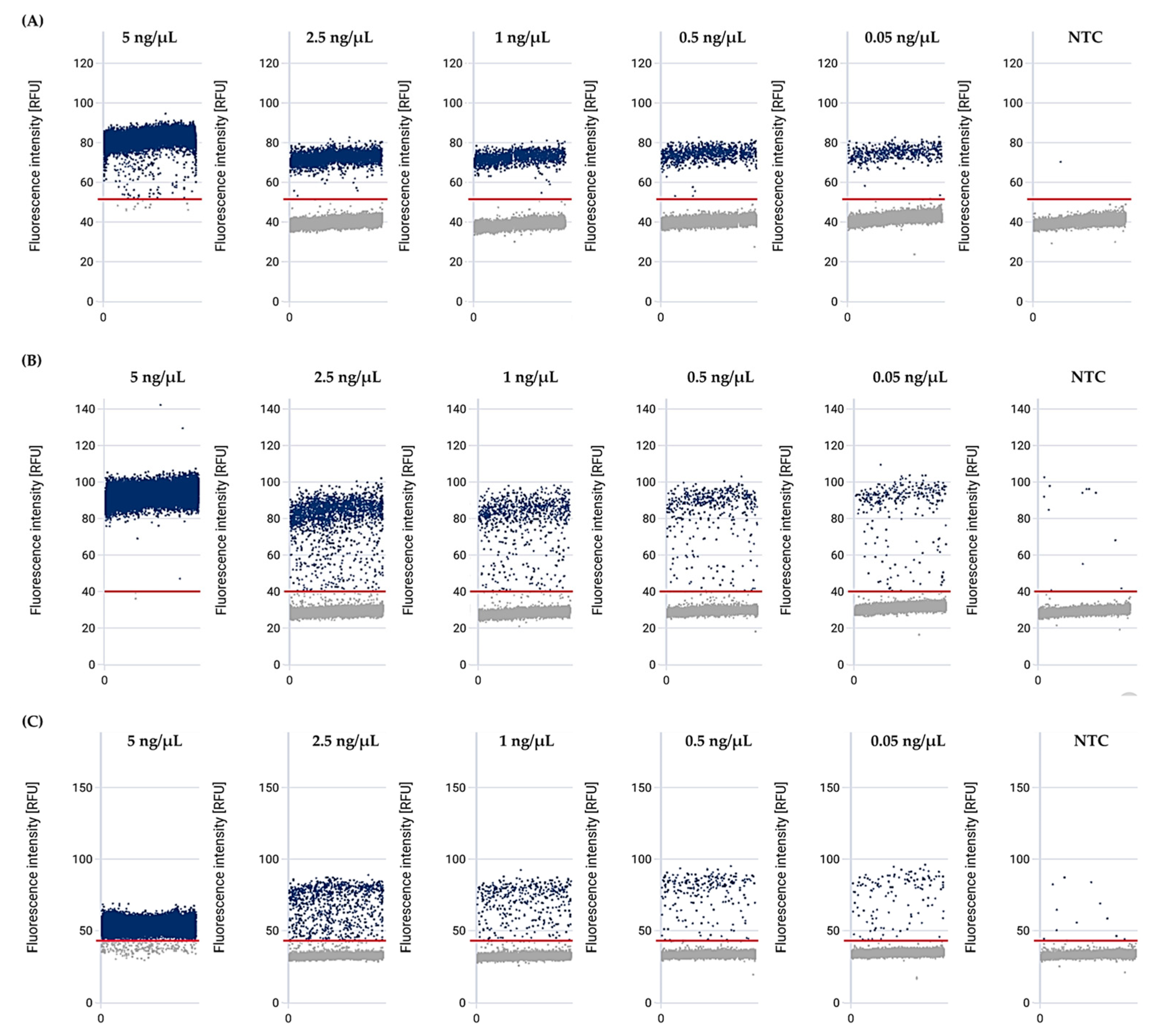
3.4. The Mex Efflux Pump Genes Can Be Detected at a Range of gDNA Concentrations in P. aeruginosa Using the effluxR Detection Assay with the mdPCR System
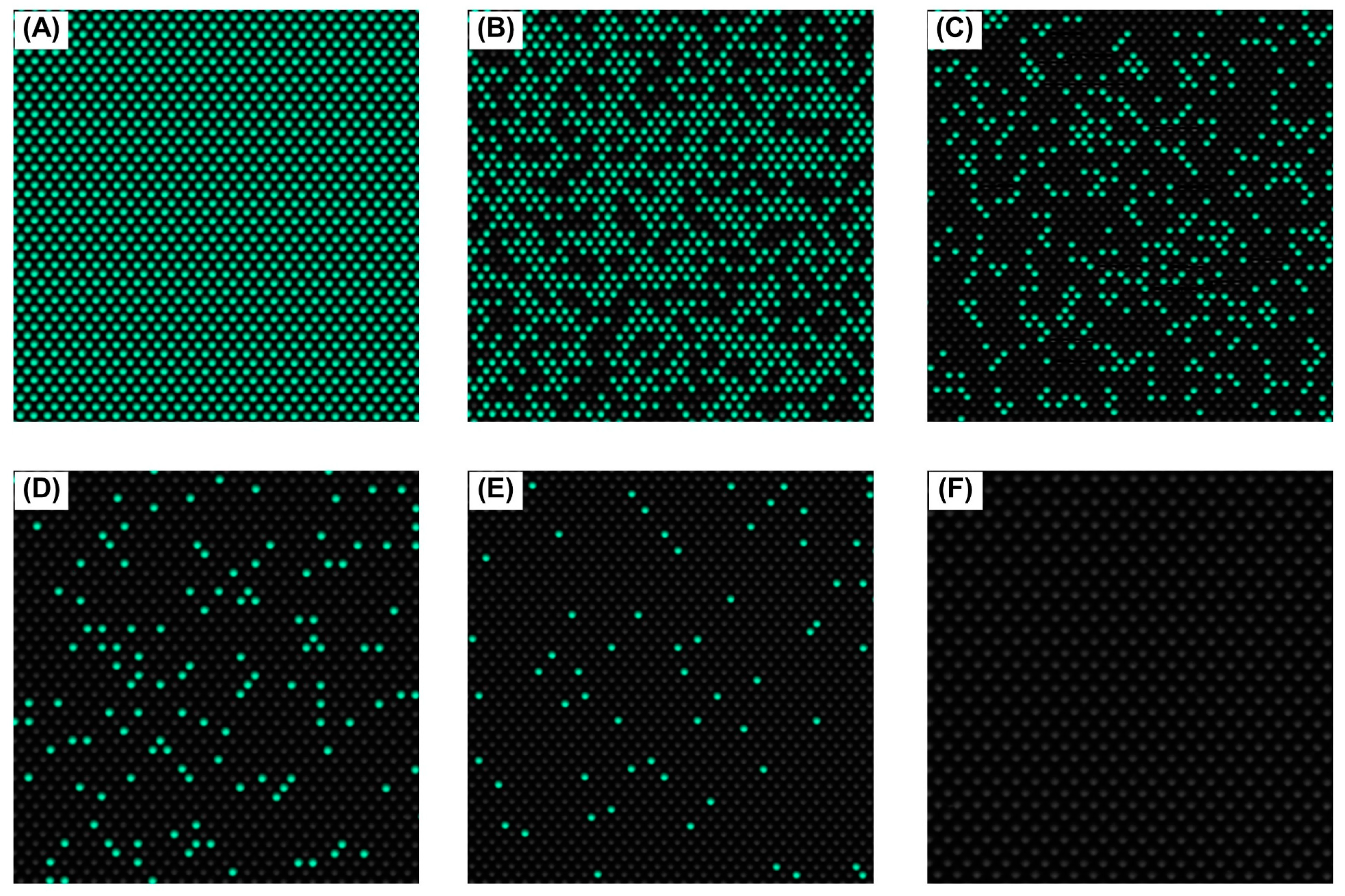
3.5. Detection Limit of the effluxR Detection Assay with the mdPCR System Is 0.001 ng/µL Equivalent to 7.04 copies/µL of the Mex Efflux Pump Genes in P. aeruginosa ATCC27853
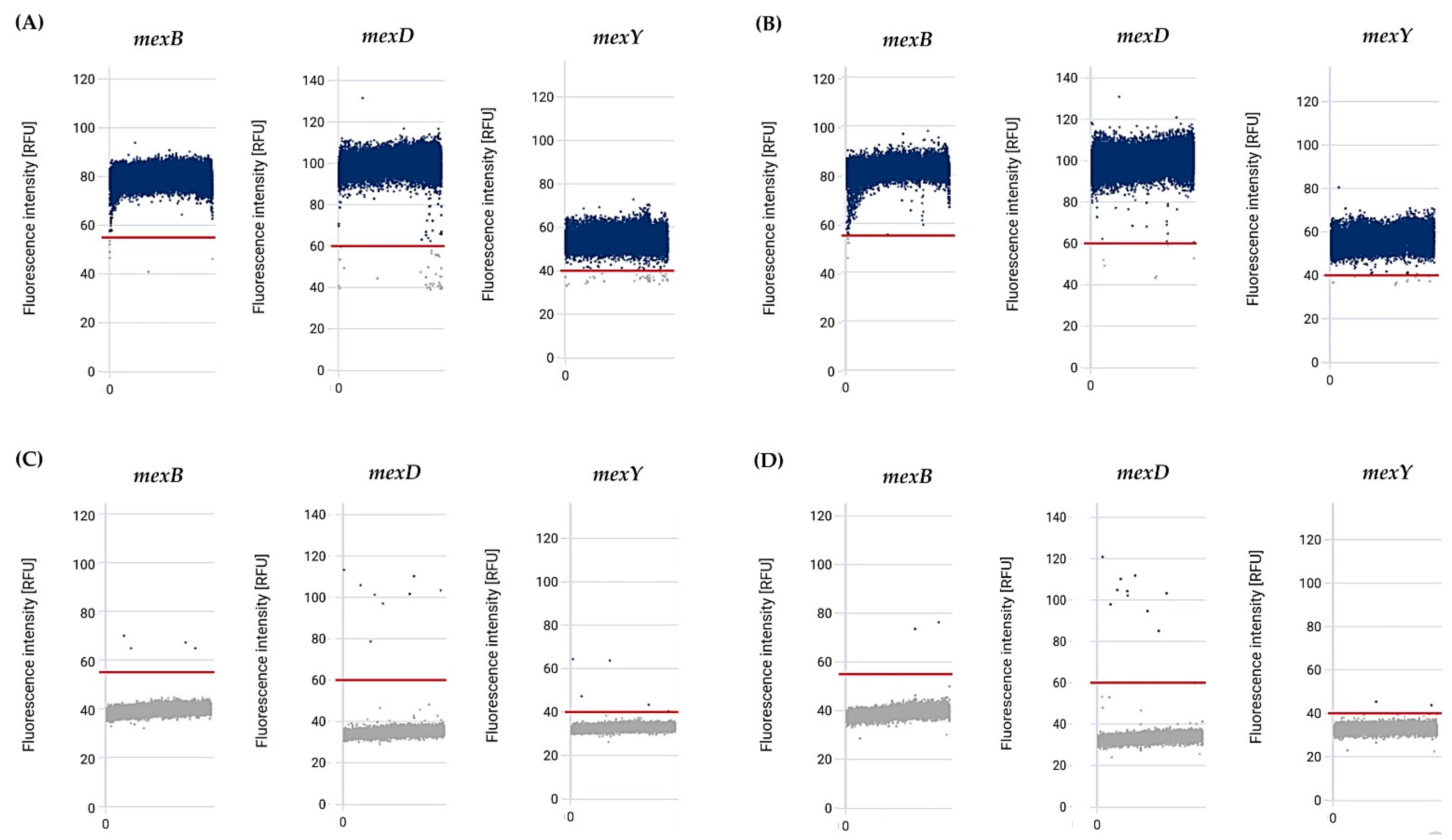
3.6. Sensitivity and Specificity of the effluxR Detection Assay with mdPCR System Were 100% for Detecting the Mex Efflux Pump Genes in the P. aeruginosa Strains
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhanel, G.G.; DeCorby, M.; Adam, H.; Mulvey, M.R.; McCracken, M.; Lagacé-Wiens, P.; Nichol, K.A.; Wierzbowski, A.; Baudry, P.J.; Tailor, F.; et al. Prevalence of antimicrobial-resistant pathogens in Canadian hospitals: Results of the Canadian Ward Surveillance Study (CANWARD 2008). Antimicrob. Agents Chemother. 2010, 54, 4684–4693. [Google Scholar] [CrossRef]
- Mahar, P.; Padiglione, A.A.; Cleland, H.; Paul, E.; Hinrichs, M.; Wasiak, J. Pseudomonas aeruginosa bacteraemia in burns patients: Risk factors and outcomes. Burns 2010, 36, 1228–1233. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.L.; Suetens, C.; Savey, A.; Palomar, M.; Hiesmayr, M.; Morales, I.; Agodi, A.; Frank, U.; Mertens, K.; Schumacher, M.; et al. Clinical outcomes of health-care-associated infections and antimicrobial resistance in patients admitted to European intensive-care units: A cohort study. Lancet Infect. Dis. 2011, 11, 30–38. [Google Scholar] [CrossRef]
- Alp, E.; Güven, M.; Yıldız, O.; Aygen, B.; Voss, A.; Doganay, M. Incidence, risk factors and mortality of nosocomial pneumonia in Intensive Care Units: A prospective study. Ann. Clin. Microbiol. Antimicrob. 2004, 3, 17. [Google Scholar] [CrossRef]
- Aliaga, L.; Mediavilla, J.D.; Cobo, F. A clinical index predicting mortality with Pseudomonas aeruginosa bacteraemia. J. Med. Microbiol. 2002, 51, 615–701. [Google Scholar] [CrossRef] [PubMed]
- Klockgether, J.; Cramer, N.; Wiehlmann, L.; Davenport, C.; Tümmler, B. Pseudomonas aeruginosa genomic structure and diversity. Front. Microbiol. 2011, 2, 150. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Outer membranes and efflux: The path to multidrug resistance in Gram-negative bacteria. Curr. Pharm. Biotechnol. 2002, 3, 77–98. [Google Scholar] [CrossRef]
- Hirsch, E.B.; Tam, V.H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon. Outcomes Res. 2010, 10, 441–451. [Google Scholar] [CrossRef]
- Kallen, A.J.; Hidron, A.I.; Patel, J.; Srinivasan, A. Multidrug resistance among gram-negative pathogens that caused healthcare-associated infections reported to the National Healthcare Safety Network, 2006–2008. Infect. Control. Hosp. Epidemiol. 2010, 31, 528–531. [Google Scholar] [CrossRef] [PubMed]
- Kerr, K.G.; Snelling, A.M. Pseudomonas aeruginosa: A formidable and ever-present adversary. J. Hosp. Infect. 2009, 73, 338–344. [Google Scholar] [CrossRef]
- Peña, C.; Cabot, G.; Gómez-Zorrilla, S.; Zamorano, L.; Ocampo-Sosa, A.; Murillas, J.; Almirante, B.; Pomar, V.; Aguilar, M.; Granados, A.; et al. Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin. Infect. Dis. 2015, 60, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Sader, H.S.; Castanheira, M.; Duncan, L.R.; Flamm, R.K. Antimicrobial susceptibility of Enterobacteriaceae and Pseudomonas aeruginosa isolates from United States Medical Centers stratified by infection type: Results from the International Network for Optimal Resistance Monitoring (INFORM) surveillance program, 2015–2016. Diagn. Microbiol. Infect. Dis. 2018, 92, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Walkty, A.; Lagace-Wiens, P.; Adam, H.; Baxter, M.; Karlowsky, J.; Mulvey, M.R.; McCracken, M.; Zhanel, G.G. Antimicrobial susceptibility of 2906 Pseudomonas aeruginosa clinical isolates obtained from patients in Canadian hospitals over a period of 8 years: Results of the Canadian Ward surveillance study (CANWARD), 2008–2015. Diagn. Microbiol. Infect. Dis. 2017, 87, 60–63. [Google Scholar] [CrossRef]
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic resistance. J. Infect. Public Health 2017, 10, 369–378. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Bouzid, D.; Zanella, M.C.; Kerneis, S.; Visseaux, B.; May, L.; Schrenzel, J.; Cattoir, V. Rapid diagnostic tests for infectious diseases in the emergency department. Clin. Microbiol. Infect. 2021, 27, 182–191. [Google Scholar] [CrossRef]
- Dreier, J.; Ruggerone, P. Interaction of antibacterial compounds with RND efflux pumps in Pseudomonas aeruginosa. Front. Microbiol. 2015, 6, 660. [Google Scholar] [CrossRef]
- Colclough, A.L.; Alav, I.; Whittle, E.E.; Pugh, H.L.; Darby, E.M.; Legood, S.W.; McNeil, H.E.; Blair, J.M. RND efflux pumps in Gram-negative bacteria; regulation, structure and role in antibiotic resistance. Future Microbiol. 2020, 15, 143–157. [Google Scholar] [CrossRef]
- Fernández, L.; Hancock, R.E.W. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef] [PubMed]
- Aghazadeh, M.; Hojabri, Z.; Mahdian, R.; Nahaei, M.R.; Rahmati, M.; Hojabri, T.; Pirzadeh, T.; Pajand, O. Role of efflux pumps: MexAB-OprM and MexXY(-OprA), AmpC cephalosporinase and OprD porin in non-metallo-β-lactamase producing Pseudomonas aeruginosa isolated from cystic fibrosis and burn patients. Infect. Genet. Evol. 2014, 24, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef]
- Venter, H.; Mowla, R.; Ohene-Agyei, T.; Ma, S. RND-type drug efflux pumps from Gram-negative bacteria: Molecular mechanism and inhibition. Front. Microbiol. 2015, 6, 377. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Tomida, J.; Kawamura, Y. MexXY multidrug efflux system of Pseudomonas aeruginosa. Front. Microbiol. 2012, 3, 408. [Google Scholar] [CrossRef] [PubMed]
- Poole, K. Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 2005, 56, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Galhano, B.S.P.; Ferrari, R.G.; Panzenhagen, P.; de Jesus, A.C.S.; Conte-Junior, C.A. Antimicrobial resistance gene detection methods for bacteria in animal-based foods: A brief review of highlights and advantages. Microorganisms 2021, 9, 923. [Google Scholar] [CrossRef]
- Seedy, F.R.E.; Samy, A.A.; Salam, H.S.H.; Khairy, E.A.; Koraney, A.A. Polymerase chain reaction detection of genes responsible for multiple antibiotic resistance Staphylococcus aureus isolated from food of animal origin in Egypt. Vet. World 2017, 10, 1205–1211. [Google Scholar] [CrossRef]
- Ahmed, W.; Smith, W.J.M.; Metcalfe, S.; Jackson, G.; Choi, P.M.; Morrison, M.; Field, D.; Gyawali, P.; Bivins, A.; Bibby, K.; et al. Comparison of RT-qPCR and RT-dPCR platforms for the trace detection of SARS-CoV-2 RNA in wastewater. ACS ES&T Water 2022, 2, 1871–1880. [Google Scholar] [CrossRef]
- Mesaros, N.; Glupczynski, Y.; Avrain, L.; Caceres, N.E.; Tulkens, P.M.; Van Bambeke, F. A combined phenotypic and genotypic method for the detection of Mex efflux pumps in Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2007, 59, 378–386. [Google Scholar] [CrossRef]
- Rattanachak, N.; Weawsiangsang, S.; Jongjitvimol, T.; Baldock, R.A.; Jongjitwimol, J. Hydroquinine possesses antibacterial activity, and at half the MIC, induces the overexpression of RND-type efflux pumps using Multiplex Digital PCR in Pseudomonas aeruginosa. Trop. Med. Int. Health 2022, 7, 156. [Google Scholar] [CrossRef]
- Kaprou, G.D.; Bergšpica, I.; Alexa, E.A.; Alvarez-Ordóñez, A.; Prieto, M. Rapid methods for antimicrobial resistance diagnostics. Antibiotics 2021, 10, 209. [Google Scholar] [CrossRef] [PubMed]
- Nakae, T.; Nakajima, A.; Ono, T.; Saito, K.; Yoneyama, H. Resistance to beta-lactam antibiotics in Pseudomonas aeruginosa due to interplay between the MexAB-OprM efflux pump and beta-lactamase. Antimicrob. Agents Chemother. 1999, 43, 1301–1303. [Google Scholar] [CrossRef] [PubMed]
- Bialvaei, A.Z.; Rahbar, M.; Hamidi-Farahani, R.; Asgari, A.; Esmailkhani, A.; Dashti, Y.M.; Soleiman-Meigooni, S. Expression of RND efflux pumps mediated antibiotic resistance in Pseudomonas aeruginosa clinical strains. Microb. Pathog. 2021, 153, 104789. [Google Scholar] [CrossRef]
- Schweizer, H.P. Efflux as a mechanism of resistance to antimicrobials in Pseudomonas aeruginosa and related bacteria: Unanswered questions. Genet. Mol. Res. 2003, 2, 48–62. [Google Scholar] [PubMed]
- Valot, B.; Guyeux, C.; Rolland, J.Y.; Mazouzi, K.; Bertrand, X.; Hocquet, D. What It takes to be a Pseudomonas aeruginosa? The core genome of the opportunistic pathogen updated. PLoS ONE 2015, 10, e0126468. [Google Scholar] [CrossRef]
- Anes, J.; McCusker, M.P.; Fanning, S.; Martins, M. The ins and outs of RND efflux pumps in Escherichia coli. Front. Microbiol. 2015, 6, 587. [Google Scholar] [CrossRef]
- Ni, R.T.; Onishi, M.; Mizusawa, M.; Kitagawa, R.; Kishino, T.; Matsubara, F.; Tsuchiya, T.; Kuroda, T.; Ogawa, W. The role of RND-type efflux pumps in multidrug-resistant mutants of Klebsiella pneumoniae. Sci. Rep. 2020, 10, 10876. [Google Scholar] [CrossRef]
- Guérin, F.; Lallement, C.; Isnard, C.; Dhalluin, A.; Cattoir, V.; Giard, J.C. Landscape of Resistance-Nodulation-Cell Division (RND)-type efflux pumps in Enterobacter cloacae complex. Antimicrob. Agents Chemother. 2016, 60, 2373–2382. [Google Scholar] [CrossRef]
- Dashtbani-Roozbehani, A.; Brown, M.H. Efflux pump mediated antimicrobial resistance by Staphylococci in health-related environments: Challenges and the quest for inhibition. Antibiotics 2021, 10, 1502. [Google Scholar] [CrossRef]
- Frayling, I.M.; Monk, E.; Butler, R. PCR-Based Methods for Mutation Detection. In Molecular Diagnostics: For the Clinical Laboratorian; Coleman, W.B., Tsongalis, G.J., Eds.; Humana Press: Totowa, NJ, USA, 2005; pp. 65–74. [Google Scholar]
- Huletsky, A.; Bergeron, M.G. Bacterial Genotypic Drug Resistance Assays. In Antimicrobial Drug Resistance: Clinical and Epidemiological Aspects; Mayers, D.L., Sobel, J.D., Ouellette, M., Kaye, K.S., Marchaim, D., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 2, pp. 1465–1499. [Google Scholar]
- Martins, M.; Santos, B.; Martins, A.; Viveiros, M.; Couto, I.; Cruz, A.; Pagès, J.M.; Molnar, J.; Fanning, S.; Amaral, L. An instrument-free method for the demonstration of efflux pump activity of bacteria. In Vivo 2006, 20, 657–664. [Google Scholar] [PubMed]
- Martins, M.; Couto, I.; Viveiros, M.; Amaral, L. Identification of efflux-mediated multi-drug resistance in bacterial clinical isolates by two simple methods. Methods Mol. Biol. 2010, 642, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Cattoir, V. Efflux-mediated antibiotics resistance in bacteria. Pathol. Biol. 2004, 52, 607–616. [Google Scholar] [CrossRef]
- Paixão, L.; Rodrigues, L.; Couto, I.; Martins, M.; Fernandes, P.; de Carvalho, C.C.C.R.; Monteiro, G.A.; Sansonetty, F.; Amaral, L.; Viveiros, M. Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng. 2009, 3, 18. [Google Scholar] [CrossRef]
- Saeidnia, S.; Abdollahi, M. Are other fluorescent tags used instead of ethidium bromide safer? DARU J. Pharm. Sci. 2013, 21, 71. [Google Scholar] [CrossRef]
- Whittle, E.E.; McNeil, H.E.; Trampari, E.; Webber, M.; Overton, T.W.; Blair, J.M.A. Efflux impacts intracellular accumulation only in actively growing bacterial cells. mBio 2021, 12, e0260821. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, O.; Warren, M.S.; Lee, A.; Galazzo, J.; Fronko, R.; Lee, M.; Blais, J.; Cho, D.; Chamberland, S.; Renau, T.; et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob. Agents Chemother. 2001, 45, 105–116. [Google Scholar] [CrossRef]

| Name | Oligonucleotide Sequences (5′ to 3′) | PCR Product Size (bp) | References |
|---|---|---|---|
| mexB | 199 | [31] | |
| F_primer | GATAGGCCCATTTTCGCGTGG | ||
| R_primer | CGATCCCGTTCATCTGCTGC | ||
| Probe | (FAM)CGCCTTGGTGATCATGCTCGCG(BHQ1) | ||
| mexD | 131 | [31] | |
| F_primer | TCATCAAGCGGCCGAACTTC | ||
| R_primer | GGTGGCGGTGATGGTGATCTG | ||
| Probe | (HEX)CTGGCCGGCCTGCTGGTCATTTC(BHQ1) | ||
| mexY | 168 | [31] | |
| F_primer | CGCAACTGACCCGCTACAAC | ||
| R_primer | CGGACAGGCGTTCTTCGAAG | ||
| Probe | (Texas Red)CGAAGCCATGCAGGCGATGGAGG(BHQ2) | ||
| 16s rRNA | 225 | [31] | |
| F_primer | CATGGCTCAGATTGAACGCTG | ||
| R_primer | GCTAATCCGACCTAGGCTCATC | ||
| Probe | (Cy5)CGAGCGGATGAAGGGAGCTTGCTC(BHQ2) | ||
| Strains | Genes | Cycle Threshold Values in Gradient Annealing/Extension Temperature (°C) | p-Value | ||||
|---|---|---|---|---|---|---|---|
| 58 | 59 | 60 | 61 | 62 | |||
| P. aeruginosa ATCC27853 | mexB | 16.50 ± 1.10 | 16.47 ± 1.13 | 16.53 ± 1.37 | 16.48 ± 1.77 | 16.99 ± 1.89 | >0.9999 |
| mexD | 16.24 ± 0.95 | 15.57 ± 0.97 | 15.99 ± 1.57 | 16.69 ± 1.04 | 17.66 ± 1.16 | >0.9999 | |
| mexY | 16.12 ± 0.98 | 15.38 ± 1.10 | 15.76 ± 1.87 | 16.38 ± 1.18 | 17.31 ± 1.45 | >0.9999 | |
| 16S rRNA | 13.17 ± 1.60 | 13.50 ± 0.33 | 14.03 ± 0.96 | 14.56 ± 1.45 | 14.67 ± 0.42 | >0.9999 | |
| P. aeruginosa ATCC BAA-2108 | mexB | 19.77 ± 0.98 | 20.61 ± 1.34 | 20.81 ± 0.80 | 21.21 ± 0.60 | 23.17 ± 1.08 | >0.9999 |
| mexD | 16.87 ± 0.62 | 16.78 ± 1.21 | 16.40 ± 0.17 | 16.20 ± 0.17 | 16.85 ± 1.50 | >0.9999 | |
| mexY | 16.75 ± 0.33 | 16.79 ± 1.20 | 16.38 ± 0.34 | 16.47 ± 0.27 | 17.42 ± 0.24 | >0.9999 | |
| 16S rRNA | 13.37 ± 0.63 | 16.99 ± 0.56 | 17.35 ± 1.38 | 18.92 ± 0.67 | 18.58 ± 1.23 | >0.9999 | |
| Strains | Target Genes | The CT Values in the Different gDNA Concentrations | p-Value | |||
|---|---|---|---|---|---|---|
| 5.0 ng/µL | 2.5 ng/µL | 1.0 ng/µL | 0.5 ng/µL | |||
| P. aeruginosa ATCC27853 | mexB | 17.97 ± 1.31 | 18.93 ± 0.92 | 22.65 ± 0.69 | 23.07 ± 1.26 | >0.9999 |
| mexD | 20.56 ± 0.93 | 19.92 ± 0.92 | 21.74 ± 0.45 | 22.50 ± 0.52 | >0.9999 | |
| mexY | 19.39 ± 0.61 | 18.98 ± 0.68 | 21.71 ± 0.80 | 22.17 ± 1.12 | >0.9999 | |
| 16S rRNA | 15.49 ± 0.90 | 17.80 ± 0.69 | 21.04 ± 0.93 | 20.80 ± 0.39 | >0.9999 | |
| P. aeruginosa ATCC BAA-2108 | mexB | 20.14 ± 0.24 | 22.98 ± 0.78 | 23.86 ± 0.22 | 27.31 ± 1.21 | >0.9999 |
| mexD | 21.23 ± 0.59 | 19.80 ± 0.22 | 20.92 ± 0.41 | 22.93 ± 0.91 | >0.9999 | |
| mexY | 19.16 ± 0.95 | 19.18 ± 0.90 | 20.61 ± 0.54 | 20.65 ± 1.54 | >0.9999 | |
| 16S rRNA | 17.44 ± 1.20 | 16.34 ± 0.35 | 19.61 ± 1.30 | 20.97 ± 0.61 | >0.9999 | |
| gDNA Concentration Samples (ng/µL) | mexB * | mexD * | mexY * | |||
|---|---|---|---|---|---|---|
| Copies/µL | 95% CI | Copies/µL | 95% CI | Copies/µL | 95% CI | |
| 0.001 | 34.81 ± 9.00 | 32.30–37.32 | 15.52 ± 2.83 | 13.76–17.28 | 7.04 ± 1.58 | 5.81–8.26 |
| 0.003 | 87.43 ± 20.37 | 83.19–91.66 | 38.32 ± 9.09 | 35.55–41.09 | 14.71 ± 1.32 | 13.04–16.38 |
| 0.005 | 183.82 ± 12.86 | 176.52–191.12 | 70.50 ± 12.04 | 66.67–74.33 | 33.00 ± 11.53 | 30.39–35.60 |
| 0.050 | 1923.07 ± 445.32 | 1891.72–1954.42 | 1721.94 ± 79.83 | 1689.22–1754.65 | 442.64 ± 198.97 | 431.44–453.83 |
| 0.500 | 7106.50 ± 44.17 | 6821.53–7391.47 | 4820.07 ± 919.77 | 4698.12–4942.02 | 2086.37 ± 144.30 | 2050.90–2121.83 |
| 1.250 | 10,184.83 ± 782.63 | 8901.54–11,468.11 | 7729.60 ± 640.49 | 7350.85–8108.35 | 4117.87 ± 643.55 | 4031.39–4204.35 |
| 2.500 | 10,388.27 ± 597.32 | 9092.85–11,683.68 | 9121.83 ± 1298.64 | 8300.86–9942.79 | 5626.67 ± 733.42 | 5465.75–5787.59 |
| NTC | 0.04 ± 0.03 | −0.02–0.09 | 0.40 ± 0.08 | 0.085–0.72 | 0.30 ± 0.04 | 0.06–0.53 |
| Cut-Off Values of the Copy Number of Genes (µg/mL) | Sensitivity | Specificity | 1—Specificity | Youden’s Index (J) |
|---|---|---|---|---|
| −0.960 | 1.000 | 0.000 | 1.000 | 0.000 |
| 0.170 | 1.000 | 0.333 | 0.667 | 0.333 |
| 0.350 | 1.000 | 0.667 | 0.333 | 0.667 |
| 3.720 | 1.000 | 1.000 | 0.000 | 1.000 |
| 10.875 | 0.952 | 1.000 | 0.000 | 0.952 |
| 15.115 | 0.905 | 1.000 | 0.000 | 0.905 |
| 24.260 | 0.857 | 1.000 | 0.000 | 0.857 |
| 33.905 | 0.810 | 1.000 | 0.000 | 0.810 |
| 36.565 | 0.762 | 1.000 | 0.000 | 0.762 |
| 54.410 | 0.714 | 1.000 | 0.000 | 0.714 |
| 78.965 | 0.667 | 1.000 | 0.000 | 0.667 |
| 135.625 | 0.619 | 1.000 | 0.000 | 0.619 |
| 313.230 | 0.571 | 1.000 | 0.000 | 0.571 |
| 1082.290 | 0.524 | 1.000 | 0.000 | 0.524 |
| 1822.505 | 0.476 | 1.000 | 0.000 | 0.476 |
| 2004.720 | 0.429 | 1.000 | 0.000 | 0.429 |
| 3102.120 | 0.381 | 1.000 | 0.000 | 0.381 |
| 4468.970 | 0.333 | 1.000 | 0.000 | 0.333 |
| 5223.370 | 0.286 | 1.000 | 0.000 | 0.286 |
| 6366.585 | 0.238 | 1.000 | 0.000 | 0.238 |
| 7418.050 | 0.190 | 1.000 | 0.000 | 0.190 |
| 8425.715 | 0.143 | 1.000 | 0.000 | 0.143 |
| 9653.330 | 0.095 | 1.000 | 0.000 | 0.095 |
| 10,286.550 | 0.048 | 1.000 | 0.000 | 0.048 |
| 10,389.270 | 0.000 | 1.000 | 0.000 | 0.000 |
| Sample No. | Bacterial Species | Present of Mex Genes | Result of effluxR Detection Assay | ||
|---|---|---|---|---|---|
| mexB | mexD | mexY | |||
| 1 | P. aeruginosa | + | + | + | Positive for three genes |
| 2 | P. aeruginosa | + | + | + | Positive for three genes |
| 3 | E. coli | - | - | - | Negative for three genes |
| 4 | P. aeruginosa | + | + | + | Positive for three genes |
| 5 | P. aeruginosa | + | + | + | Positive for three genes |
| 6 | P. aeruginosa | + | + | + | Positive for three genes |
| 7 | S. aureus | - | - | - | Negative for three genes |
| 8 | E. cloacae | - | - | - | Negative for three genes |
| 9 | P. aeruginosa | + | + | + | Positive for three genes |
| 10 | P. aeruginosa | + | + | + | Positive for three genes |
| 11 | P. aeruginosa | + | + | + | Positive for three genes |
| 12 | P. aeruginosa | + | + | + | Positive for three genes |
| 13 | E. cloacae | - | - | - | Negative for three genes |
| 14 | P. aeruginosa | + | + | + | Positive for three genes |
| 15 | P. aeruginosa | + | + | + | Positive for three genes |
| 16 | P. aeruginosa | + | + | + | Positive for three genes |
| 17 | P. aeruginosa | + | + | + | Positive for three genes |
| 18 | P. aeruginosa | + | + | + | Positive for three genes |
| 19 | P. aeruginosa | + | + | + | Positive for three genes |
| 20 | K. pneumoniae | - | - | - | Negative for three genes |
| 21 | P. aeruginosa | + | + | + | Positive for three genes |
| 22 | P. aeruginosa | + | + | + | Positive for three genes |
| 23 | P. aeruginosa | + | + | + | Positive for three genes |
| 24 | P. aeruginosa | + | + | + | Positive for three genes |
| 25 | P. aeruginosa | + | + | + | Positive for three genes |
| 26 | P. aeruginosa | + | + | + | Positive for three genes |
| 27 | P. aeruginosa | + | + | + | Positive for three genes |
| 28 | S. aureus | - | - | - | Negative for three genes |
| 29 | P. aeruginosa | + | + | + | Positive for three genes |
| 30 | P. aeruginosa | + | + | + | Positive for three genes |
| 31 | S. aureus | - | - | - | Negative for three genes |
| 32 | P. aeruginosa | + | + | + | Positive for three genes |
| 33 | P. aeruginosa | + | + | + | Positive for three genes |
| 34 | P. aeruginosa | + | + | + | Positive for three genes |
| 35 | E. coli | - | - | - | Negative for three genes |
| 36 | P. aeruginosa | + | + | + | Positive for three genes |
| 37 | P. aeruginosa | + | + | + | Positive for three genes |
| 38 | P. aeruginosa | + | + | + | Positive for three genes |
| 39 | P. aeruginosa | + | + | + | Positive for three genes |
| 40 | K. pneumoniae | - | - | - | Negative for three genes |
| 41 | P. aeruginosa | + | + | + | Positive for three genes |
| 42 | P. aeruginosa | + | + | + | Positive for three genes |
| 43 | P. aeruginosa | + | + | + | Positive for three genes |
| 44 | P. aeruginosa | + | + | + | Positive for three genes |
| 45 | E. coli | - | - | - | Negative for three genes |
| 46 | K. pneumoniae | - | - | - | Negative for three genes |
| 47 | P. aeruginosa | + | + | + | Positive for three genes |
| 48 | P. aeruginosa | + | + | + | Positive for three genes |
| 49 | P. aeruginosa | + | + | + | Positive for three genes |
| 50 | P. aeruginosa | + | + | + | Positive for three genes |
| 51 | P. aeruginosa | + | + | + | Positive for three genes |
| 52 | P. aeruginosa | + | + | + | Positive for three genes |
| 53 | P. aeruginosa | + | + | + | Positive for three genes |
| 54 | P. aeruginosa | + | + | + | Positive for three genes |
| 55 | P. aeruginosa | + | + | + | Positive for three genes |
| 56 | P. aeruginosa | + | + | + | Positive for three genes |
| 57 | P. aeruginosa | + | + | + | Positive for three genes |
| 58 | S. aureus | - | - | - | Negative for three genes |
| 59 | P. aeruginosa | + | + | + | Positive for three genes |
| 60 | P. aeruginosa | + | + | + | Positive for three genes |
| 61 | P. aeruginosa | + | + | + | Positive for three genes |
| 62 | P. aeruginosa | + | + | + | Positive for three genes |
| 63 | P. aeruginosa | + | + | + | Positive for three genes |
| 64 | P. aeruginosa | + | + | + | Positive for three genes |
| 65 | P. aeruginosa | + | + | + | Positive for three genes |
| 66 | P. aeruginosa | + | + | + | Positive for three genes |
| 67 | P. aeruginosa | + | + | + | Positive for three genes |
| 68 | E. coli | - | - | - | Negative for three genes |
| 69 | P. aeruginosa | + | + | + | Positive for three genes |
| 70 | P. aeruginosa | + | + | + | Positive for three genes |
| 71 | P. aeruginosa | + | + | + | Positive for three genes |
| 72 | P. aeruginosa | + | + | + | Positive for three genes |
| 73 | P. aeruginosa | + | + | + | Positive for three genes |
| 74 | K. pneumoniae | - | - | - | Negative for three genes |
| 75 | P. aeruginosa | + | + | + | Positive for three genes |
| 76 | P. aeruginosa | + | + | + | Positive for three genes |
| 77 | K. pneumoniae | - | - | - | Negative for three genes |
| 78 | P. aeruginosa | + | + | + | Positive for three genes |
| 79 | P. aeruginosa | + | + | + | Positive for three genes |
| 80 | P. aeruginosa | + | + | + | Positive for three genes |
| 81 | P. aeruginosa | + | + | + | Positive for three genes |
| 82 | P. aeruginosa | + | + | + | Positive for three genes |
| 83 | P. aeruginosa | + | + | + | Positive for three genes |
| 84 | P. aeruginosa | + | + | + | Positive for three genes |
| Total | 69 | 69 | 69 | ||
| Percentage | 100 | 100 | 100 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rattanachak, N.; Weawsiangsang, S.; Baldock, R.A.; Jaifoo, T.; Jongjitvimol, T.; Jongjitwimol, J. A Novel and Quantitative Detection Assay (effluxR) for Identifying Efflux-Associated Resistance Genes Using Multiplex Digital PCR in Clinical Isolates of Pseudomonas aeruginosa. Methods Protoc. 2023, 6, 96. https://doi.org/10.3390/mps6050096
Rattanachak N, Weawsiangsang S, Baldock RA, Jaifoo T, Jongjitvimol T, Jongjitwimol J. A Novel and Quantitative Detection Assay (effluxR) for Identifying Efflux-Associated Resistance Genes Using Multiplex Digital PCR in Clinical Isolates of Pseudomonas aeruginosa. Methods and Protocols. 2023; 6(5):96. https://doi.org/10.3390/mps6050096
Chicago/Turabian StyleRattanachak, Nontaporn, Sattaporn Weawsiangsang, Robert A. Baldock, Theerasak Jaifoo, Touchkanin Jongjitvimol, and Jirapas Jongjitwimol. 2023. "A Novel and Quantitative Detection Assay (effluxR) for Identifying Efflux-Associated Resistance Genes Using Multiplex Digital PCR in Clinical Isolates of Pseudomonas aeruginosa" Methods and Protocols 6, no. 5: 96. https://doi.org/10.3390/mps6050096






