Abstract
The greenfin horse-faced filefish (Thamnaconus septentrionalis) is susceptible to recurrent Amyloodinium ocellatum (AO) infestation over the grow-out production cycle. This parasite breeds mainly on the gills, causing hypoxia in the fish body, and leading to many deaths. The host-parasite response drives a complex immune reaction, which is poorly understood. To generate a model for host-parasite interaction and the pathogenesis of AO in greenfin horse-faced filefish, an RNA-seq approach, differential gene expression, GO, and KEGG analyses were employed. Overall, 624 new genes and 2076 differentially expressed genes (DEGs) were detected, including 942 upregulated and 1134 downregulated genes in the gills. Compared with the control group, the expression of leptin a, GTPase IMAP family member 4, and NLR family CARD domain-containing protein 3 was significantly higher in the AO-infected group. Conversely, cell wall integrity and stress response component 1-like, and hepcidin-like were significantly downregulated in the gills of AO-infected fish. GO and KEGG enrichment analysis indicated that DEGs were significantly enriched in signaling pathways associated with viral protein interaction with cytokine and cytokine receptor and cytokine–cytokine receptor interaction. Collectively, this transcriptomic study provides novel molecular insights into the pathology caused by AO infestation and alternative theories for future research implementing strategies to control and manage AO.
1. Introduction
Amyloodinium ocellatum (AO) is an ectoparasite protozoan that usually parasitizes the skin and gills of seawater fishes, resulting in sometimes nearly 100% mortality in just a few days [1,2]. Amyloodinium ocellatum has recently been considered the most consequential parasite in the marine aquaculture industry. Many types of marine fish are susceptible to AO infection, such as the seabass (Dicentrarchus labrax), silver pomfret (Pampus argenteus), and gilthead bream (Sparus aurata) [3,4,5]. The lifecycle of AO is triphasic, consisting of a parasitic trophont, a reproductive tomont, and an infective motile dinospore stage [6]. Under favorable environmental conditions (optimal temperature range: 24–28 °C), AO completes its entire lifecycle within a week [2]. During an infection outbreak, the trophont separates from the host to form a tomont, which could asexually reproduce up to 256 dinospores. After adhering to the new host, the motile dinospore is quickly transformed into a trophont [7,8].
Previous research suggests that AO can survive salinities ranging from 3 to 50 PSU (practical salinity units) and that reproduction is inhibited at temperatures lower than 15 °C [7,8]. Because of strong adaptive and parasitic capacity (both euryhaline and eurythermal), AO can infect many marine animals. In mariculture fishes, the parasite can be detected in gills or skin using molecular and microscopy techniques [9]. Copper sulphate, ferrous sulphate, and formalin are the most widely used compounds for controlling AO. In addition, chloroquine phosphate, hydrogen peroxide, freshwater, ozone, ultraviolet rays, and some biological control methods (such as wrasse, goby, and brine shrimp) could prevent and control AO infection [6,10,11,12]. Another recent study showed that of the 16 plant-derived compounds (2′,4′-dihydroxychalcone; 7-hydroxyflavone; artemisinin; camphor (1R); diallyl sulfide; esculetin; eucalyptol; garlicin 80%; harmalol hydrochloride dihydrate; palmatine chloride; piperine; resveratrol; rosmarinic acid; sclareolide; tomatine, and umbelliferone), only two (2′,4′-dihydroxychalcone and tomatine) could inhibit the motility of viable AO dinospores on the gill cell line G1B [13]. Chemical bath treatments are the most feasible treatments, but chemicals from aquaculture systems may contaminate the environment. Understanding the genetic changes and related signal pathways during infection is the basis for preventing and controlling AO.
Disease control is critical to intensive farming of marine fishes. Transcriptome sequencing is the most widely used, fastest, and most effective method for studying internal host response to pathogen infection. This technology has been widely applied in aquatic animal research. However, most previous studies focused on interacting with viral and bacterial pathogens [14], and only a few studies have investigated the interaction between the host and eukaryotic parasite [15,16]. AO infection can be an example to map parasite infection, and better understand how AO affects these fishes. Regrettably, AO infects most marine fish, but transcriptome-based AO infection studies have been limited to a few fish species [2,5,17]. Thus, these studies did not provide adequate information to consider other kinds of fish.
The greenfin horse-faced filefish (Thamnaconus septentrionalis) is a type of Tetraodontiformes widely distributed in the Indo-West Pacific Ocean [18,19]. Until the end of the 20th century, it was an important fishery resource in China. However, recently, it has been protected because of a sharp decline in its population. Despite breakthroughs in artificial breeding of the greenfin horse-faced filefish, whole genome data has only been recently analyzed [19]. During its breeding period, juvenile fish are very susceptible to AO infection, resulting in many deaths. Few studies have investigated AO infection, which is a major bottleneck to the greenfin horse-faced filefish breeding industry [5,18].
Considering the lack of research to support effective control of AO, preventing parasitic infection of the greenfin horse-faced filefish is a challenge. We first confirmed AO-infected and uninfected greenfin horse-faced filefish by observation and PCR detection. High-throughput sequencing was then performed using the AO-infected and uninfected fish gill tissues to obtain transcriptomes involved in immunity, metabolism, and growth. This study can deepen our understanding of signaling pathways involved in host response to AO infection and the underlying molecular mechanism to control parasites and reduce host mortality.
2. Materials and Methods
2.1. AO Infection and Sample Collection
A batch of spontaneous spawning greenfin horse-faced filefish was cultured at Ganyu County, Jiangsu, China. Gill tissue samples from infected and non-infected fish were collected from greenfin horse-faced filefish juveniles (60-day-old) during a severe AO outbreak that occurred naturally in August 2021. In this experiment, samples were first identified using microscopy and PCR. AO-specific primers 18SF (5′–GACCTTGCCCGAGAGGG–3′) and 18SR (5′–GGTGTTAAGATTCACCACACTTTCC–3′) were used to detect the parasites in this study [6]. AO-infected and uninfected fish were initially cultured in separate tanks (1000-L tanks containing filtered seawater), and uninfected fish did not experience mass AO-infection deaths as AO-infected fish did at the end of the culture. Due to uncertainty on the exact time of disease onset, the time-point of sample collection was 2 days after infection being noted. The body weight and body length of this batch of fish were AO-infected (9.7 ± 0.14 g and 4.7 ± 0.42cm) and control (9.8 ± 0.12 g and 4.7 ± 0.39 cm), respectively. Water temperature was maintained at 22 ± 0.5 °C, pH at 7.8 ± 0.2, dissolved oxygen concentration at 5.6 ± 0.5 mg/L, and salinity at 26 ± 0.5%. Healthy fish were fed 6# particle size (1.1–1.4 mm) (Santong Bio-engineering (Weifang) CO., Ltd., Weifang, China), with a daily feeding rate of 2–3% of their body weight.
2.2. DNA Extraction and PCR Detection
DNA extraction was done using the TIANamp Marine Animals DNA Kit (TIANGEN BIOTECH, Beijing, China) per the manufacturer’s instructions. PCR amplification of the DNA using the AO 18SF/R primer set was carried out in a final volume of 50 μL. PCR reaction was performed using a Veriti 96-well thermal cycler (Applied Biosystems) with a reaction mixture containing 100 ng DNA, 100 ng of each primer, and 1.25 Units HotStart Taq DNA Polymerase (TaKaRa, Shiga, Japan) under the following conditions: initial denaturation at 94 °C for 5 min followed by 35 cycles of initial denaturation (94 °C for 1 min), annealing (58 °C for 1 min), extension (72 °C for 1 min), and a final elongation (72 °C for 5 min). Then, 10 μL of the PCR product was analyzed by 1.5% agarose gel electrophoresis stained with ethidium bromide and visualized with a UV transilluminator.
2.3. RNA Extraction and Library Construction
Greenfin horse-faced filefish were anesthetized with tricaine methanesulphonate (MS-222; 100 mg/L) and on ice dissected to collect gill tissue samples. Each sample was immediately frozen in liquid nitrogen and stored at −80 °C until RNA extraction. Total RNA was extracted with TRIzol reagent (Invitrogen, CA, USA) following the manufacturer’s instructions. An Agilent 2100 bioanalyzer (Agilent Technologies) was used to determine the integrity and quality of total RNA. For RNA library construction and deep sequencing, equal quantities of RNA (5 μg) from each fish in the control and infection groups were dissolved in RNase-free water and produced the control and infection samples, respectively. Samples 1–3 were each collected from three AO-infected fish whereas Samples 4–6 were each from three healthy fish. After extracting total RNA, library preparations were sequenced using the HiSeq 2500 sequencing system (Illumina, CA, USA) in the paired-end mode technology by Gene Denovo Biotechnology Co. (Guangzhou, China).
2.4. Filtering Reads and Alignment with Ribosome RNA (rRNA)
To obtain high-quality clean reads and eliminate raw reads contaminated with adapter sequences or low-quality bases, reads were further filtered using fastp version 0.18.0 [20]. The parameters used were as follows: (1) removing reads containing adapters; (2) removing reads containing more than 10% of unknown nucleotides (N); and (3) removing low-quality reads containing more than 50% of low-quality (Q-value ≤ 20) bases. The short reads alignment tool Bowtie2 version 2.2.8 [21] was used for mapping reads to the greenfin horse-faced filefish ribosomal RNA (rRNA) database; rRNA mapped reads were then removed. The remaining clean reads were assembled and used for gene abundance calculation.
2.5. Alignment with Reference Genome
An index of the reference genome was built from draft genome data [19], and paired-end clean reads were mapped to the reference genome using HISAT2.2.4 [22] with default “-rna-strandness RF” and other parameters. In AO-infected samples, host and parasite mRNAs could not be distinguished.
2.6. Quantification of Gene Expression Abundance and Differentially Expressed Genes (DEGs)
Mapped reads from each sample were assembled using StringTie v1.3.1 in a reference-based approach [23]. An FPKM (fragment per kilobase of transcript per million mapped reads) for each transcription region was calculated to quantify its expression abundance and variations using StringTie software v1.3.1. Differential expression analysis of RNAs was performed using DESeq2 [24] software for comparisons between two different groups and edgeR [25] for comparisons between two samples. Genes/transcripts with false discovery rate (FDR) below 0.05 and absolute fold change ≥ 2 were considered differentially expressed genes/transcripts (DEGs).
2.7. Relationship Analysis of Samples
Correlation analysis was performed using R (http://www.rproject.org/, accessed on 13 April 2022). A correlation coefficient closer to 1 indicated better repeatability between two parallel experiments. Principal component analysis (PCA) with the R package models was performed in this experiment.
2.8. GO Enrichment and Pathway Enrichment Analysis
Gene ontology enrichment analysis generates all GO terms significantly enriched in DEGs compared to the genome background and filters DEGs corresponding to biological functions. All DEGs were first mapped to GO terms in the Gene Ontology database (http://www.geneontology.org/, accessed on 13 May 2022), then gene numbers were calculated for every term, and significantly enriched GO terms in DEGs compared with the genome background were defined using a hypergeometric test. Enrichment analysis of significantly expressed GO terms was performed using the parent-child-intersection method with Benjamini–Hochberg multiple testing corrections in Ontologizer 2.0 [26]. Pathway analysis was based on KEGG central public pathway-related database [27]. The threshold was set with an FDR-corrected p-value < 0.05.
3. Results
3.1. Identification and Disease Signs of AO Infection in Greenfin Horse-Faced Filefish
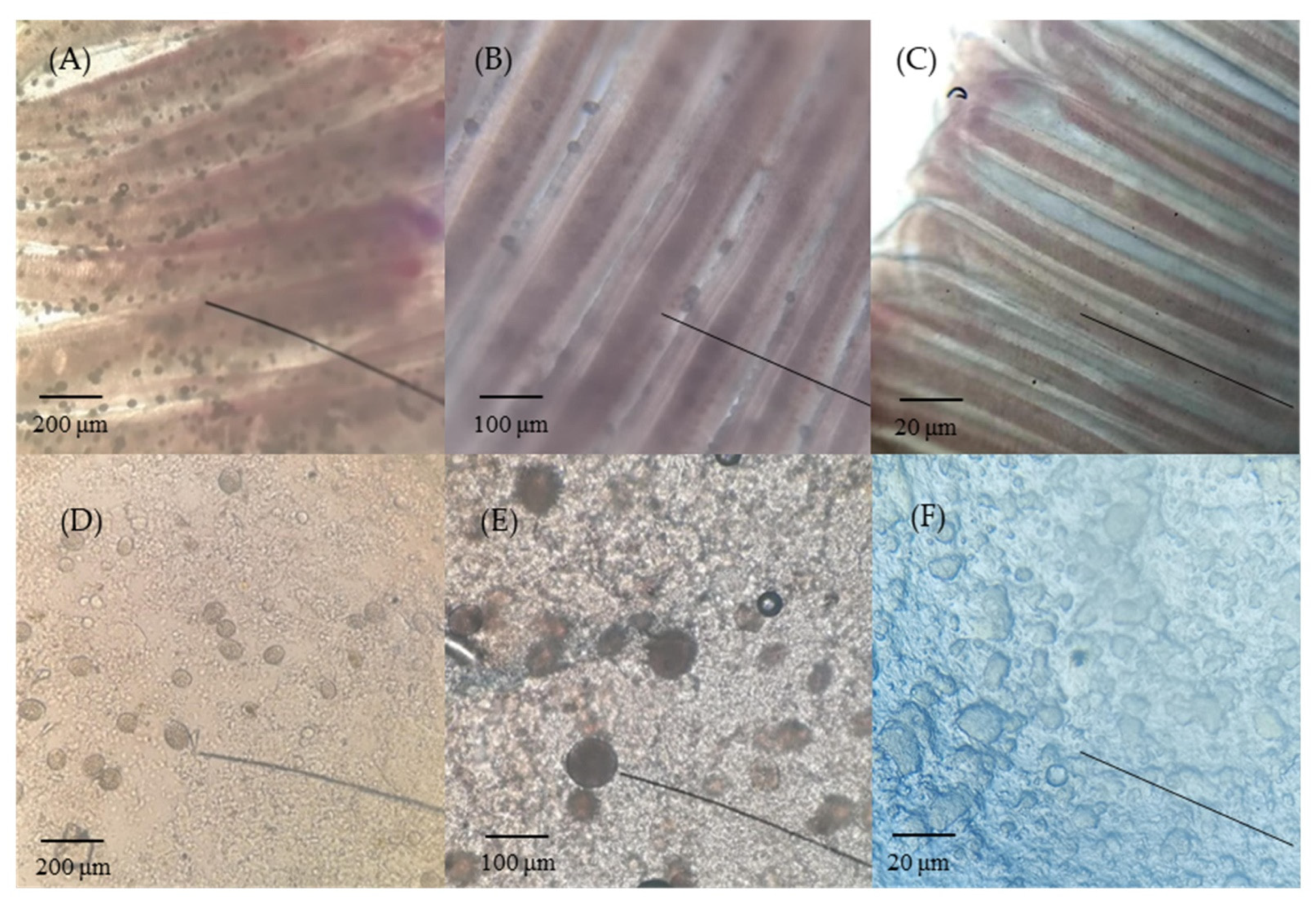
Amyloodinium ocellatum-infected greenfin horse-faced filefish were differentiated from those in the control group based on appearance, microscopic observation, and PCR detection results. Amyloodinium ocellatum-infected greenfin horse-faced filefish lost balance, swam sideways, and had low appetite. As the infection accelerated, the fish exhibited faster breathing, struggling until death, and were quickly cannibalized by healthy fish. Under an ordinary optical microscope, many translucent AO trophozoites could be observed on the gills and skin mucus of AO-infected greenfin horse-faced filefish (Figure 1). The identity histological analysis of AO-infected fish was further verified using a PCR based on genomic DNA extracted from the gills of ailing fish. The target band (248 bp) was detected in DNA from AO-infected fish but not in the control DNA samples (Figure S1).
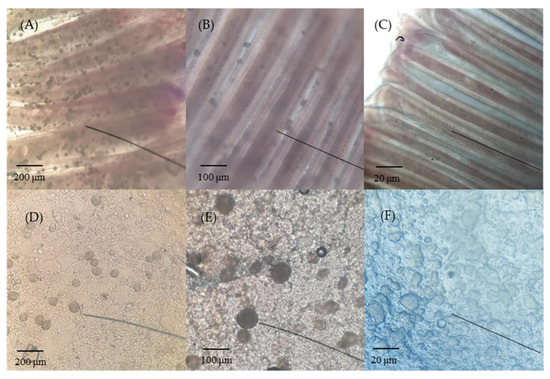
Figure 1.
Gill and skin mucus of AO-infected greenfin horse-faced filefish. (A) Gills showing severe infection (40 ×); (B) gill tissues of AO-infected (100 ×); (C) gill tissues of control (40 ×); (D) skin showing severe infection (40 ×); (E) skin tissues of AO-infected (100 ×); and (F) skin tissues of control (40 ×). Bars = 200 μm (A,D), 100 μm (B,E), and 20 μm (C,F).
3.2. Transcriptome Assembly and Sequence Description
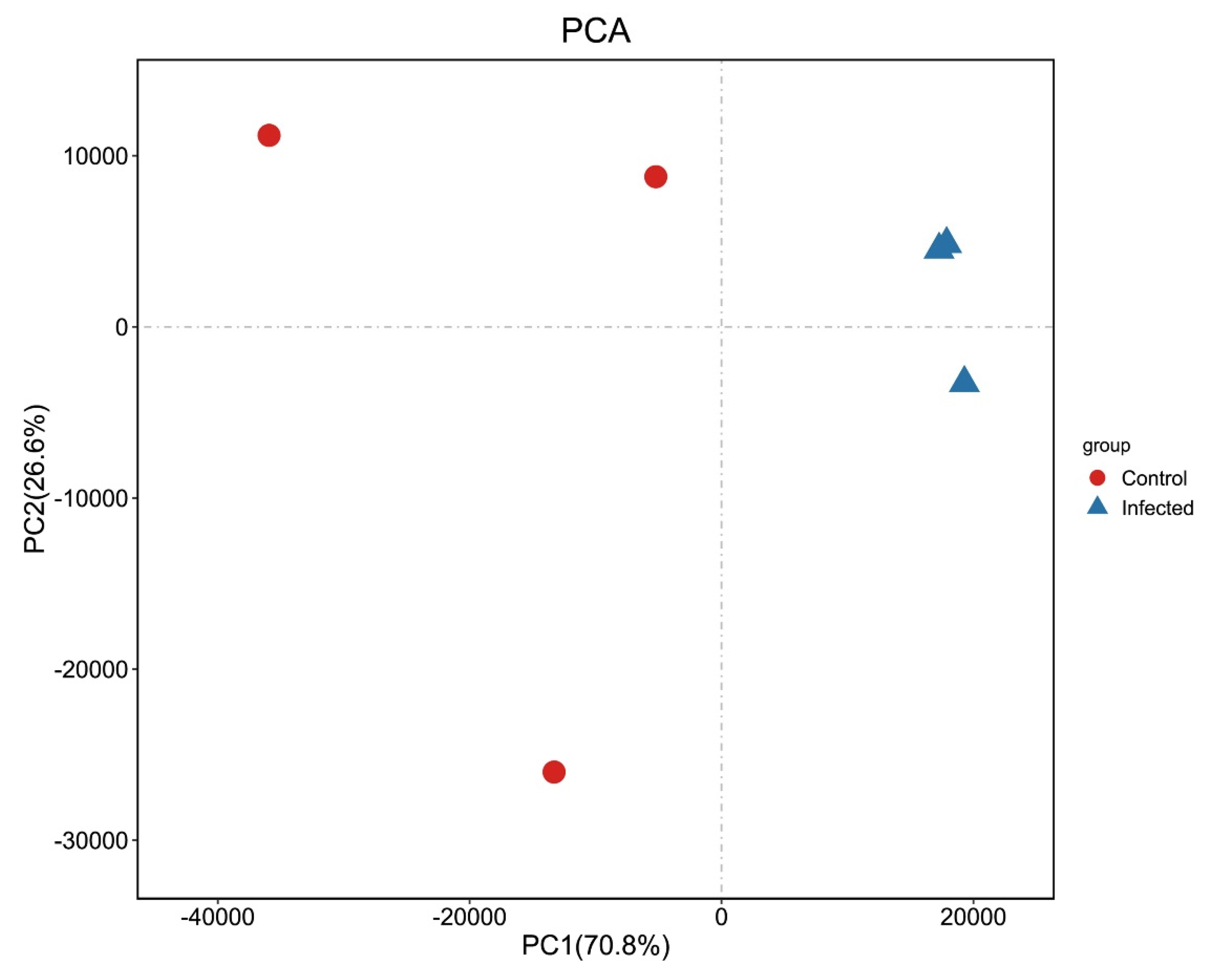
Reads from control and AO-infected (n = 3) gill tissues were analyzed using Illumina sequencing. From the six libraries constructed, 39,302,198–55,153,762 raw reads were generated, and 99.60–99.66% clean reads were obtained. The Q20 was 97.61-97.68% (Table 1), and 86.18–89.21 % of the reads were mapped to the greenfin horse-faced filefish reference genome (Table 1). The PCA results (Figure 2) revealed the relationships among them. In addition, the transcriptome of this experiment was annotated to obtain new genes by aligning the reference genome of greenfin horse-faced filefish. In total, 624 new genes were identified potentially derived from fish and the AO parasite. In addition, our transcriptome data contained transcript expression information in a specific period. By reassembling these data, we found some genes that were not annotated in the original reference genome.

Table 1.
Summary of the greenfin horse-faced filefish transcriptome assembly.
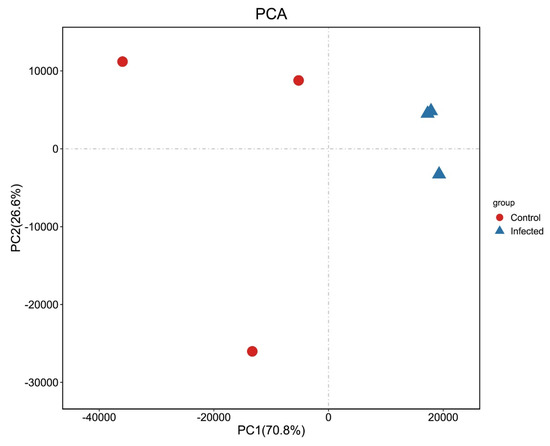
Figure 2.
PCA of transcriptome samples. Abscissa PC1 and ordinate PC2 represent the scores of the first and second principal components, respectively. Scatter points of different colors represent samples belonging to different groups.
3.3. Differentially Expressed Genes after AO Infection
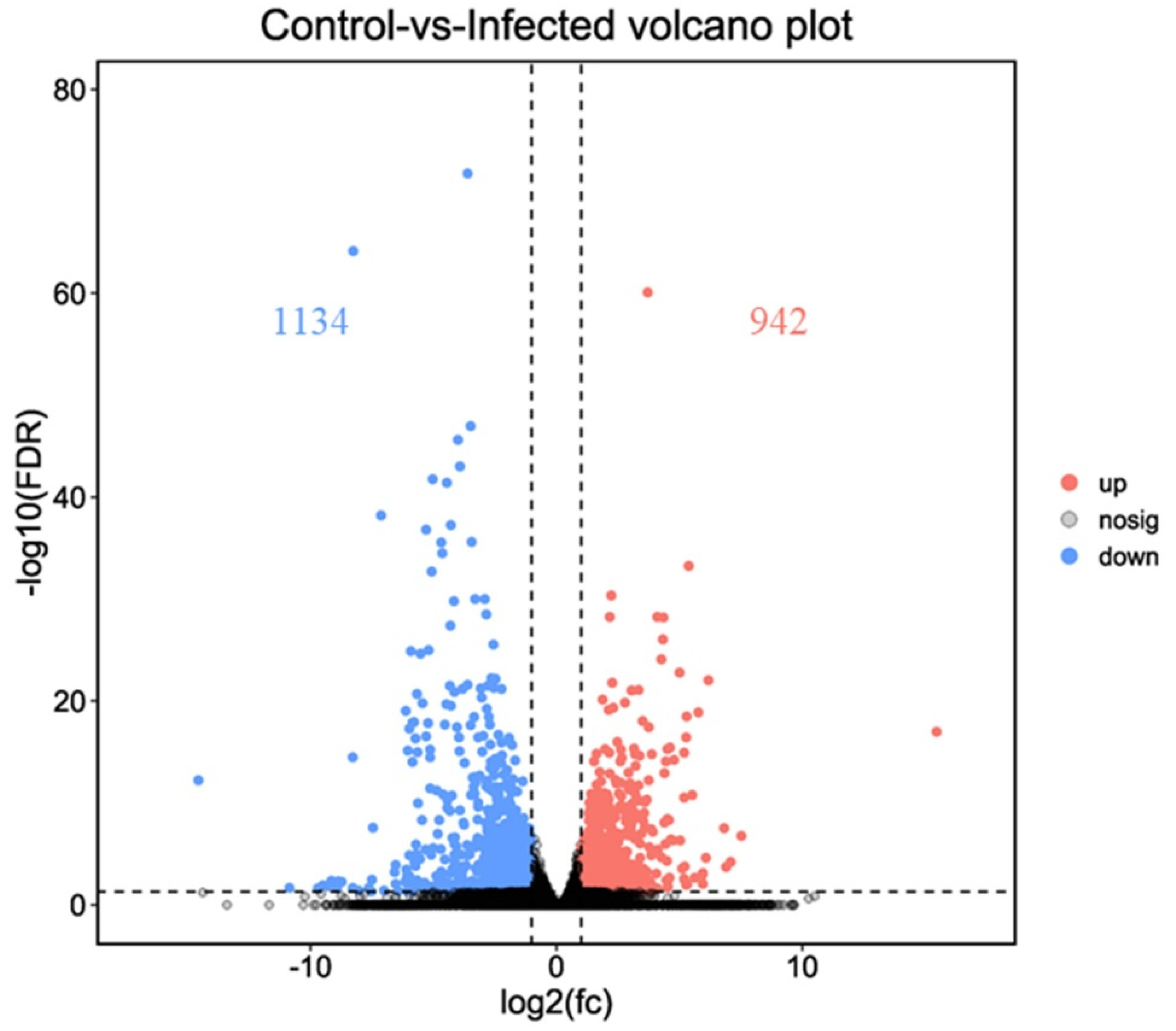
A comparison of gene expression levels between control and AO infection groups revealed 2076 DEGs in gills (p < 0.01). These included 942 upregulated and 1134 downregulated genes (Figure 3 and Figure 4). Compared with the control group, the expression of leptin a, GTPase IMAP family member 4, and NLR family CARD domain-containing protein 3 was significantly higher in the AO-infected group. Conversely, cell wall integrity and stress response component 1-like and hepcidin-like were significantly downregulated in the gills of AO-infected fish. Furthermore, two genes (ccl21 (c-c motif chemokine 21-like) and ankrd1 (ankyrin repeat domain-containing protein 1)) were upregulated and four genes (tnfsf12 (tumor necrosis factor ligand superfamily member 12-like), f11 (coagulation factor xi-like), tlr5 (Toll-like receptor 5), and CD40 (tumor necrosis factor receptor superfamily member 5)) were downregulated in fish, respectively. These data and DEGs provide important insights into host responses to AO infection (Table S1).
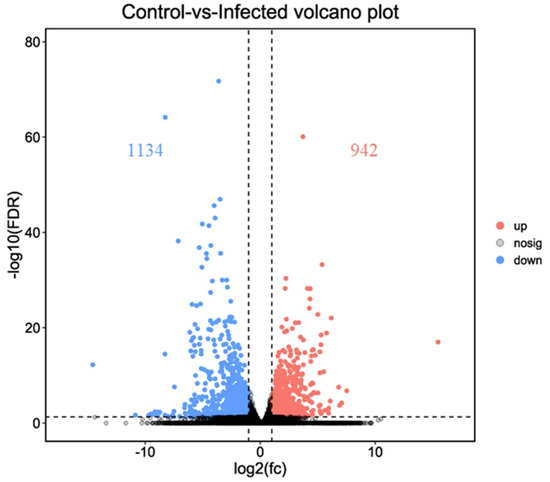
Figure 3.
Volcano plot of the differentially expressed genes of greenfin horse-faced filefish (upregulated differentially expressed genes (red); downregulated differentially expressed genes (blue); non-differentially expressed genes (black)).
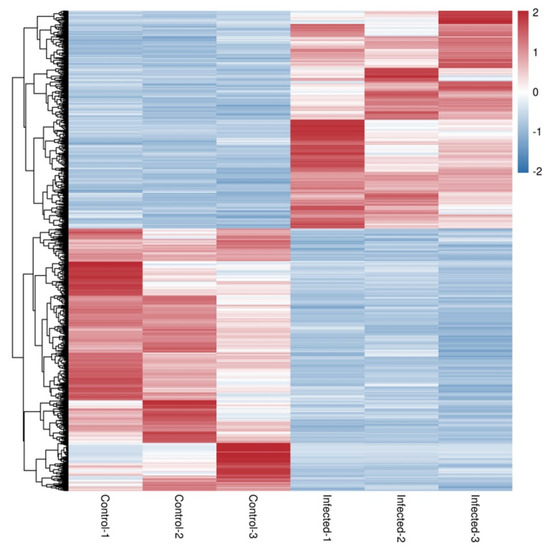
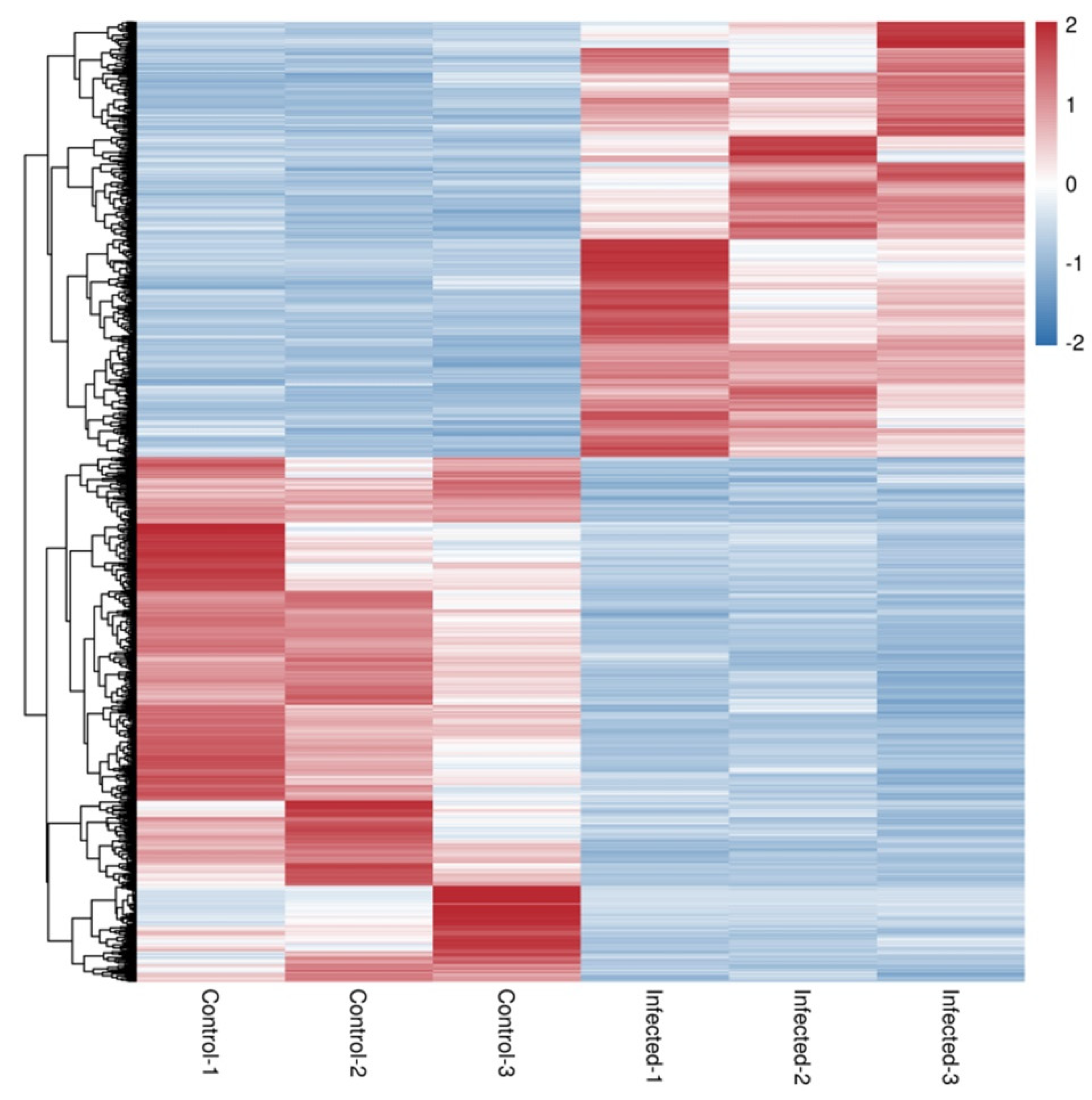
Figure 4.
Heatmap of the differential gene expression. RNA-seq heatmap of the expression profile of the differentially expressed genes in control 1–3 and infected 1–3. The differential gene expression was presented under a false discovery rate (FDR) < 0.05. Overexpressed genes are shown in red and downregulated genes are shown in blue.
3.4. GO Annotated after AO Infection
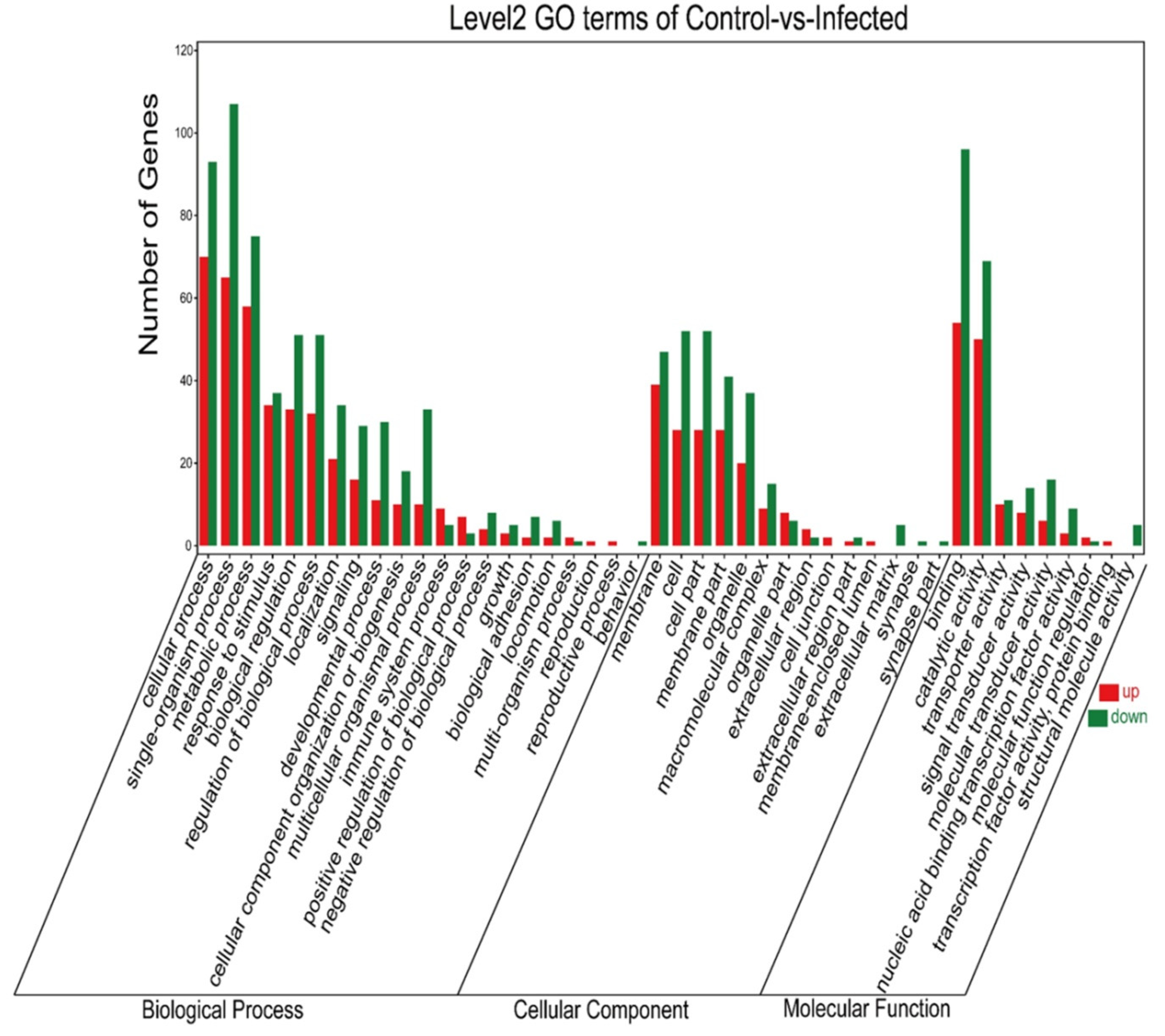
Genes differentially expressed in the gills were mainly annotated into three GO categories: biological process, cellular component, and molecular function. Among upregulated genes, the top five DEGs for the biological process (cellular process, single-organism process, metabolic process, response to stimulus, and biological regulation), cellular component (membrane, cell, cell part, membrane part, and organelle), and molecular function (binding, catalytic activity, transporter activity, signal transducer activity, and molecular transducer activity) were annotated. Likewise, among the downregulated genes annotated, the top five DEGs in the biological process, cellular component, and molecular function are shown in Figure 5.
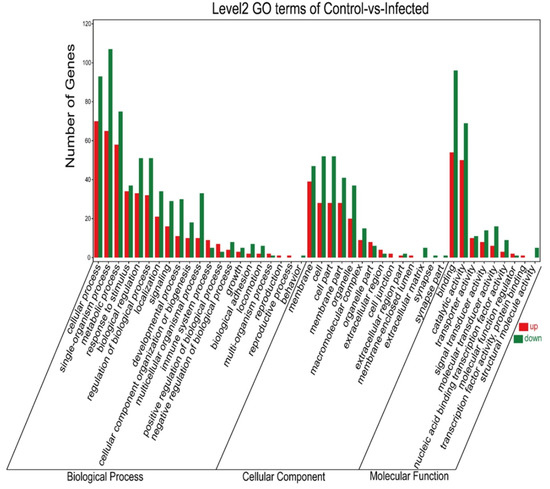
Figure 5.
GO enrichment classification histogram. The abscissa is the secondary GOterm; the ordinate is the number of differential genes in the term; red means upregulated; green means downregulated.
3.5. KEGG Pathway Enrichment of DEGs
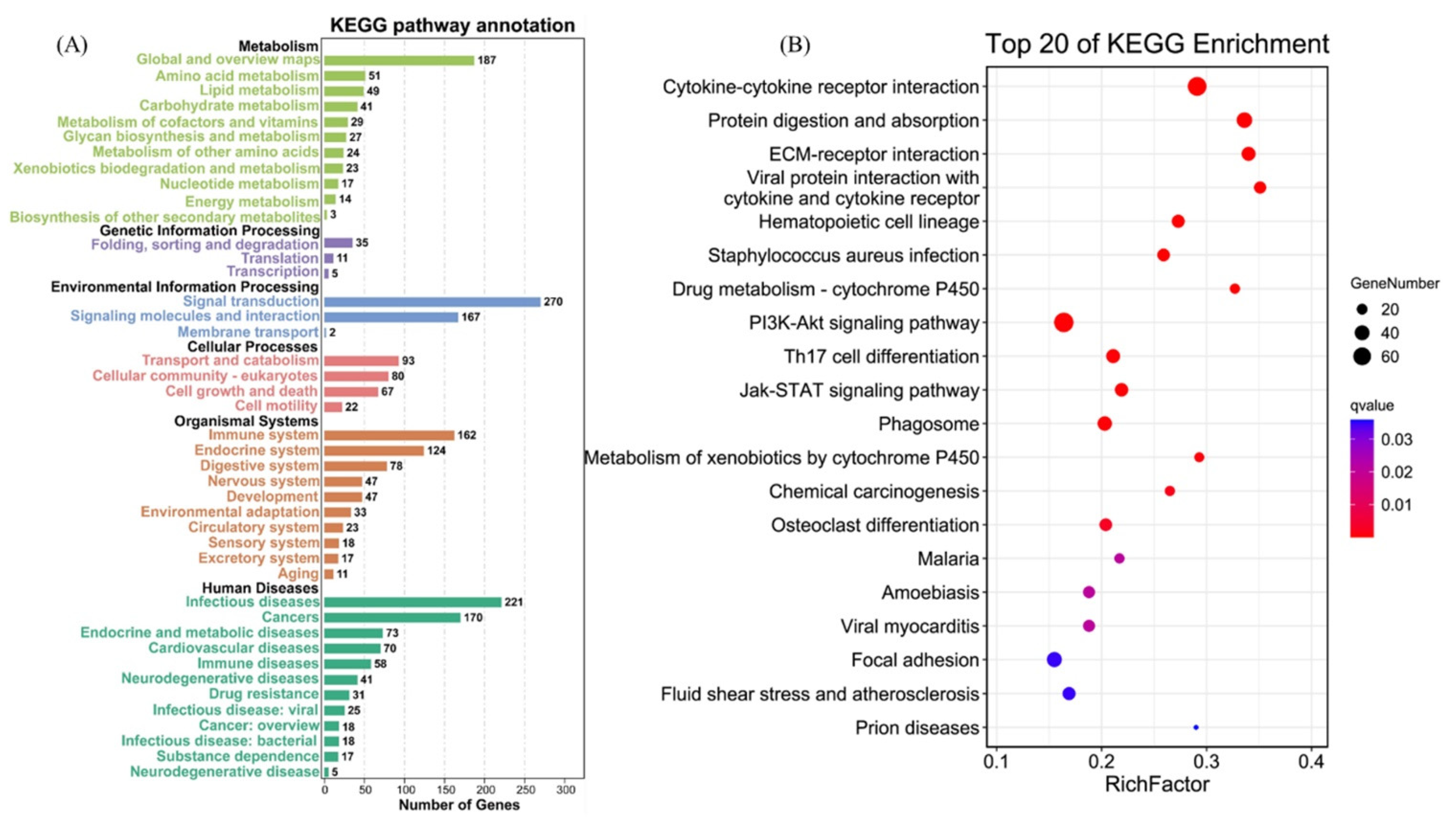
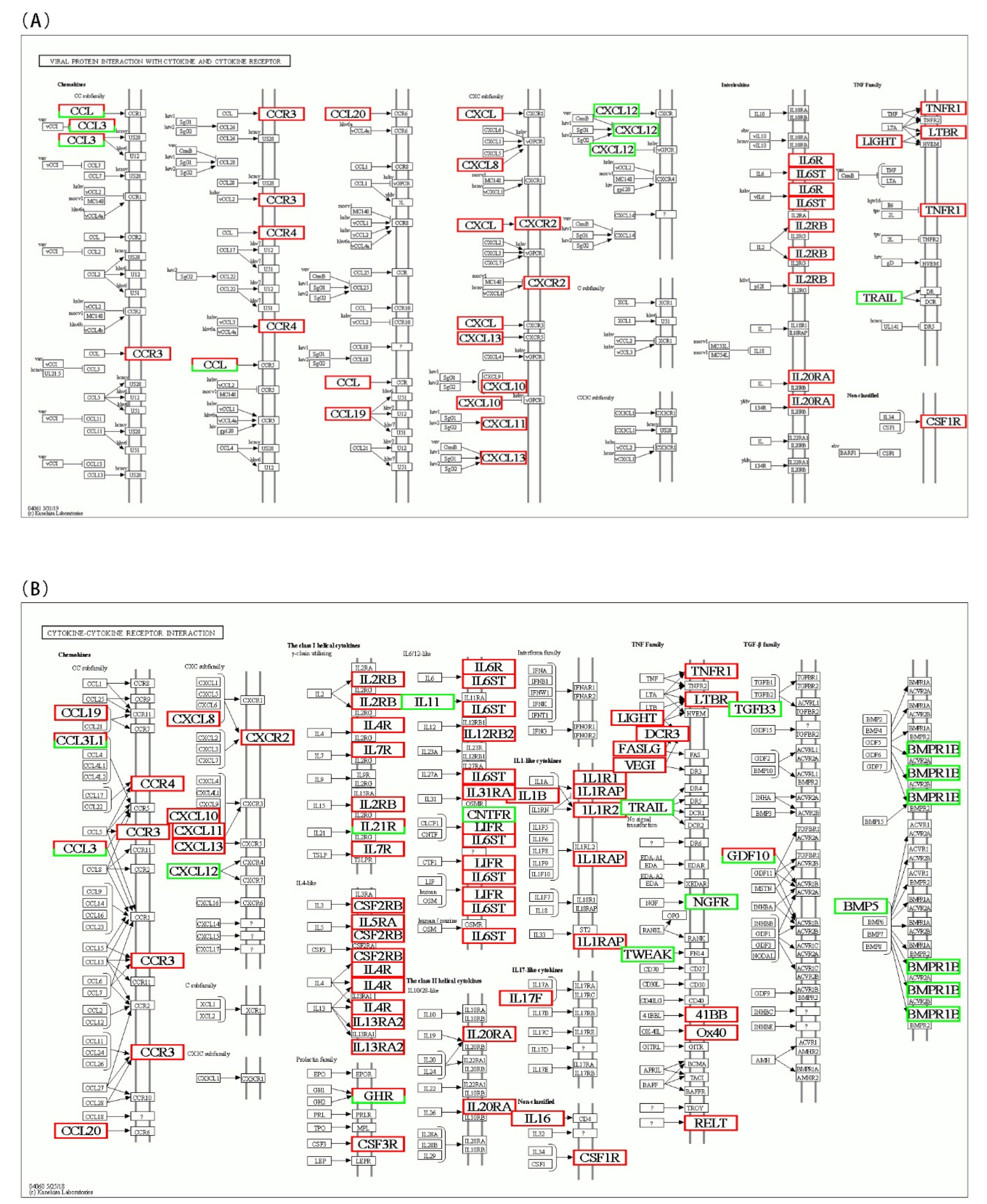
To identify the interaction between DEGs in AO infection, 2076 DEGs were mapped to 300 KEGG pathways. Figure 6A shows the KEGG pathway annotation, which included six aspects (metabolism, genetic information, environmental information processing, cellular processes, organismal systems, and human diseases). The six most annotated aspects were global and overview maps (187), folding, sorting degradation (35), signal transduction (270), transport and catabolism (93), immune system (152), and infectious diseases (221). This annotation was based on the top 20 enriched pathways and p-value < 0.05 (Figure 6B), viral protein interaction with cytokine and cytokine receptor (ko04061), ECM–receptor interaction (ko04512), protein digestion and absorption (ko04974), drug metabolism–cytochrome P450 (ko00982), and cytokine–cytokine receptor interaction (ko04060) in gills, which appeared to play a vital role in parasitic AO infection. The variations in the number of genes involved in the viral protein interaction with cytokine and cytokine receptor and cytokine–cytokine receptor interaction after AO infection are presented in Figure 7A,B.
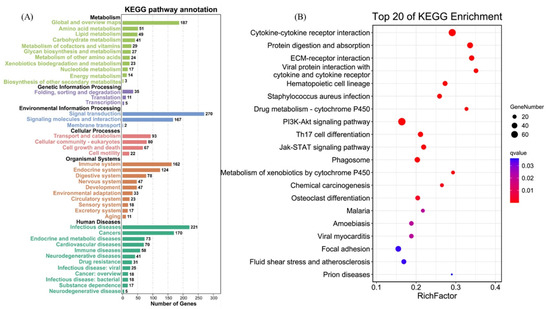
Figure 6.
KEGG pathway enrichment analysis of DEGs. (A) KEGG pathway annotation and classification of DEGs in gills. (B) Scatterplot of top 20 KEGG enrichment pathways of DEGs.
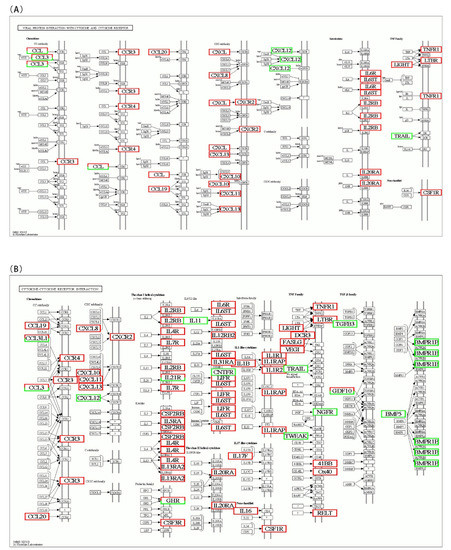
Figure 7.
Differentially expressed genes identified by KEGG as involved in the (A) viral protein interaction with cytokine and cytokine receptor (ko04061) and (B) cytokine–cytokine receptor interaction (ko04060) after AO infection. The color of the boxes indicates the fold change in each gene. When a gene was increased after AO infection, it was colored red. If a gene was downregulated, it was colored green. Genes with white boxes were not detected in the analysis.
4. Discussion
Amyloodinium ocellatum infection leads to many sudden deaths, especially in the juvenile fish stage. For many years, AO infection has been reported in some fish but not in greenfin horse-faced filefish [3,4,5]. In this experiment, infected samples were first identified in the filefish using microscopy and PCR. Then, transcriptomes from infected and control gill tissues were sequenced to obtain DEGs for GO and KEGG enrichment analysis. These analyses and findings provide broader biological insights into greenfin horse-faced filefish–AO interactions.
Amyloodinium ocellatum spores can be observed on the gills and skin mucus under the microscope, aiding the diagnosing and treatment of this disease. However, further diagnostic methods should be used to distinguish AO infection from related diseases, such as Cryptonucleus irritans and Ichthyophthiriasis [28,29,30]. Moreover, the microscopy has a certain lag, as AO spores can only be detected when the infection progresses to a certain stage, limiting very early treatment and prevention of this infection. The results showed a bright band at the expected size of 248 bp in the tissue DNA of AO-infected fish. Molecular diagnostic techniques, such as PCR and loop isothermal-mediated methods, are most effective, detecting AO in all three stages of the parasite’s life cycle (trophont, tomont, dinospore) [6,9]. The detection limit of the AO-LAMP assay was 10 fg, exceptionally higher than that of conventional PCR (1 pg) [9].
Transcriptome sequencing of some fish revealed several up- and downregulated genes in which AO-infected fish exhibited more upregulated genes than downregulated [4,5]. The present study found more downregulated genes (1,134) than upregulated genes (942). Nevertheless, two genes (ccl21 and ankrd1) were upregulated in both European sea bass and silver pomfret, and one gene (tnfsf12) and three genes (f11, tlr5, and CD40) were downregulated in European sea bass and silver pomfret, respectively, consistent with the results of transcriptome data analysis in this experiment [4,5]. The gill is the primary target tissue of AO infection, which can cause physiological changes in the gill. The discovery of these differentially expressed genes will be beneficial to further studies exploring the molecular mechanism of parasitic infection in fish.
Through genome alignment and annotation, we obtained 624 new genes, further deepening the current understanding of the greenfin horse-faced filefish’s genomic information and providing valuable insights into AO infection. We found that hepcidin-like (MTSRG.11446) was the most downregulated gene among the new gene annotations. Hepcidin reduces the iron that is required for pathogen growth, but is upregulated in bacteria- or parasite-infected salmonids and sea bass [31,32]. We found eight transcripts annotated as hepcidin in the greenfin horse-faced filefish transcriptome. Of these, EVM0001859, EVM0010144, EVM0012177, and EVM0012796 were upregulated, whereas MTSRG.11444-11447 was downregulated. Hepcidin-mediated low serum iron levels function as a host defense mechanism, which evolved to limit iron availability for pathogen growth and development [33]. From our results, hepcidin played a role in greenfin horse-faced filefish after AO infection, but further studies are needed to assess this role.
Among the most significantly upregulated genes, leptin a, an important hormone involved in mediating animal feeding, reproduction, and energy expenditure, was most highly expressed. It can maintain the body’s energy homeostasis by suppressing appetite and promoting metabolism [34]. During AO infection, the appetite of the greenfin horse-faced filefish decreased significantly, which coincided with their death. We speculate that this could be caused by the upregulation of leptin a. In zebrafish under hypoxia stress, leptin a was also upregulated [35]. The greenfin horse-faced filefish has a tiny mouth gape. During AO infection, many spores occur on the gill filaments, which cause hypoxia and induce high expression of leptin a, further suppressing appetite, which leads to the death of greenfin horse-faced filefish.
In pathway enrichment, we found that viral protein interaction with cytokine and cytokine receptor (ko04061) and cytokine–cytokine receptor interaction (ko04060) were significantly enriched and induced changes in many cytokine family genes in both pathways. Many chemokine genes were altered when fishes (European sea bass or silver pomfret) were infected [4,5]. The immune response induced by all upregulated chemokines probably enhances resistance against AO pathogen in marine fish. The CC chemokine plays a significant role in wound healing due to parasite invasion. We also found that some signaling pathways, such as ECM–receptor interaction (ko04512), protein digestion and absorption (ko04974), and drug metabolism–cytochrome P450 (ko00982) were significantly enriched. The enrichment of these pathways indicates that AO infection could cause dyspnea, trigger hypoxic stress, affect food intake and lipid metabolism, and induce apoptosis [5,36].
Biochemical processes involve complex interactions between cellular components [37]. Amyloodinium ocellatum-infected fish revealed significant differential expression of genes implicated in cellular, single-organism, and metabolic processes. Many DEGs were annotated in the cellular process, which relies on intricate molecular mechanisms to function in extraordinarily diverse intra- and extra-cellular environments [38]. For terms associated with molecular function, genes involved in binding and catalytic activity were significantly enriched, including the significantly downregulated gene cell wall integrity and stress response component 1-like. This plays a role in the additive effect on mineralization and alters the skeletal metabolism during development [39]. Likewise, it can be seen that Th17 cell differentiation, malaria, amoebiasis, and Staphylococcus aureus infection pathways are enriched in the AO infection. Alterations in many IL family genes may induce Th17 cell differentiation, stimulating an immune response [14,40]. Malaria, amoebiasis, and Staphylococcus aureus infection pathways are essential components of the response to infectious diseases. The enrichment of these pathways may be the biological basis for the large-scale infection spread caused by AO. Further investigations on its molecular mechanisms will undoubtedly provide new clues and targets for the prevention and treatment of AO infections in production.
5. Conclusions
This study is the first contribution to greenfin horse-faced filefish disease research using transcriptome sequencing, aiming to expand the current understanding of essential growth, immunity, metabolism, and other processes after AO infection. These findings provide new insights into the genetic breeding, disease prevention and control, and healthy farming of the greenfin horse-faced filefish.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fishes7050252/s1, Table S1: Representative DEGs after AO infection. Include sheet 1 (representative genes), sheet 2 (DEGs), sheet 3 (GO terms of DEGs), and sheet 4 (KEGG pathways of DEGs). Figure S1: Agarose gel electrophoresis of PCR products.
Author Contributions
Conceptualization, L.-G.Y. and Y.-Q.W.; methodology, Y.W. and W.-B.X.; formal analysis, L.-G.Y., Y.W. and W.-B.X.; investigation, L.-G.Y., B.-B.Z., B.Q., N.Y., X.-F.S., Y.-F.Y., X.-S.W. and Y.-Q.W.; data curation, L.-G.Y. and Y.-Q.W.; writing—original draft preparation, L.-G.Y. and Y.W.; writing—review and editing, L.-G.Y., Y.W., W.-B.X., B.-B.Z. and Y.-Q.W.; funding acquisition, L.-G.Y., B.-B.Z., B.Q. and Y.-Q.W. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Central Public-interest Scientific Institution Basal Research Fund, CAFS (Grant No. 2016M08 and 2018M07).
Data Availability Statement
The data that support the findings of this study have been deposited into the CNGB Sequence Archive (CNSA) of China National GeneBank DataBase (CNGBdb) with accession number CNP0003149.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Noga, E.J. Amyloodinium ocellatum. In Fish Parasites Pathobiology and Protection; Woo, P.T.K., Buchman, K., Eds.; CAB International: London, UK, 2012; p. 383. [Google Scholar]
- Byadgi, O.; Marroni, F.; Dirks, R.; Massimo, M.; Volpatti, D.; Galeotti, M.; Beraldo, P. Transcriptome Analysis of Amyloodinium ocellatum Tomonts Revealed Basic Information on the Major Potential Virulence Factors. Genes 2020, 11, 1252. [Google Scholar] [CrossRef]
- Alvarezpellitero, P.; Eiras, J.; Segner, H.; Wahli, T.; Kapoor, B.G. Diseases caused by flagellates. Fish Dis. 2008, 2, 435–582. [Google Scholar]
- Byadgi, O.; Beraldo, P.; Volpatti, D.; Massimo, M.; Bulfon, C.; Galeotti, M. Expression of infection-related immune response in European sea bass (Dicentrarchus labrax) during a natural outbreak from a unique dinoflagellate Amyloodinium ocellatum. Fish Shellfish Immunol. 2019, 84, 62–72. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, J.; Li, Y.; Zhang, M.; Jacques, K.J.; Gu, W.; Sun, Y.; Sun, J.; Yang, Y.; Xu, S.; et al. Immune response of silver pomfret (Pampus argenteus) to Amyloodinium ocellatum infection. J. Fish Dis. 2021, 44, 2111–2123. [Google Scholar] [CrossRef]
- Levy, M.; Poore, M.; Colorni, A.; Noga, E.; Vandersea, M.; Litaker, R.W. A highly specific PCR assay for detecting the fish ectoparasite Amyloodinium ocellatum. Dis. Aquat. Org. 2007, 73, 219–226. [Google Scholar] [CrossRef]
- Paperna, I. Reproduction cycle and tolerance to temperature and salinity of Amyloodinium ocellatum (Brown, 1931) (Dinoflagellida). Ann. Parasitol. Hum. Comp. 1984, 59, 7–30. [Google Scholar] [CrossRef]
- Noga, E.J. Propagation in cell culture of the dinoflagellate amyloodinium, an ectoparasite of marine fishes. Science 1987, 236, 1302–1304. [Google Scholar] [CrossRef]
- Picón-Camacho, S.M.; Thompson, W.P.; Blaylock, R.B.; Lotz, J.M. Development of a rapid assay to detect the dinoflagellate Amyloodinium ocellatum using loop-mediated isothermal amplification (LAMP). Veter- Parasitol. 2013, 196, 265–271. [Google Scholar] [CrossRef]
- Oestmann, D.J.; Lewis, D.H.; Zettler, B.A. Communications: Clearance of Amyloodinium ocellatum Dinospores by Artemia salina. J. Aquat. Anim. Health 1995, 7, 257–261. [Google Scholar] [CrossRef]
- Fajer-Ávila, E.J.; la Parra, I.A.-D.; Aguilar-Zarate, G.; Contreras-Arce, R.; Zaldívar-Ramírez, J.; Betancourt-Lozano, M. Toxicity of formalin to bullseye puffer fish (Sphoeroides annulatus Jenyns, 1843) and its effectiveness to control ectoparasites. Aquaculture 2003, 223, 41–50. [Google Scholar] [CrossRef]
- Owatari, M.S.; Sterzelecki, F.C.; da Silva, C.S.; Magnotti, C.; Mouriño, J.L.P.; Cerqueira, V.R. Amyloodiniosis in farmed Sardinella brasiliensis (Steindachner, 1879): Case report of successful therapeutic control with copper sulfate. Parasitol. Int. 2020, 76, 102091. [Google Scholar] [CrossRef]
- Tedesco, P.; Beraldo, P.; Massimo, M.; Fioravanti, M.L.; Volpatti, D.; Dirks, R.; Galuppi, R. Comparative Therapeutic Effects of Natural Compounds Against Saprolegnia spp. (Oomycota) and Amyloodinium ocellatum (Dinophyceae). Front. Veter Sci. 2020, 7, 83. [Google Scholar] [CrossRef]
- Sudhagar, A.; Kumar, G.; El-Matbouli, M. Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review. Int. J. Mol. Sci. 2018, 19, 245. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, T.; Chen, B.; Bai, H.; Bai, Y.; Zhao, J.; Pu, F.; Wu, Y.; Chen, L.; Shi, Y.; et al. Identification and Expression Analysis of Long Non-coding RNA in Large Yellow Croaker (Larimichthys crocea) in Response to Cryptocaryon irritans Infection. Front. Genet. 2020, 11, 590475. [Google Scholar] [CrossRef]
- Xie, X.; Jiang, Y.; Miao, R.; Huang, J.; Zhou, L.; Kong, J.; Yin, F. The gill transcriptome reveals unique antimicrobial features that protect Nibea albiflora from Cryptocaryon irritans infection. J. Fish Dis. 2021, 44, 1215–1227. [Google Scholar] [CrossRef]
- Byadgi, O.; Massimo, M.; Dirks, R.P.; Pallavicini, A.; Bron, J.E.; Ireland, J.H.; Volpatti, D.; Galeotti, M.; Beraldo, P. Innate immune-gene expression during experimental amyloodiniosis in European seabass (Dicentrarchus labrax). Veter- Immunol. Immunopathol. 2021, 234, 110217. [Google Scholar] [CrossRef]
- Kang, K.H.; Kang, H.K.; Zong, T.C.; Kim, J.M.; Zhi, F.Z. Cryopreservation of filefish (Thamnaconus septentrionalis gunther, 1877) sperm. Aquac. Res. 2015, 35, 1429–1433. [Google Scholar] [CrossRef]
- Bian, L.; Li, F.; Ge, J.; Wang, P.; Chang, Q.; Zhang, S.; Li, J.; Liu, C.; Liu, K.; Liu, X.; et al. Chromosome-level genome assembly of the greenfin horse-faced filefish (Thamnaconus septentrionalis) using Oxford Nanopore PromethION sequencing and Hi-C technology. Mol. Ecol. Resour. 2020, 20, 1069–1079. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Bauer, S.; Grossmann, S.; Vingron, M.; Robinson, P.N. Ontologizer 2.0--A multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics 2008, 24, 1650–1651. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 29–34. [Google Scholar] [CrossRef]
- Fringuelli, E.; Gordon, A.W.; Rodger, H.; Welsh, M.D.; A Graham, D. Detection of Neoparamoeba perurans by duplex quantitative Taqman real-time PCR in formalin-fixed, paraffin-embedded Atlantic salmonid gill tissues. J. Fish Dis. 2012, 35, 711–724. [Google Scholar] [CrossRef]
- Cervera, L.; González-Fernández, C.; Arizcun, M.; Cuesta, A.; Chaves-Pozo, E. Severe Natural Outbreak of Cryptocaryon irritans in Gilthead Seabream Produces Leukocyte Mobilization and Innate Immunity at the Gill Tissue. Int. J. Mol. Sci. 2022, 23, 937. [Google Scholar] [CrossRef]
- Massimo, M.; Volpatti, D.; Galeotti, M.; Bron, J.E.; Beraldo, P. News Insights into the Host-Parasite Interactions of Amyloodiniosis in European Sea Bass: A Multi-Modal Approach. Pathogens 2022, 11, 62. [Google Scholar] [CrossRef]
- Xu, G.; Huang, T.; Gu, W.; Zhang, Y.; Yao, Z.; Zhao, C.; Wang, B. Characterization, expression, and functional analysis of the hepcidin gene from Brachymystax lenok. Dev. Comp. Immunol. 2018, 89, 131–140. [Google Scholar] [CrossRef]
- Neves, J.V.; Caldas, C.; Vieira, I.; Ramos, M.F.; Rodrigues, P.N. Multiple hepcidins in a teleost fish, Dicentrarchus labrax: Different hepcidins for different roles. J. Immunol. 2015, 195, 2696–2709. [Google Scholar] [CrossRef]
- Drakesmith, H.; Prentice, A.M.; Parducci, L.; Edwards, M.E.; Bennett, K.D.; Alm, T.; Elverland, E.; Tollefsrud, M.M.; Jørgensen, T.; Houmark-Nielsen, M.; et al. Hepcidin and the Iron-Infection Axis. Science 2012, 338, 768–772. [Google Scholar] [CrossRef]
- Zhang, Y.; Chua, S., Jr. Leptin function and regulation. Compr. Physiol. 2017, 8, 351–369. [Google Scholar]
- Yu, R.M.K.; Chu, D.L.H.; Tan, T.-F.; Li, V.W.T.; Chan, A.K.Y.; Giesy, J.P.; Cheng, S.H.; Wu, R.S.S.; Kong, R.Y.C. Leptin-Mediated Modulation of Steroidogenic Gene Expression in Hypoxic Zebrafish Embryos: Implications for the Disruption of Sex Steroids. Environ. Sci. Technol. 2012, 46, 9112–9119. [Google Scholar] [CrossRef]
- Zhang, D.; Mohammed, H.; Ye, Z.; Rhodes, M.A.; Thongda, W.; Zhao, H.; Jescovitch, L.N.; Fuller, S.A.; Davis, D.A.; Peatman, E. Transcriptomic profiles of Florida pompano (Trachinotus carolinus) gill following infection by the ectoparasite Amyloodinium ocellatum. Fish Shellfish Immunol. 2022, 125, 171–179. [Google Scholar] [CrossRef]
- Han, H.-H.; Sedgwick, A.C.; Shang, Y.; Li, N.; Liu, T.; Li, B.-H.; Yu, K.; Zang, Y.; Brewster, J.T.; Odyniec, M.L.; et al. Protein encapsulation: A new approach for improving the capability of small-molecule fluorogenic probes. Chem. Sci. 2019, 11, 1107–1113. [Google Scholar] [CrossRef]
- Vilar, J.M.; Saiz, L. Control of gene expression by modulated self-assembly. Nucleic Acids Res. 2011, 39, 6854–6863. [Google Scholar] [CrossRef]
- Lie, K.K.; Kvalheim, K.; Rasinger, J.D.; Harboe, T.; Nordgreen, A.; Moren, M. Vitamin A and arachidonic acid altered the skeletal mineralization in Atlantic cod ( Gadus morhua ) larvae without any interactions on the transcriptional level. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2016, 191, 80–88. [Google Scholar] [CrossRef]
- Wang, T.; Secombes, C.J. The cytokine networks of adaptive immunity in fish. Fish Shellfish. Immunol. 2013, 35, 1703–1718. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).