Muscle Transcriptome Analysis Reveals Molecular Mechanisms of Superior Growth Performance in Kuruma Shrimp, Marsupenaeus japonicus
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
2.3. RNA Extraction and cDNA Synthesis
2.4. Library Sequencing, Assembly, and Functional Annotation
2.5. Differentially Expressed Gene Analysis
2.6. Homologous Cloning and Sequence Analysis
2.7. Real-Time PCR Verification
2.8. Single Nucleotide Polymorphism (SNP) Detection
3. Results
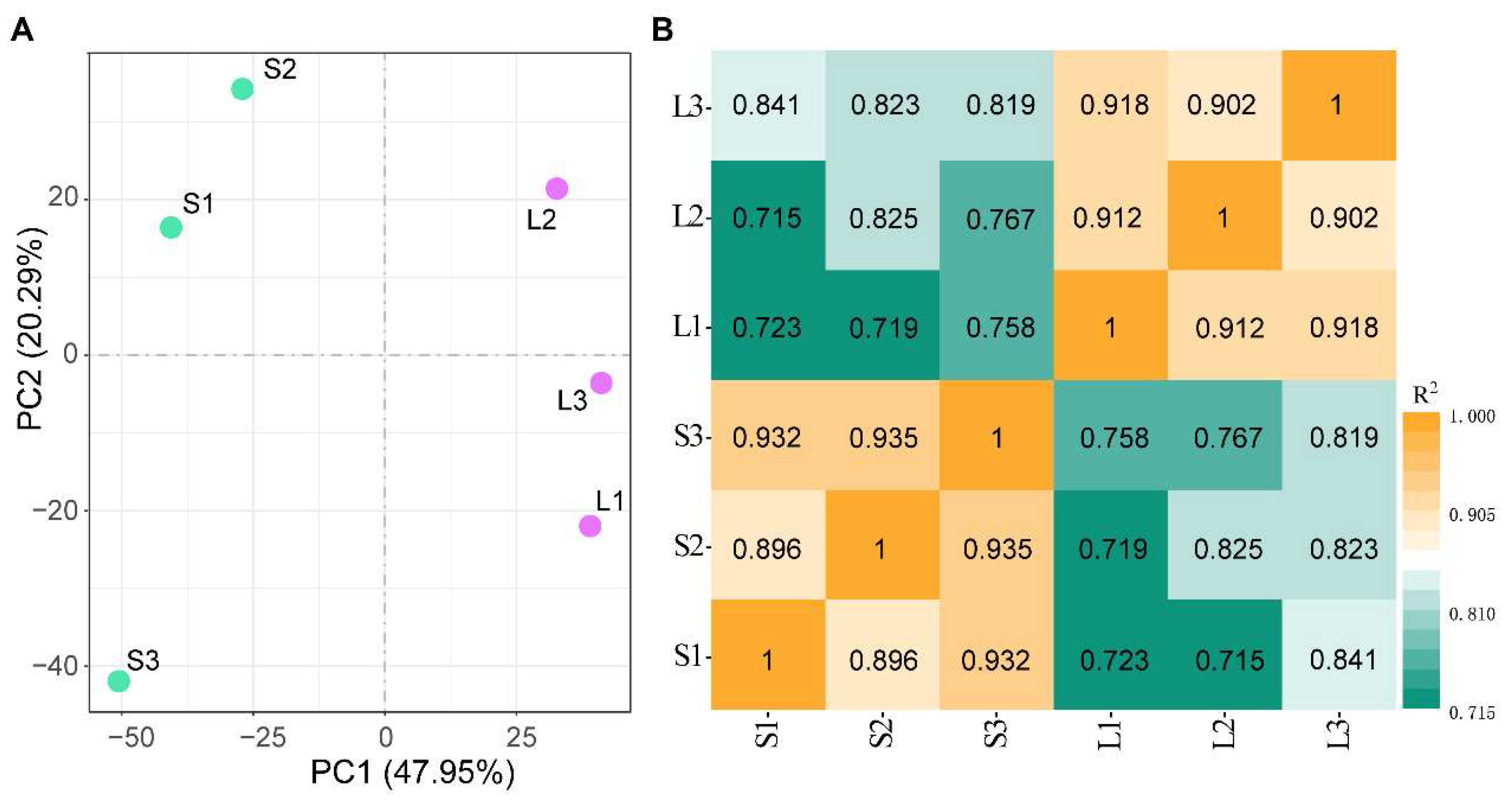
3.1. Transcriptome Sequencing and Assembly
3.2. Identification of Differentially Expressed Genes
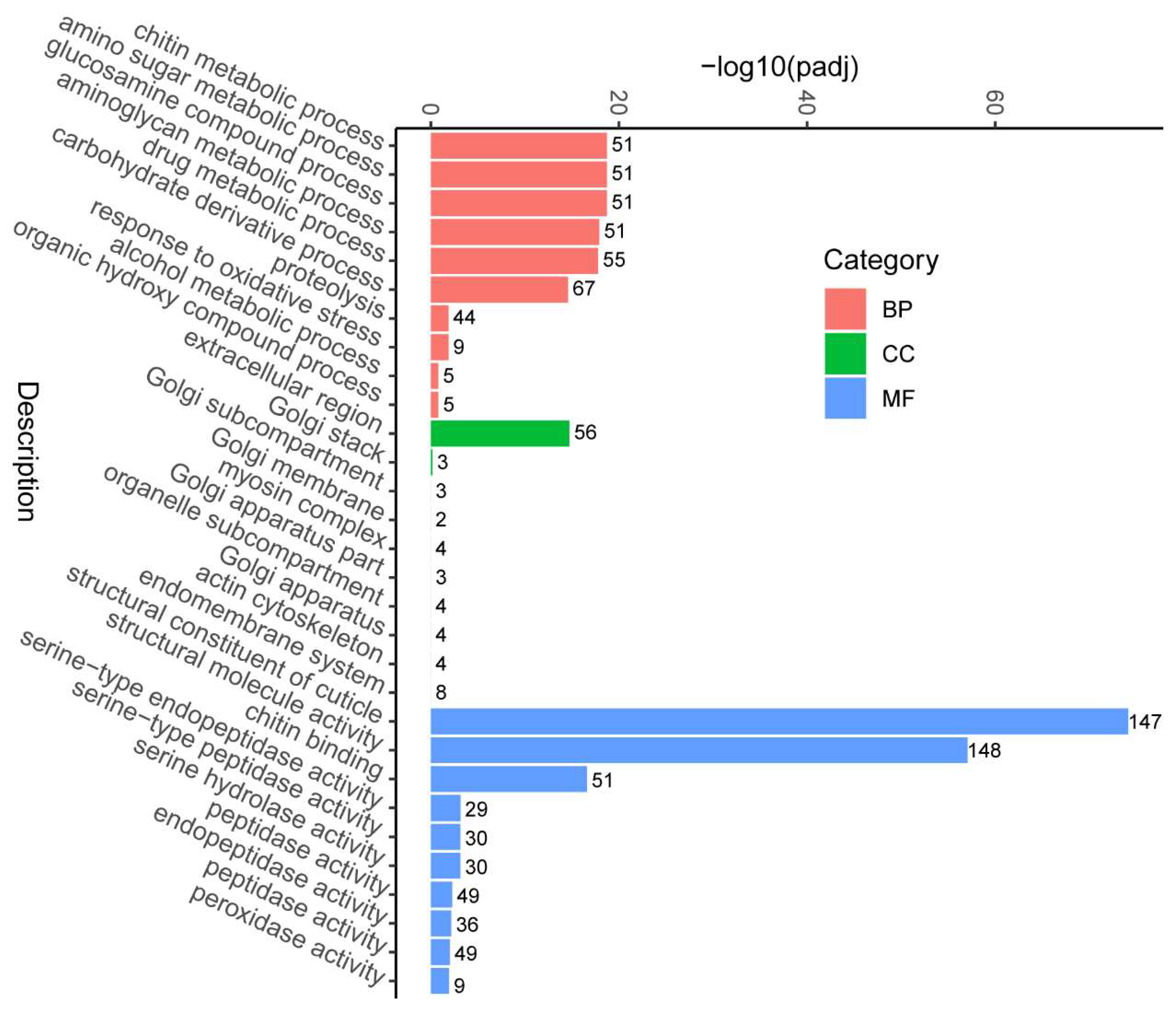
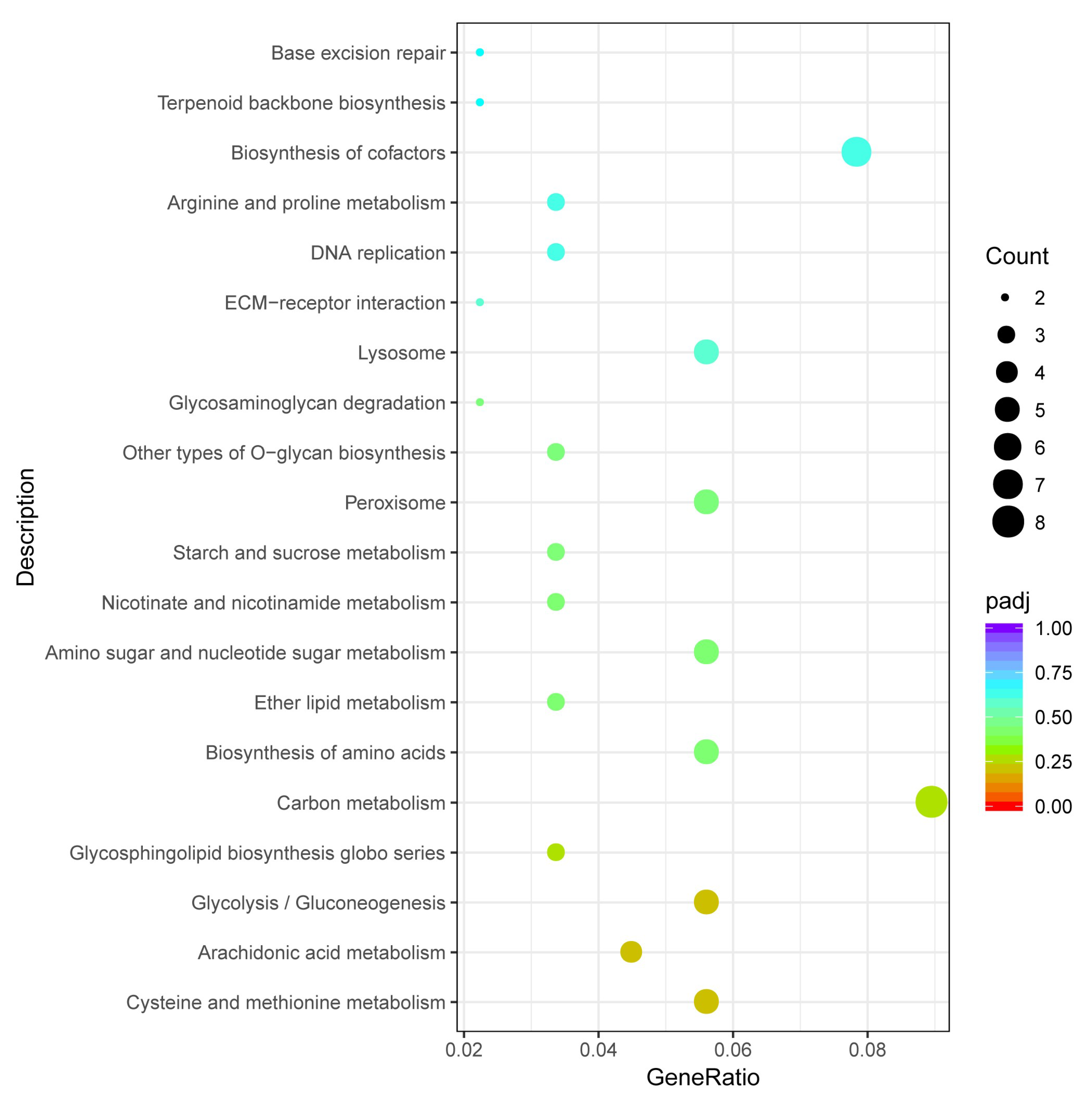
3.3. Enrichment Analysis of Differentially Expressed Genes
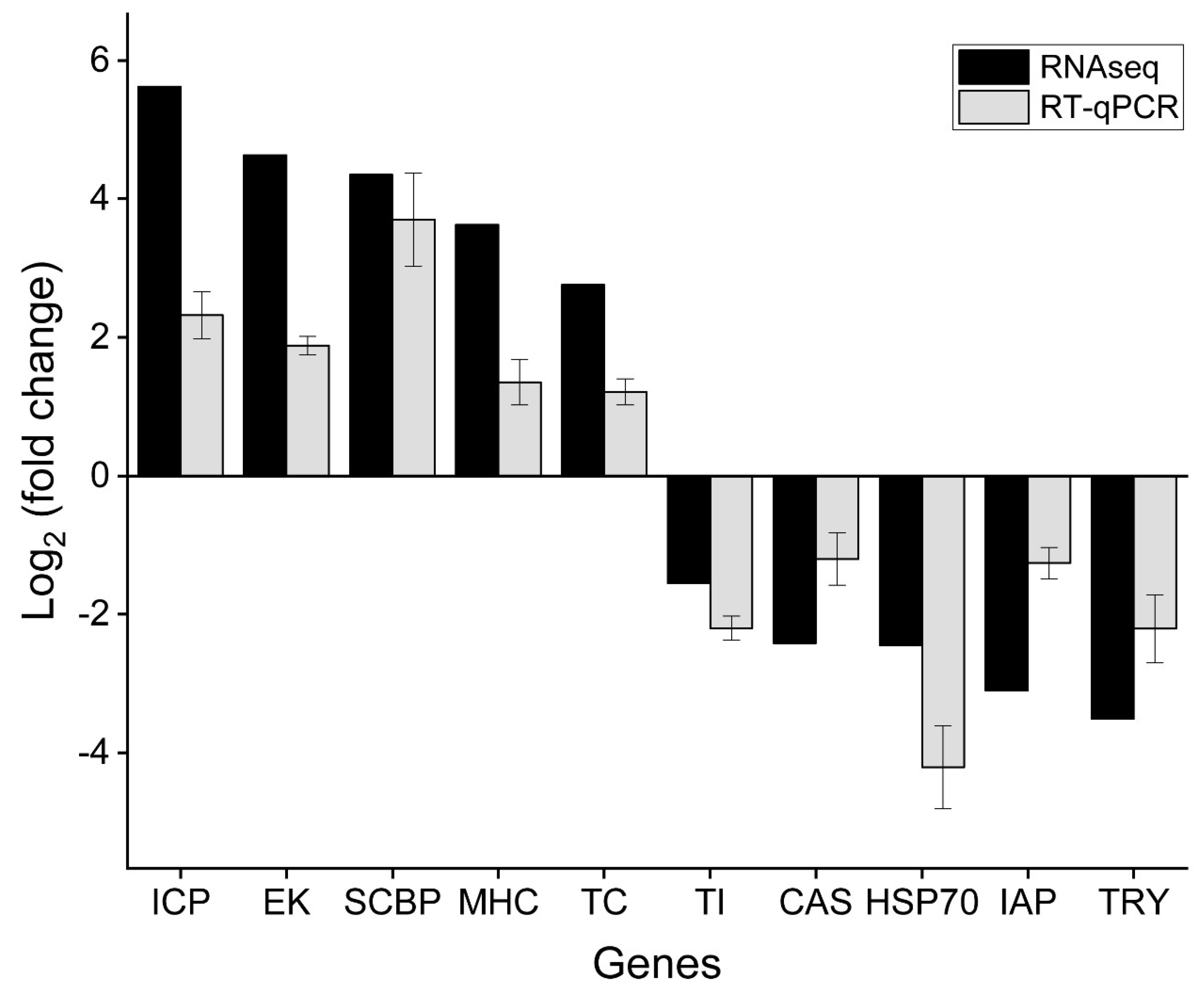
3.4. Quantitative PCR to Verify Differentially Expressed Genes
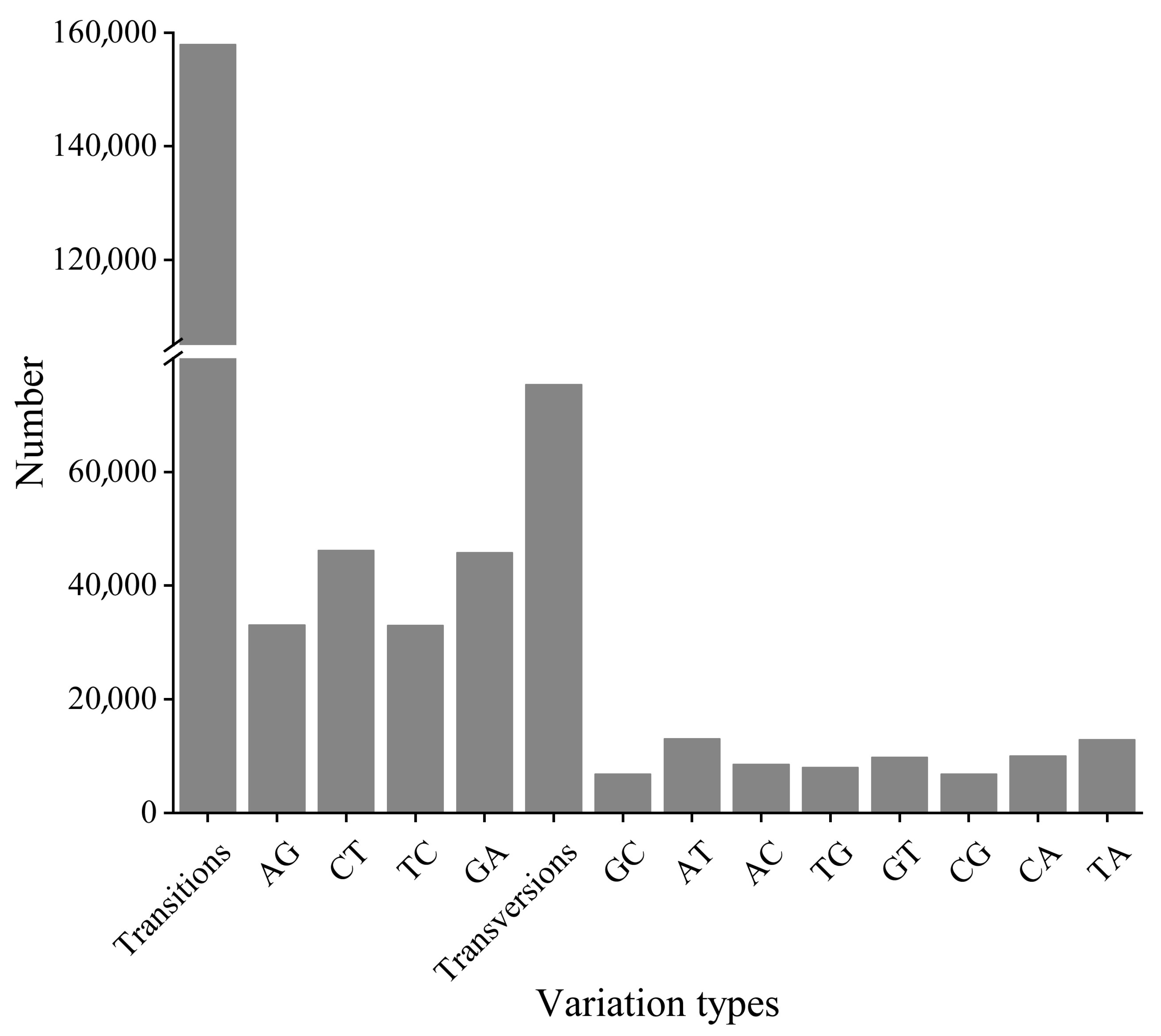
3.5. Single-Nucleotide Polymorphism (SNP) Analysis
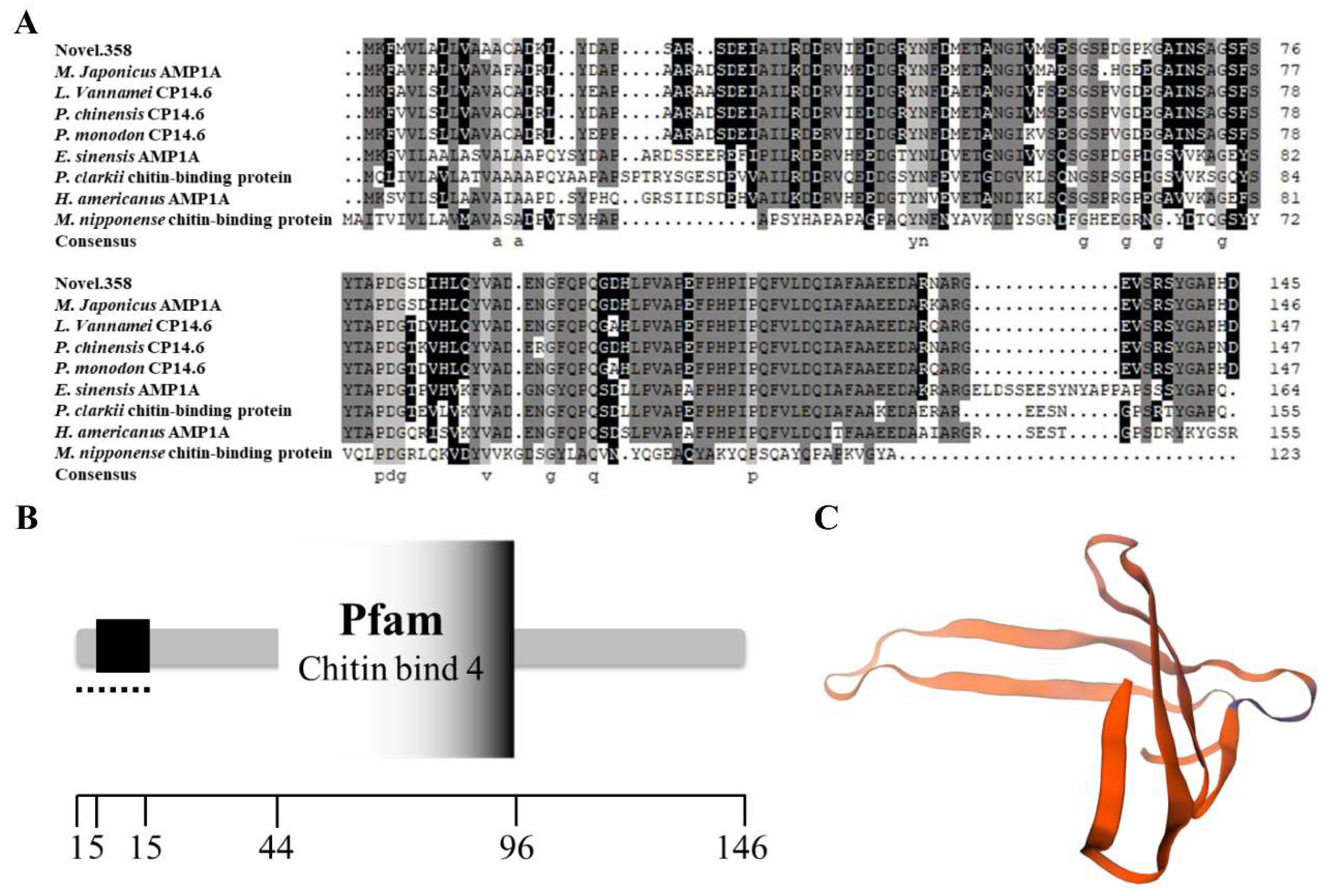
3.6. Structure and Phylogenetic Analysis of Cuticle Protein Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Tsoi, K.H.; Ma, K.Y.; Wu, T.H.; Fennessy, S.T.; Chu, K.H.; Chan, T.Y. Verification of the cryptic species Penaeus pulchricaudatus in the commercially important kuruma shrimp P. japonicus (Decapoda: Penaeidae) using molecular taxonomy. Invertebr. Syst. 2014, 28, 476–490. [Google Scholar] [CrossRef]
- Wang, P.; Chen, B.; Zheng, J.; Cheng, W.; Zhang, H.; Wang, J.; Su, Y.; Xu, P.; Mao, Y. Fine-scale population genetic structure and parapatric cryptic species of kuruma shrimp (Marsupenaeus japonicus), along the northwestern Pacific coast of China. Front. Genet. 2020, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- He, Y.Q.; Su, Y.Q.; Mao, Y.; Wang, J. Genetic diversity analysis of microsatellite DNA in different varieties of Marsupenaneus japonicus. J. Fish. China 2012, 36, 1520–1528. [Google Scholar] [CrossRef]
- Wang, P.; Mao, Y.; Su, Y.; Wang, J. Comparative analysis of transcriptomic data shows the effects of multiple evolutionary selection processes on codon usage in Marsupenaeus japonicus and Marsupenaeus pulchricaudatus. BMC Genom. 2021, 22, 1–14. [Google Scholar] [CrossRef]
- Duan, Y.; Zhang, J.; Dong, H.; Wang, Y.; Liu, Q.; Li, H. Effect of desiccation and resubmersion on the oxidative stress response of the kuruma shrimp Marsupenaeus japonicus. Fish Shellfish Immunol. 2016, 49, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, D.; Duncan, P. Effect of high water temperature on the survival, moulting and food consumption of Penaeus (Marsupenaeus) japonicus (Bate, 1888). Aquac. Res. 2001, 32, 305–313. [Google Scholar] [CrossRef]
- FAO (Food and Agriculture Organization of the United Nations). Global Production Statistics. 2018. Available online: http://www.fao.org/fishery/statistics/global-aquaculture-production/query/en (accessed on 30 March 2018).
- Zhao, J.; He, Z.; Chen, X.; Huang, Y.; Xie, J.; Qin, X.; Ni, Z.; Sun, C. Growth trait gene analysis of kuruma shrimp (Marsupenaeus japonicus) by transcriptome study. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100874. [Google Scholar] [CrossRef]
- Cheng, W.; Zhang, H.; Wang, P.; Wei, Y.; Chen, C.; Hou, Y.; Deng, X.; Li, S.; Sun, S.; Cai, Q. The multiple influences of natural farming environment on the cultured population behavior of kuruma prawn, Penaeus japonicus. Animals 2022, 12, 3383. [Google Scholar] [CrossRef]
- Kim, S.-K.; Guo, Q.; Jang, I.-K. Effect of biofloc on the survival and growth of the postlarvae of three penaeids (Litopenaeus vannamei, Fenneropenaeus chinensis, and Marsupenaeus japonicus) and their biofloc feeding efficiencies, as related to the morphological structure of the third maxilliped. J. Crustac. Biol. 2015, 35, 41–50. [Google Scholar]
- Jung, H.; Lyons, R.E.; Hurwood, D.A.; Mather, P.B. Genes and growth performance in crustacean species: A review of relevant genomic studies in crustaceans and other taxa. Rev. Aquac. 2013, 5, 77–110. [Google Scholar] [CrossRef]
- Shuang, Z.; Xuange, L.; Pengfei, W.; Peng, X.; Lei, Z.; Lei, Z.; Qindong, T.; Zhi, C.; Guifeng, L. Muscle transcriptome of Siniperca chuatsi with different weights from a full-sib family. J. Fish. Sci. China 2020, 27, 53–61. [Google Scholar]
- Lanmei, W.; Wenbin, Z.; Jianjun, F.; Zaijie, D. De novo transcriptome analysis and comparison of the FFRC No. 2 strain common carp (Cyprinus carpio) associated with its muscle growth. J. Fish. China 2021, 45, 79–87. [Google Scholar]
- Wang, Z.; Liang, F.; Huang, R.; Deng, Y.; Li, J. Identification of the differentially expressed genes of Pinctada maxima individuals with different sizes through transcriptome analysis. Reg. Stud. Mar. Sci. 2019, 26, 100512. [Google Scholar] [CrossRef]
- Huang, J.; You, W.; Luo, X.; Ke, C. iTRAQ-based identification of proteins related to muscle growth in the Pacific abalone, Haliotis discus hannai. Int. J. Mol. Sci. 2017, 18, 2237. [Google Scholar] [CrossRef] [PubMed]
- Yunjie, Y.; Xia, L.; Xianhong, M.; Sheng, L.; Baolong, C.; Jie, K. Screening of genes related to muscle growth under the myostatin regulation by RNA-seq in Fenneropenaeus chinensis. Prog. Fish. Sci. 2021, 42, 55–63. [Google Scholar]
- Guo, X.F.; Zhou, Y.L.; Liu, M.; Wang, Z.W.; Gui, J.F. Integrated application of Iso-seq and RNA-seq provides insights into unsynchronized growth in red swamp crayfish (Procambarus clarkii). Aquac. Rep. 2022, 22, 101008. [Google Scholar] [CrossRef]
- Uengwetwanit, T.; Uawisetwathana, U.; Arayamethakorn, S.; Khudet, J.; Chaiyapechara, S.; Karoonuthaisiri, N.; Rungrassamee, W. Multi-omics analysis to examine microbiota, host gene expression and metabolites in the intestine of black tiger shrimp (Penaeus monodon) with different growth performance. PeerJ 2020, 8, e9646. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, Z.; Solberg, M.F.; Chen, Z.; Wei, M.; Zhu, F.; Jia, C.; Meng, Q.; Zhang, Z. Comparative transcriptome analysis of mixed tissues of black porgy (Acanthopagrus schlegelii) with differing growth rates. Aquac. Res. 2021, 52, 5800–5813. [Google Scholar] [CrossRef]
- Zhao, L.; He, K.; Xiao, Q.; Liu, Q.; Luo, W.; Luo, J.; Fu, H.; Li, J.; Wu, X.; Du, J. Comparative transcriptome profiles of large and small bodied large-scale loaches cultivated in paddy fields. Sci. Rep. 2021, 11, 4936. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Z.; Dai, X. Transcriptome analysis of growth retardation and normal Macrobrachium rosenbergii. Genom. Appl. Biol. 2021, 40, 89–100. (In Chinese) [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Amit, I. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2013, 29, 644–652. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Smyth, G.K.; Wei, S. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar]
- Simon, A.; Wolfgang, H. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, 106. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2011, 40, 302–305. [Google Scholar] [CrossRef]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Sigrist, C.J.A.; Lorenzo, C.; Edouard, D.C.; Langendijk-Genevaux, P.S.; Virginie, B.; Amos, B.; Nicolas, H. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010, 38, 161–166. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S.J. Primer Premier 5. Biotech Softw. Internet Rep. Comput. Softw. J. Sci. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cingolani, P. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 1–13. [Google Scholar] [CrossRef]
- Liew, M.; Pryor, R.; Palais, R.; Meadows, C.; Erali, M.; Lyon, E.; Wittwer, C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 2004, 50, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Skov, M.W.; Hartnoll, R.G. Comparative suitability of binocular observation, burrow counting and excavation for the quantification of the mangrove fiddler crab Uca annulipes (H. Milne Edwards). Adv. Decapod Crustac. Res. 2001, 154, 201–212. [Google Scholar]
- El Haj, A.J.; Harrison, P.; Whiteley, N.M. Regulation of muscle gene expression in Crustacea over the moult cycle. Symp. Soc. Exp. Biol. 1992, 46, 151–165. [Google Scholar]
- Haj, A.J.E.; Whiteley, N.M. Molecular regulation of muscle growth in Crustacea. J. Mar. Biol. Assoc. UK 1997, 77, 95–106. [Google Scholar] [CrossRef]
- Mykles, D.L. The mechanism of fluid absorption at ecdysis in the American lobster, Homarus americanus. J. Exp. Biol. 2015, 84, 89–102. [Google Scholar] [CrossRef]
- Tobacman, L.S. Troponin revealed: Uncovering the structure of the thin filament on-off switch in striated muscle. Biophys. J. 2021, 120, 1–9. [Google Scholar] [CrossRef]
- Cao, T.X.; Thongam, U.; Jin, J.P. Invertebrate troponin: Insights into the evolution and regulation of striated muscle contraction. Arch. Biochem. Biophys. 2019, 666, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, Y.; Ozawa, H. Biochemical and physicochemical characteristics of the major muscle proteins from fish and shellfish. Fish. Sci. 2020, 86, 729–740. [Google Scholar] [CrossRef]
- Wang, J.; Ge, Q.; Li, J.; Chen, Z.; Li, J. Isolation and characterization of three skeletal troponin genes and association with growth-related traits in Exopalaemon carinicauda. Mol. Biol. Rep. 2019, 46, 705–718. [Google Scholar] [CrossRef]
- Xing, C.F.; Xiong, J.Y.; Xie, S.M.; Guo, H.X.; Hua, S.S.; Yao, Y.J.; Zhu, J.W.; Yan, B.L.; Shen, X.; Gao, H.; et al. Comparative transcriptome and gut microbiota analysis of Exopalaemon carinicauda with different growth rates from a full-sib family. Aquac. Rep. 2023, 30, 101580. [Google Scholar] [CrossRef]
- Heissler, S.M.; Sellers, J.R. Kinetic adaptations of myosins for their diverse cellular functions. Traffic 2016, 17, 839–859. [Google Scholar] [CrossRef]
- Harzsch, S.; Kreissl, S. Myogenesis in the thoracic limbs of the American lobster. Arthropod Struct. Dev. 2010, 39, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Andruchov, O.; Andruchova, O.; Wang, Y.; Galler, S. Dependence of cross-bridge kinetics on myosin light chain isoforms in rabbit and rat skeletal muscle fibres. J. Physiol. 2006, 571, 231–242. [Google Scholar] [CrossRef]
- Ren, X.; Yu, X.; Gao, B.; Li, J.; Liu, P. iTRAQ-based identification of differentially expressed proteins related to growth in the swimming crab, Portunus trituberculatus. Aquac. Res. 2017, 48, 3257–3267. [Google Scholar] [CrossRef]
- Zhang, X.; Yuan, J.; Li, F.; Xiang, J. Chitin synthesis and degradation in crustaceans: A genomic view and application. Mar. Drugs 2021, 19, 153. [Google Scholar] [CrossRef]
- Abehsera, S.; Weil, S.; Manor, R.; Sagi, A. The search for proteins involved in the formation of crustacean cuticular structures. Hydrobiologia 2018, 825, 29–45. [Google Scholar] [CrossRef]
- Mrak, P.; Bogataj, U.; Štrus, J.; Žnidaršič, N. Cuticle morphogenesis in crustacean embryonic and postembryonic stages. Arthropod Struct. Dev. 2017, 46, 77–95. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, H. The crustacean cuticle: Structure, composition and mineralization. Front. Biosci. 2012, 4, 711–720. [Google Scholar] [CrossRef]
- Vincent, J.F. Arthropod cuticle: A natural composite shell system. Compos. Part A Appl. Sci. Manuf. 2002, 33, 1311–1315. [Google Scholar] [CrossRef]
- Willis, J.H.; Papandreou, N.C.; Iconomidou, V.A.; Hamodrakas, S.J. Cuticular proteins. In Insect Molecular Biology and Biochemistry; Academic Press: San Diego, CA, USA, 2012; pp. 134–166. [Google Scholar]
- Cornman, R.S. Molecular evolution of Drosophila cuticular protein genes. PLoS ONE 2009, 4, e8345. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, X.; Xie, K.; Liu, Q.; Li, Y.; Tan, X.; Dong, H.; Li, X.; Dong, Z.; Xia, Q. Chitin and cuticle proteins form the cuticular layer in the spinning duct of silkworm. Acta Biomater. 2022, 145, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Balabanidou, V.; Grigoraki, L.; Vontas, J. Insect cuticle: A critical determinant of insecticide resistance. Curr. Opin. Insect Sci. 2018, 27, 68–74. [Google Scholar] [CrossRef]
- Cesar, J.R.; Zhao, B.; Yang, J. Analysis of expressed sequence tags from abdominal muscle cDNA library of the pacific white shrimp Litopenaeus vannamei. Animal 2008, 2, 1377–1383. [Google Scholar] [CrossRef]
- Yang, F.; Li, X.; Li, S.; Xiang, J.; Li, F. A novel cuticle protein involved in WSSV infection to the Pacific white shrimp Litopenaeus vannamei. Dev. Comp. Immunol. 2019, 102, 103491. [Google Scholar] [CrossRef]
- Santos, C.A.; Andrade, S.C.S.; Teixeira, A.K.; Farias, F.; Guerrelhas, A.C.; Rocha, J.L.; Freitas, P.D. Transcriptome differential expression analysis reveals the activated genes in Litopenaeus vannamei shrimp families of superior growth performance. Aquaculture 2021, 531, 735871. [Google Scholar] [CrossRef]
- Jundong, C.; Pengyun, D.; Xuguang, L.; Keran, B. Cloning and expression analysis of epidermal protein gene EsCAP from Chinese Mitten crab. Jiangsu Agric. Sci. 2021, 49, 74–80. [Google Scholar]
- Coblentz, F.E.; Shafer, T.H.; Roer, R.D. Cuticular proteins from the blue crab alter in vitro calcium carbonate mineralization. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 1998, 121, 349–360. [Google Scholar] [CrossRef]
- Glazer, L.; Sagi, A. On the involvement of proteins in the assembly of the crayfish gastrolith extracellular matrix. Invertebr. Reprod. Dev. 2012, 56, 57–65. [Google Scholar] [CrossRef]
- Andersen, S.O. Insect cuticular sclerotization: A review. Insect Biochem. Mol. Biol. 2010, 40, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Persson, P.; Watanabe, T. Molecular cloning of the crustacean DD4 cDNA encoding a Ca2+-binding protein. Biochem. Biophys. Res. Commun. 2000, 276, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Takagi, Y.; Ozaki, N.; Kogure, T.; Watanabe, T. A crustacean Ca2+-binding protein with a glutamate-rich sequence promotes CaCO3 crystallization. Biochem. J. 2004, 384, 159–167. [Google Scholar] [CrossRef]
- Inoue, H.; Ohira, T.; Ozaki, N.; Nagasawa, H. Cloning and expression of a cDNA encoding a matrix peptide associated with calcification in the exoskeleton of the crayfish. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2003, 136, 755–765. [Google Scholar] [CrossRef]
- Inoue, H.; Ohira, T.; Ozaki, N.; Nagasawa, H. A novel calcium-binding peptide from the cuticle of the crayfish, Procambarus clarkii. Biochem. Biophys. Res. Commun. 2004, 318, 649–654. [Google Scholar] [CrossRef]

| Sample | Raw Reads | Clean Reads | Clean Bases (G) | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|
| S1 | 46,763,772 | 45,912,332 | 6.89 | 98.03 | 94.4 | 50.12 |
| S2 | 48,942,932 | 47,858,272 | 7.18 | 98.26 | 94.85 | 50.04 |
| S3 | 45,487,750 | 44,475,408 | 6.67 | 98.03 | 94.52 | 51.44 |
| L1 | 48,220,794 | 47,140,442 | 7.07 | 98.48 | 95.41 | 51.04 |
| L2 | 47,360,730 | 46,243,766 | 6.94 | 98.28 | 94.82 | 48.69 |
| L3 | 44,954,984 | 43,991,266 | 6.6 | 98.46 | 95.5 | 52.72 |
| Gene_id | Gene_Description | Log2 (Fold Change) | padj |
|---|---|---|---|
| Hic_asm_3.1371 | Cuticle protein AMP4 | 8.005 | 0.026 |
| Hic_asm_3.1791 | Chitin binding Peritrophin-A domain | 7.675 | 0.020 |
| Hic_asm_29.1989 | Insect cuticle protein | 5.620 | 0.001 |
| Hic_asm_3.3781 | Transmembrane protease serine 11D | 5.533 | 0.045 |
| Hic_asm_21.381 | Tubulin alpha-1 chain | 5.078 | 0.004 |
| Hic_asm_5.353 | Ecdysteroid kinase | 4.628 | 0.001 |
| Hic_asm_4.2410 | Myosin heavy chain C | 3.630 | 0.002 |
| Hic_asm_7.2362 | Sarcoplasmic calcium-binding protein | 4.349 | 0.005 |
| Hic_asm_28.2387 | Mitochondrial basic amino acids transporter | 3.939 | 0.006 |
| Hic_asm_37.790 | Troponin C | 2.759 | 0.002 |
| Hic_asm_16.1606 | Mitochondrial enolase superfamily member 1 | 2.656 | 0.024 |
| Hic_asm_27.1758 | phosphatidylinositol 4,5-bisphosphate phosphodiesterase | 2.618 | 0.020 |
| Hic_asm_13.2114 | Mitochondrial carnitine/acylcarnitine carrier protein | 2.082 | 0.000 |
| Hic_asm_12.2116 | Mitochondrial dicarboxylate carrier | 2.061 | 0.048 |
| Hic_asm_30.362 | Coactosin-like protein | 1.965 | 0.025 |
| Hic_asm_22.2436 | Carbonyl reductase (NADPH) 3 | −4.978 | 0.002 |
| Hic_asm_35.393 | Trypsin-1 | −3.509 | 0.047 |
| Hic_asm_12.273 | Inhibitor of apoptosis protein | −3.098 | 0.001 |
| Hic_asm_3.2630 | Methyl farnesoate epoxidase | −3.064 | 0.002 |
| Hic_asm_32.1003 | Alpha-amylase | −2.876 | 0.020 |
| Hic_asm_26.1575 | Superoxide dismutase (Cu-Zn) | −2.805 | 0.024 |
| Hic_asm_38.1026 | Actin-2, muscle-specific | −2.536 | 0.000 |
| Hic_asm_14.203 | zinc-RING finger domain | −2.520 | 0.017 |
| Hic_asm_8.3322 | Heat shock 70 kDa protein | −2.441 | 0.017 |
| Hic_asm_16.604 | Crustacyanin-A2 subunit | −2.414 | 0.005 |
| Hic_asm_1.2118 | Ubiquitin carboxyl-terminal hydrolase 22 | −2.334 | 0.020 |
| Hic_asm_25.2408 | Glutathione S-transferase D7 | −2.394 | 0.006 |
| Hic_asm_19.605 | Pyruvate kinase | −1.968 | 0.026 |
| Hic_asm_36.203 | Superoxide dismutase (Mn), mitochondrial | −1.656 | 0.019 |
| Hic_asm_17.2603 | Troponin I | −1.549 | 0.002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Yu, F.; Li, X.; Xie, S.; Wang, L.; Zhu, J.; Zhou, X.; Zhou, X.; Yan, B.; Gao, H.; et al. Muscle Transcriptome Analysis Reveals Molecular Mechanisms of Superior Growth Performance in Kuruma Shrimp, Marsupenaeus japonicus. Fishes 2023, 8, 350. https://doi.org/10.3390/fishes8070350
Wang P, Yu F, Li X, Xie S, Wang L, Zhu J, Zhou X, Zhou X, Yan B, Gao H, et al. Muscle Transcriptome Analysis Reveals Molecular Mechanisms of Superior Growth Performance in Kuruma Shrimp, Marsupenaeus japonicus. Fishes. 2023; 8(7):350. https://doi.org/10.3390/fishes8070350
Chicago/Turabian StyleWang, Panpan, Fei Yu, Xinyang Li, Shumin Xie, Lei Wang, Jiawei Zhu, Xinlei Zhou, Xinyi Zhou, Binlun Yan, Huan Gao, and et al. 2023. "Muscle Transcriptome Analysis Reveals Molecular Mechanisms of Superior Growth Performance in Kuruma Shrimp, Marsupenaeus japonicus" Fishes 8, no. 7: 350. https://doi.org/10.3390/fishes8070350
APA StyleWang, P., Yu, F., Li, X., Xie, S., Wang, L., Zhu, J., Zhou, X., Zhou, X., Yan, B., Gao, H., & Xing, C. (2023). Muscle Transcriptome Analysis Reveals Molecular Mechanisms of Superior Growth Performance in Kuruma Shrimp, Marsupenaeus japonicus. Fishes, 8(7), 350. https://doi.org/10.3390/fishes8070350






